Abstract
Cytochrome P450 2A13-catalyzed α-hydroxylation is a critical step in the activation of the tobacco carcinogens 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) and (S)-N′-nitrosonornicotine [(S)-NNN]. In the enzyme's active site, a single polar residue, Asn297, can influence substrate binding, orientation, and metabolism. We determined the effects of N297A mutation on enzyme kinetics and specificity for NNK, NNN, and coumarin metabolism. [5-3H]-NNK, [5-3H]-(S)-NNN, [14C]coumarin, and radioflow high-performance liquid chromatography analysis were used to quantify metabolites. Cytochrome P450 (P450) 2A13 N297A catalyzed NNK α-hydroxylation, with a 3-fold preference for methylene versus methyl hydroxylation, similar to wild type. Docking studies using the P450 2A13 crystal structure predicted that when the pyridine ring of NNK cannot hydrogen bond to residue 297 it tilts and orients NNK in positions unfavorable for α-hydroxylation. The N297A mutation resulted in a 5- and 4-fold decrease in catalytic efficiency of NNK and NNN metabolism, respectively, primarily because of increased Km values. The N297A mutation strikingly affected coumarin metabolism. The ratio of coumarin 7-hydroxylation to coumarin 3,4-epoxidation is approximately equal for wild-type enzyme, whereas the ratio was 1:9 for the N297A mutant. Coumarin 3,4-epoxidation was significantly underestimated unless the epoxide was trapped and quantified as its glutathione conjugate. The Km value for this reaction was 4-fold greater for the mutant enzyme; the Vmax value increased nearly 40-fold. The observed shift toward coumarin 3,4-epoxidation is consistent with docking studies. In summary, Asn297 in P450 2A13 is important for orienting NNK and coumarin in the active site, changing this residue to Ala results in altered enzyme kinetics for NNK, NNN, and coumarin.
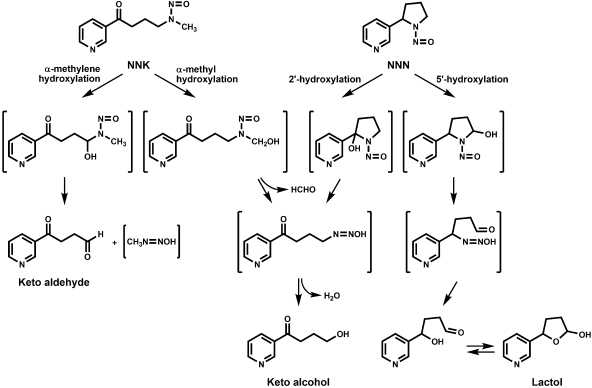
The tobacco-specific nitrosamines (S)-N′-nitrosonornicotine [(S)-NNN] and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) are carcinogens found in unburned tobacco and cigarette smoke. Both NNN and NNK were recently classified as human carcinogens (International Agency for Research on Cancer, 2007). NNN induces esophageal and nasal tumors in rodents, and NNK is a potent lung carcinogen in rodents (Hecht, 1998). Both NNN and NNK require P450-catalyzed metabolism to exert their carcinogenic effects (Hecht, 1998). Metabolic activation occurs by hydroxylation of the carbons α to the nitroso moiety. For NNN, hydroxylation occurs at the 2′- and 5′-carbon positions to ultimately form 4-oxo-4-(3-pyridyl)-1-butanol (keto alcohol), 5-(3-pyridyl)-2-hydroxytetrahydrofuran (lactol), and reactive diazohydroxide intermediates (Fig. 1). Hydroxylation of NNK occurs at the α-methyl and α-methylene carbon positions, forming keto alcohol, 4-oxo-4-(3-pyridyl)-1-butanone (keto aldehyde), and two diazohydroxides that are capable of pyridyloxobutylating or methylating DNA (Fig. 1).
Fig. 1.
NNN and NNK α-hydroxylation.
CYP2A13 is the most efficient human P450 catalyst of NNK α-hydroxylation (Jalas et al., 2005). It is a 300-fold more efficient catalyst than the hepatic enzyme CYP2A6, despite 93.5% sequence identity between these enzymes. The X-ray crystal structures for CYP2A6 and CYP2A13 have been reported previously (Yano et al., 2005; Smith et al., 2007). The active sites of CYP2A6 and CYP2A13 share several similar characteristics. These include a cluster of phenylalanine residues that line the “roof” of the active site and the presence of a single polar residue, Asn297. Docking NNK into the crystal structures of CYP2A13 and CYP2A6 predicts, because of a slightly different active site volume and shape, that the orientation of NNK in CYP2A13 is more favorable for α-hydroxylation (Smith et al., 2007). In both enzymes, a hydrogen bond between NNK and Asn297 seems to affect substrate orientation. With CYP2A6, the hydrogen bond forms between Asn297 and the carbonyl oxygen of NNK, and with CYP2A13 it is between Asn297 and the pyridine nitrogen; only the latter orientation is compatible with α-hydroxylation.
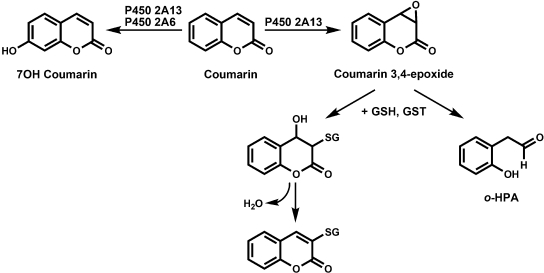
CYP2A6 and CYP2A13 are both efficient catalysts of coumarin metabolism (Honkakoski and Negishi, 1997; von Weymarn and Murphy, 2003). CYP2A6 exclusively catalyzes the 7-hydroxylation of coumarin. The crystal structure of CYP2A6 was solved with coumarin in the active site, and a hydrogen bond interaction between Asn297 and the carbonyl oxygen of coumarin contributes to the orientation of coumarin for 7-hydroxylation (Yano et al., 2005). CYP2A13 catalyzes both the 7-hydroxylation and 3,4-epoxidation of coumarin with similar efficiency. The 3,4-epoxidation of coumarin has been quantified as the formation of o-hydroxyphenylacetaldehyde (o-HPA), which is generated nonenzymatically from the unstable coumarin 3,4-epoxide (Fig. 2) (Born et al., 1997; Vassallo et al., 2004). We report here on the effect of an N297A mutation on the specificity of CYP2A13-catalyzed coumarin metabolism.
Fig. 2.
Coumarin 7-hydroxylation and 3,4-epoxidation pathways.
In humans, CYP2A6 and CYP2A13 are important catalysts of NNN and NNK α-hydroxylation (Jalas et al., 2005; Wong et al., 2005a). The high efficiency of CYP2A13 for NNK metabolic activation combined with its expression in respiratory tissues, including nasal mucosa, lung, and trachea, suggests that CYP2A13 plays a critical role in tobacco-specific nitrosamine-induced carcinogenesis (Su et al., 2000; Jalas et al., 2005; Wong et al., 2005b; Zhu et al., 2006). The goal of the present study was to determine the importance of the active site residue Asn297 on CYP2A13 function, specifically on metabolism of the tobacco-specific nitrosamines (S)-NNN and NNK.
Materials and Methods
Caution: NNK and NNN are International Agency for Research on Cancer class I human carcinogens. Both are carcinogenic in laboratory animals and should be handled with care.
Chemicals and Reagents. Lactol, keto alcohol, 4-oxo-1-(3-pyridyl)-1-butanone, (S)-NNN, and [5-3H](S)-NNN were generous gifts from Dr. Stephen Hecht (Cancer Center, Minneapolis, MN). [5-3H]NNK (10.5–11 Ci · mmol–1) was purchased from Moravek Biochemicals (Brea, CA) and purified by HPLC to >99% purity before use. [U-14C-benzyl]Coumarin (31 mCi · mmol–1; 99.5% purity) was purchased from Amersham Pharmacia Biotech (Piscataway, NJ). o-HPA was a gift from Dr. Louis Lehman-McKeeman (Procter & Gamble, Cincinnati, OH). All other chemicals and reagents were purchased from Sigma-Aldrich (St. Louis, MO).
Site-Directed Mutagenesis. Full-length CYP2A13 (with the second codon changed from leucine to alanine and four histidine residues added to the C terminus) in the pKK233-2 plasmid (Pharmacia, Uppsala, Sweden) was a gift from Dr. Emily Scott (University of Kansas, Lawrence, KS). The QuikChange (Stratagene, La Jolla, CA) method was used to introduce the single-nucleotide mutation. Primers used for site-directed mutagenesis were the following: 5′-gtgatgaccaccctggccctcttctttgcgggc-3′ (forward primer, mutated bases are underlined) and 5′-cccgcaaagaagagggccagggtggtcatcacc-3′ (reverse primer, mutated bases are underlined). The single N297A mutation was confirmed by DNA sequencing.
Protein Expression and Purification. Rat NADPH-P450 oxidoreductase (reductase) was expressed in Escherichia coli and purified as described previously (Hanna et al., 1998). Wild-type CYP2A13 and CYP2A13 N297A were expressed in TOPP-3 E. coli cells (Stratagene) at 30°C with 190-rpm shaking and with an induction time of 72 h using the protocol described previously (Von Weymarn et al., 2005). Cells were harvested and purified using a modified method from previously described protocols (Soucek, 1999; Von Weymarn et al., 2004).
Protein Characterization and Enzyme Reconstitution. Enzyme expression and purity was visualized by SDS-polyacrylamide gel electrophoresis with Gel Code Blue staining (Pierce Chemical, Rockford, IL). Expression of CYP2A13 and CYP2A13 N297A was confirmed by Western blot using anti-CYP2A6 antibody (BD Biosciences, San Jose, CA) that also detects CYP2A13 (Wong et al., 2005b). Cytochrome P450 content was determined for each enzyme preparation by dithionite-reduced CO difference spectra obtained using an Olis DW-2000 UV-Vis spectrophotometer (Olis Inc., Bogard, GA) (Omura and Sato, 1964). Enzymes were reconstituted with reductase and dilauroyl-l-α-phosphatidylcholine (0.2 μg/pmol P450) for 45 min at 4°C just before use. The ratio of P450 to reductase was 1:4 for coumarin metabolism and 1:2 for NNK and NNN metabolism.
Coumarin Metabolism. Total coumarin metabolism was determined using [U-14C-benzyl]coumarin and radioflow HPLC analysis. The method used was a modification of that described previously (von Weymarn and Murphy, 2003). Reconstituted CYP2A13 or CYP2A13 N297A (40 pmol) was each incubated with 65 μM[U-14C-benzyl]coumarin (31 mCi · mmol–1), 100 mM Tris buffer, pH 7.4, an NADPH-generating system (0.4 mM NADP+, 100 mM glucose 6-phosphate, and 0.4 units · ml–1 glucose-6-dehydrogenase) in a final volume of 150 μl for 10 to 60 min at 37°C. Reactions were terminated by addition of 15 μl of 15% trichloroacetic acid. All analyses were conducted in triplicate. Products were analyzed by radioflow HPLC with a Phenomenex Gemini C18 (5 μm; 250 × 4.60 mm) column. A flow rate of 0.8 ml · min–1 was used. An isocratic system of 72% A (1% acetic acid in water) and 28% B (1% acetic acid in methanol) was used. As the column aged, B was decreased to 25%. Radioactive detection was accomplished using a β-RAM radioflow detector (IN/US Systems, Tampa, FL), with a scintillant flow rate of 2.4 ml · min–1 (Monoflow 5; National Diagnostics, Atlanta, GA). 14C-Labeled products were coinjected with o-HPA, 8-hydroxycoumarin, 7-hydroxycoumarin, and 3-hydroxycoumarin standards detected by absorbance at 270 nm. Initially, 4-hydroxycoumarin, 5-hydroxycoumarin, and 6-hydroxycoumarin were also evaluated as potential products based on retention times.
Coumarin 3,4-epoxidation was quantified by trapping the epoxide as a GSH conjugate. The method used is analogous to that used previously to quantify naphthalene epoxidation (Shultz et al., 1999). Reactions were carried out as described above, with the addition of 1 to 5 mM GSH and 2.5 to 20 units of glutathione transferase (GST) (from equine liver; Sigma-Aldrich). Using [14C]coumarin, conditions were determined under which the 3,4-epoxide was trapped by the addition of GSH (1.5 mM) and GST (5 units), as determined by the decrease in detection of o-HPA, and no further increase in the GSH conjugate peak with increasing amounts of GST. With CYP2A13 N297A, 87% of the 3,4-epoxide was trapped, whereas with CYP2A13 o-HPA was no longer detectable when GSH and GST were added to the reaction mix. To determine the kinetic parameters of coumarin 7-hydroxylation and 3,4-epoxidation, reconstituted CYP2A13 (7.5 pmol) or CYP2A13 N297A (2–5 pmol) enzyme was incubated in the presence of GST and GSH with 1 to 500 μM coumarin under the conditions described above. Metabolites were analyzed by HPLC with UV detection at 310 nm (λmax for coumarin). By comparing absorbance at 310 nm to radioactivity in reactions carried out with [14C]coumarin, the ratio of the absorbance of 7-hydroxycoumarin relative to the coumarin GSH conjugate was determined to be 1.2. A standard curve of peak area versus 7-hydroxycoumarin was used to determine metabolite concentrations. HPLC conditions were those described above.
[3H](S)-NNN and [3H]NNK Metabolism. Reactions were carried out and the products of [5-3H]-(S)-NNN α-hydroxylation were quantified by radioflow HPLC as described previously (Schlicht et al., 2007). The concentrations of [5-3H]-(S)-NNN used were 10 to 1000 μM (specific activities ranged from 0.01 to 0.5 Ci · mmol–1). Reactions were conducted in duplicate for each sample. The NNK α-hydroxylation method was slightly modified from previously described methods (Jalas et al., 2003; Dicke et al., 2005). [3H]NNK was incubated with CYP2A13 (10 pmol) and CYP2A13 N297A (25 pmol) for 30 min at 37°C. Reactions were 0.2 ml in total volume and were conducted in duplicate. Substrate concentrations used ranged from 0.5 to 100 μM for CYP2A13 and from 10 to 300 μM for CYP2A13 N297A, specific activities ranged from 0.05 to 10 Ci · mmol–1. Sodium bisulfite (5 mM) was present in reaction mixtures to trap keto aldehyde. NNK metabolites were analyzed by radioflow HPLC as described previously (Dicke et al., 2005).
LC/MS/MS Analysis of Coumarin Metabolites. Products of coumarin metabolism were collected from the HPLC system described above and concentrated under N2. Reaction products and hydroxycoumarin standards were analyzed using a TSQ Quantum Ultra AM triple quadrupole mass spectrometer operated with electrospray ionization (ESI) (Thermo Fisher Scientific, Waltham, MA). Sample (4–8 μl) was loaded onto a Zorbax SB-C18 (5 μm; 0.5 × 150 mm) column with a flow rate of 10 μl · min–1 of 70% A (1% acetic acid in water) and 30% B (1% acetic acid in methanol) using a Waters nanoAcquity UPLC solvent delivery module (Waters, Milford, MA). The hydroxycoumarins were analyzed by ESI/MS/MS, with selection for the protonated molecular ion at m/z 163. The ESI source was set in positive ion mode; voltage, 3.3 kV; and capillary tube, 250°C. The argon collision gas pressure was 1.0 mTorr; collision energy, 20 to 25 V; scan time, 0.5 s; and peak width of Q1 and Q3, 0.7. A full scan was performed for detection of a coumarin GSH conjugate, with m/z 452 [M + H]+, and product spectra were also generated. In addition, the coumarin GSH conjugate was analyzed in negative ESI mode and the full product scan of the parent ion m/z 450 [M – H]– was evaluated. The ESI source parameters were voltage, 3.5 kV; and capillary tube, 250°C. The argon collision gas pressure was 1.0 mTorr; collision energy, 30 V; scan time, 0.55 s; and peak width of Q1 and Q3, 0.7.
Data Analysis. Kinetic parameters, Km and Vmax, for coumarin 7-hydroxylation, coumarin 3-hydroxylation, coumarin 3,4-epoxidation, (S)-NNN 5′-hydroxylation, and NNK α-hydroxylation were determined using the EZ-fit 5 kinetics software from Perrella Scientific (Amherst, NH). Statistical differences between mean Km or Vmax values were determined using a χ2 test.
Computer Modeling and Docking. Coordinates from the X-ray crystal structure for CYP2A13 (Protein Data Bank 2P85) (Smith et al., 2007) were obtained from the Protein Data Bank (http://www.rcsb.org) and used to model CYP2A13 and CYP2A13 N297A in MAESTRO version 3.5 (Schrodinger, LLC, New York, N Y). NNK, coumarin, and (S)-NNN were generated and minimized in MAESTRO before docking. Docking studies were conducted using the program GLIDE within MAESTRO to view the top 25 possible binding orientations (of 10,000 dockings) for the substrates within active sites of CYP2A13 and CYP2A13 N297A. Substrates were allowed to dock flexibly within the active sites.
Results
CYP2A13 and the single-residue mutant enzyme CYP2A13 N297A were expressed and purified. Protein expression levels were not hindered by the introduction of the Ala297 site-mutation as confirmed by Western blotting. Proper incorporation of heme was verified with CO-difference spectra.
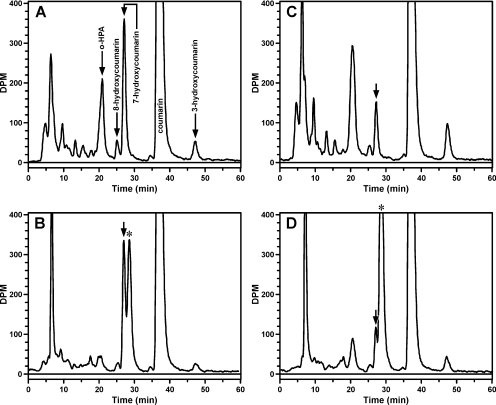
The relative metabolism of [14C]coumarin by wild-type CYP2A13 and the N297A mutant were analyzed by radioflow HPLC (Fig. 3). The major metabolites generated by the wild-type enzyme were o-HPA, a product of coumarin 3,4-epoxidation and 7-hydroxycoumarin (Fig. 3A). These two metabolites were present in approximately equal amounts. In contrast, o-HPA was a 3-fold more abundant metabolite of CYP2A13 N297A-catalyzed coumarin metabolism than was 7-hydroxycoumarin (Fig. 3C). In addition, other radioactive products, eluting before 10 min and at 48 min, accounted for a significant percentage of the radioactivity. The later-eluting metabolite coeluted with 3-hydroxycoumarin. Its identity as 3-hydroxycoumarin was confirmed by LS/MS/MS analysis of the collected peak. None of the early eluting peaks coeluted with available standards, including o-hydroxyphenyl acetic acid, 6,7-dihydroxycoumarin, and all hydroxycoumarin metabolites. We hypothesized these peaks may be secondary metabolites of the coumarin 3,4-epoxide or products of the conjugation of the 3,4-epoxide with a nucleophile or nucleophiles present in the enzyme reaction mixture. A minor product of both CYP2A13- and CYP2A13 N297A-catalyzed metabolism coeluted with 8-hydroxycoumarin.
Fig. 3.
Radioflow HPLC analysis of 65 μM [14C]coumarin metabolism by CYP2A13 (10 pmol; 30 min) (A and B) or CYP2A13 N297A (5 pmol; 20 min) (C and D). Reactions were carried out in the absence (A and C) or presence (B and D) of GSH and GST. Arrows in A indicate retention times of metabolite standards. The arrow in B to D indicate the retention time of 7-hydroxycoumarin. The asterisk (*) is the GSH/GST-dependent peak.
To confirm that the early eluting metabolite(s) are product(s) of coumarin 3,4-epoxidation, and to more accurately quantify the 3,4-epoxidation of coumarin, a method was developed that trapped coumarin 3,4-epoxide as its GSH conjugate. Conditions were established under which 87% or greater of the o-HPA formed was no longer detected as a product (Fig. 3, B and D). Efficient trapping of coumarin 3,4-epoxide required GSH concentrations of at least 1 mM and excess GST, because the reaction was sensitive to the amount of GST available.
Under these conditions, not only did little o-HPA remain but also the majority of the radioactivity eluting before 12 min was no longer detected. In the presence of GSH and GST, the size of the early eluting radioactive peak was the same with either wild-type or mutant enzyme. The addition of GSH and GST to the enzyme reactions resulted in the appearance of a new metabolite peak eluting 2 min later than 7-hydroxycoumarin. This peak was presumed to be a GSH conjugate of coumarin 3,4-epoxide (Fig. 2). The ratio of this peak to 7-hydroxycoumarin was 1.3:1 for CYP2A13 and 9:1 for CYP2A13 N297A (Fig. 3, B and D). These values are significantly greater than the ratio of o-HPA to 7-hydroxycoumarin obtained in the absence of GSH and GST (Fig. 3, A and C); consistent with our hypothesis that quantifying o-HPA alone did not accurately reflect the extent of coumarin 3,4-epoxidation.
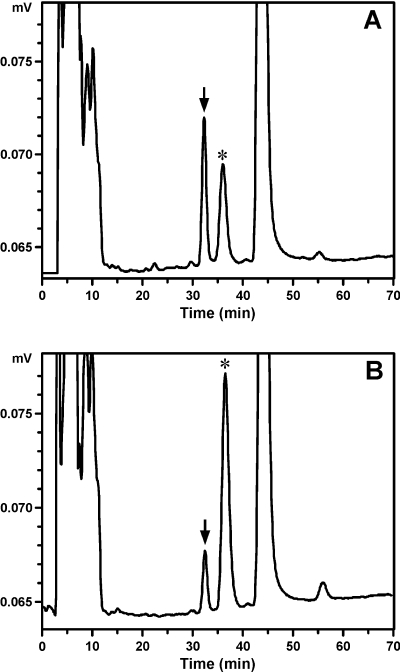
Using the conditions established with [14C]coumarin and radioflow HPLC analysis, a UV-HPLC method was developed for the detection of 7-hydroxycoumarin and the coumarin GSH conjugate. To determine the rate of coumarin 3,4-epoxidation, the coumarin GSH conjugate was quantified by HPLC with UV detection at 310 nm. Note that the absorbance of 7-hydroxycoumarin at 310 nm is 1.2-fold greater than the coumarin GSH conjugate. Sample chromatograms for the UV-HPLC analysis of coumarin 7-hydroxylation and 3,4-epoxidation by CYP2A13 and CYP2A13 N297A are illustrated in Fig. 4.
Fig. 4.
HPLC analysis of 20 μM coumarin metabolism by CYP2A13 (7.5 pmol; 20 min) (A) or CYP2A13 N297A (2 pmol; 20 min) (B) in the presence of GSH and GST. The arrow indicates the retention time of 7-hydroxycoumarin, and the asterisk (*) is the GSH/GST-dependent peak.
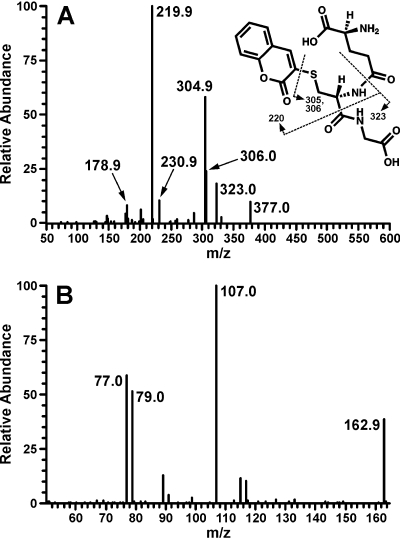
The presumed GSH conjugate, 7-hydroxycoumarin, and the metabolite that coeluted with 3-hydroxycoumarin were collected and analyzed by LC/MS/MS. Both negative and positive ion tandem mass spectrometry were used to identify the GSH conjugate. The product ion spectra from m/z 450 [M – H]– contained m/z 128, 143, 177, 179, 210, 254, and 272. The m/z 177 is γ-glutamyl-S-(3-coumarin)cysteiy glycine, and the other fragments are generated from the glutathione moiety (Vassallo et al., 2003). Mass spectral analysis with ESI in the positive mode detected a molecular ion of m/z 452 [M + H]+. The major ions in the product spectrum from m/z 452 were m/z 220; the 1-aminothioether of coumarin, m/z 323, (3-coumarin)cysteinyl glycine, and m/z, 305/306, (3-coumarin)cysteinyl glycine, minus NH3 (Fig. 5). The identities of the 7-hydroxycoumarin and 3-hydroxycoumarin metabolites were confirmed by comparison of product spectra from m/z 163 [M + H]+ to authentic standards. The major product ion was m/z 107 (Fig. 5).
Fig. 5.
ESI/MS/MS positive ion spectra of the collected coumarin-GSH conjugate [M + H]+, m/z 452 (A), and the late-eluting coumarin metabolite 3-hydroxycoumarin, m/z 163 (B). Spectra are identical to authentic standards.
The kinetic parameters of coumarin 7-hydroxylation, 3,4-epoxidation, and 3-hydroxycoumarin were determined for CYP2A13 N297A and CYP2A13 (Table 1). Kinetic parameters were determined in the presence of GSH and GST to better quantify 3,4-epoxidation. The Km values for these reactions were approximately 4-fold greater for the mutant enzyme. However, the Vmax values increased 3-fold for coumarin 7-hydroxylation, 19-fold for 3-hydroxylation, and 40-fold for 3,4-epoxidation. The catalytic efficiency of coumarin 3,4-epoxidation increased 10-fold, whereas coumarin 7-hydroxylation decreased by 25%. It is interesting to note that 3-hydroxycoumarin was still detected in the presence of GSH and GST, which would be expected if coumarin 3-hydroxylation occurs independently from the 3,4-epoxication pathway. The catalytic efficiency of coumarin 3-hydroxylation was 6-fold greater for the mutant enzyme. Coumarin 7-hydroxylation by wild-type CYP2A13 accounted for approximately 35% of coumarin metabolism by the 3 pathways, but only 9% of that by the mutant enzyme. Coumarin 8-hydroxylation was catalyzed by wild-type and the mutant enzyme, but it accounted for less than 3% of the total coumarin metabolism.
TABLE 1.
Kinetic parameters for CYP2A13 and CYP2A13 N297A reactions
Values for Km and Vmax represent mean ± S.E. of two to five independent experiments.
| Reactiona | CYP2A13 wt Km | CYP2A13 wt Vmax | CYP2A13 N297A Km | CYP2A13 N297A Vmax |
|---|---|---|---|---|
| μMb | pmol/min/pmol P450 | μMb | pmol/min/pmol P450 | |
| Coumarin 7-hydroxylation | 3.7 ± 0.39 | 0.59 ± 0.01 | 16.6 ± 0.9 | 1.94 ± 0.03 |
| Coumarin 3,4-epoxidation | 3.8 ± 0.43 | 0.25 ± 0.006 | 14.8 ± 1.1 | 9.25 ± 0.09 |
| Couamarin 3-hydroxylation | 6.3 ± 1.5 | 0.06 ± 0.003 | 18.9 ± 0.3 | 1.12 ± 0.03 |
| NNK α-hydroxylationc | 11.9 ± 3.2 | 0.85 ± 0.07 | 66.7 ± 8.1 | 0.87 ± 0.09 |
| (S)-NNN 5′-hydroxylation | 30.7 ± 10.1 | 1.28 ± 0.11 | 108 ± 22.1 | 1.12 ± 0.12 |
Coumarin concentrations were 1, 2, 5, 10, 20, 40, 100, 200, and 500 μM; NNK concentrations were 0.5, 2, 5, 10, 25, 50, 75, 100, 200, and 300 μM; and (S)-NNN concentrations were 10, 25, 50, 100, 250, 500, 750, and 1000 μM. Products were detected by UV-HPLC (coumarin) or radioflow-HPLC (NNK and NNN). Details are described under Materials and Methods.
Km values for all substrates were significantly different for CYP2A13 N297A compared with CYP2A13 (P < 0.03).
α-Hydroxylation was quantified as the sum of methyl and methylene hydroxylation that occurred in a ratio of 1:3 at all NNK concentrations.
The effect of the N297A mutation on CYP2A13-catalyzed α-hydroxylation of (S)-NNN and NNK was also examined. Metabolism of (S)-NNN by CYP2A13 and the CYP2A13 N297A resulted in generation of lactol, the product of NNN 5′-hydroxylation. The product of 2′-hydroxylation, keto alcohol, was not detected for either enzyme. The kinetic parameters for the 5′-hydroxylation of (S)-NNN are presented in Table 1. Only modest differences in the kinetic parameters for wild-type CYP2A13 and the N297A mutant were observed. The Km value for the mutant was ∼3-fold greater than that of the wild type, and the Vmax value was not significantly different. The N297A mutation had a somewhat greater effect on the efficiency of CYP2A13-catalyzed NNK α-hydroxylation (Table 1); there was no change in product distribution. Both CYP2A13 enzymes generated products of α-methyl and α-methylene hydroxylation in a ratio of 1:3. The Km value for total NNK α-hydroxylation by CYP2A13 N297A was 6-fold higher than for the wild-type enzyme; the Vmax value was unchanged.
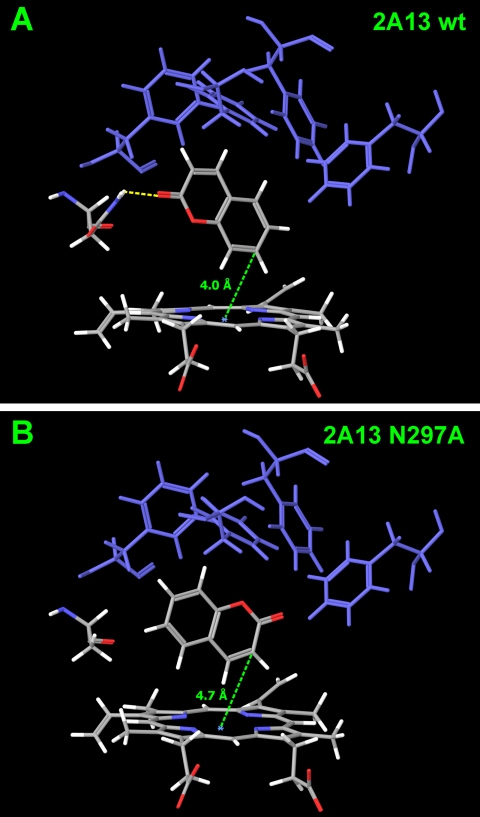
To further investigate the role of Asn297 on substrate orientation, coumarin, (S)-NNN, and NNK were computationally docked into the active site of CYP2A13 and CYP2A13 N297A. Docking studies generally complemented the enzyme kinetic analyses. When coumarin was docked in CYP2A13, it equally favored orientations for 7-hydroxylation and 3,4-epoxidation. Approximately 40% of the poses predicted a hydrogen bond forming between Asn297 and the carbonyl oxygen of coumarin, positioning the 7′-carbon over the heme iron, as close as 3.9 Å (Fig. 6). In the other major poses (40%), the coumarin molecule was flipped so that the double bound was positioned for 3,4-epoxidation by CYP2A13. In contrast, when coumarin was docked into the CYP2A13 N297A mutant, only 3,4-epoxidation was predicted (Fig. 6B).
Fig. 6.
Model of coumarin docked in the active site of CYP2A13 (A) and CYP2A13 N297A (B).
Docking of (S)-NNN resulted in similar predictions for either the wild-type or mutant CYP2A13. Orientations favorable for (S)-NNN 5′-hydroxylation were predicted 88% of the time in wild-type CYP2A13 and 75% of the time in mutant CYP2A13. Hydrogen bond formation was indicated (6 of 25 poses) in CYP2A13 between the nitrogen of the pyridine ring of NNN and Asn297. However, positioning of the pyridine nitrogen toward residue 297 was favored regardless of residue type or presence of the hydrogen bond interaction, predicting 5′-hydroxylation of (S)-NNN. This observation suggests less importance for the Asn297-substrate interaction in (S)-NNN substrate orientation.
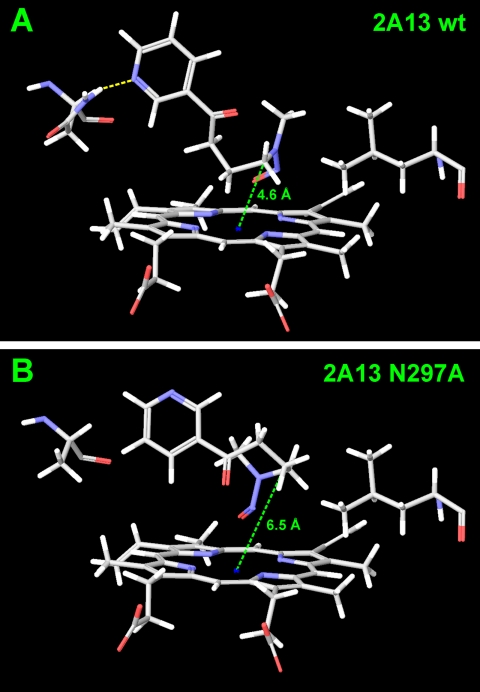
NNK docked into the active site of wild-type CYP2A13 with a consistent orientation. The pyridine nitrogen was always positioned toward Asn297 with hydrogen bonds occurring in 10 of 25 poses (Fig. 7A). The alkyl chain of NNK was positioned such that the methyl group was extended toward Leu366. Fourteen of the poses placed the methylene carbon closest to the heme iron (as close as 3.8 Å). The methyl carbon was placed closest to the heme iron in 11 of the poses (as close as 3.4 Å). Substitution of the Asn297 residue with Ala resulted in disruption of this consistent orientation for NNK (Fig. 7B). The pyridine nitrogen was twisted away from residue 297 in many of the poses (7 of 25 poses). In addition, the methyl moiety of NNK curled up to point toward the pyridine ring (12 of 25 poses).
Fig. 7.
Model of NNK docked in the active site of CYP2A13 (A) and CYP2A13 N297A (B).
Discussion
CYP2A13 is an efficient catalyst of tobacco-specific nitrosamine metabolism. The amino acids critical to CYP2A13-catalyzed metabolism of (S)-NNN and NNK α-hydroxylation have not been fully elucidated. Residue Asn297 has been implicated as an important residue for substrate orientation in CYP2A6. In this study, a single site mutation N297A was used to investigate the importance of the active site residue Asn297 on CYP2A13-catalyzed coumarin metabolism and α-hydroxylation of (S)-NNN and NNK.
We reported previously that CYP2A13 efficiently catalyzed the 3,4-epoxidation of coumarin as measured by the formation of o-HPA (von Weymarn and Murphy, 2003). However, it became obvious in carrying out the studies reported here that o-HPA formation did not accurately reflect the rate of coumarin 3,4-epoxidation. Therefore, to quantitatively measure coumarin 3,4-epoxidation, GSH and GST were included in the reaction, and the rate of coumarin GSH conjugate formation was determined. Using this methodology, the ratio of coumarin 7-hydroxylation to 3,4-epoxidation by wild-type CYP2A13 was 1.0 to 1.3 compared with a ratio of 1.0 to 0.7 we reported previously (von Weymarn and Murphy, 2003).
The N297A mutation had a striking impact on coumarin metabolism, significantly altering the metabolite product distribution. The CYP2A13 N297A mutant catalyzed coumarin 3,4-epoxidation with 10-fold greater efficiency than wild-type enzyme. This shift to 3,4-epoxidation is consistent with the preferred substrate orientation predicted by docking coumarin in the active site of the N297A mutant. With wild-type CYP2A13, coumarin may form a hydrogen bond with the carbonyl oxygen positioning the molecule for 7-hydroxylation. However, a second orientation stabilized by a cluster of phenyl alanines in the active site positions coumarin for 3,4-epoxidation. In the mutant enzyme, this orientation predominates, thereby favoring the 3,4-epoxidation of coumarin.
In contrast to the increase in total coumarin metabolism that was observed with CYP2A13 N297A compared with CYP2A13, both NNN and NNK metabolism by CYP2A13 N297A was significantly decreased. The catalytic efficiency of (S)-NNN α-hydroxylation decreased 4-fold, because of an increase in the Km value. The N297A mutation had little impact on substrate orientation of (S)-NNN in silico. Docking of (S)-NNN was similar in CYP2A13 and the N297A mutant, positioning (S)-NNN for 5′-hydroxylation in the active sites of both enzymes. These data suggest the hydrogen bond to Asn297 is not critical to NNN metabolism by CYP2A13, but it seems to improve enzyme affinity for NNN. The impact of the N297A mutation on NNK α-hydroxylation was greater. Catalytic efficiency was decreased 6-fold relative to wild-type CYP2A13. Docking studies reported here and previously indicated that residue Asn297 is important for the orientation of NNK in the active site of CYP2A13 (Smith et al., 2007). Changing this residue from an Asn to Ala caused NNK to dock in poses unfavorable for α-methyl and α-methylene hydroxylation.
In CYP2A6, mutation of residue Asn297 has been shown to influence substrate binding and metabolism. As observed in CYP2A13, mutation of Asn297 in CYP2A6 results in altered coumarin 7-hydroxylation. An N297S mutant was determined to have ∼4-fold decreased catalytic efficiency for coumarin 7-hydroxylation (Kim et al., 2005). This mutation also decreased binding affinity for coumarin by nearly 30-fold. The rate of coumarin 3,4-epoxidation was not measured in this study; however, given the dramatically decreased binding affinity of coumarin it is unlikely the N297S CYP2A6 mutant catalyzed this reaction. Two additional mutants, N297H and N297Q have also been shown to have decreased catalytic efficiency for coumarin 7-hydroxylation compared with wild-type CYP2A6 (Nakamura et al., 2001). Recently, the crystal structure for the CYP2A6 N297Q mutant was reported (Sansen et al., 2007). Although the glutamine substitution had little effect on the overall size of the active site, it did cause altered protein folding interactions between the B′-C helix region and helix I. In addition, no hydrogen bond interactions were able to form between Gln297 and substrates such as coumarin.
The ability of Asn297 to hydrogen bond with substrates has been demonstrated in both CYP2A6 and CYP2A13 crystal structures. This interaction was shown to orient indole in the active site of CYP2A13 and to position coumarin for 7-hydroxylation in CYP2A6. The importance of Asn297 for CYP2A13 substrate orientation and catalysis was confirmed by the present study.
Acknowledgments
We thank Jessica van Lengerich for assistance in carrying out the coumarin 3,4-epoxidation assays and Yuk Sham for assistance with P450 docking experiments, carried out in the Minnesota Supercomputing Institute (Minneapolis, MN).
This work was supported in part by the National Institutes of Health National Cancer Institute [Grant R01-CA84529]; the National Institutes of Health National Institute on Drug Abuse [Grant F30-DA020968], predoctoral fellowship; and the National Institutes of Health National Cancer Institute [Grant P30-CA77598], mass spectrometry in the Analytical Biochemistry Core.
J.Z.B. and K.E.S. contributed equally to this work.
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
doi:10.1124/dmd.108.025072.
ABBREVIATIONS: (S)-NNN, (S)-N′-nitrosonornicotine; NNK, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone; keto alcohol, 4-oxo-4-(3-pyridyl)-1-butanol; keto aldehyde, 4-oxo-4-(3-pyridyl)-1-butanone; lactol, 5-(3-pyridyl)-2-hydroxytetrahydrofuran; P450, cytochrome P450; o-HPA, o-hydroxyphenylacetaldehyde; HPLC, high-performance liquid chromatography; GSH, glutathione; GST, glutathione transferase; LC/MS/MS, liquid chromatography-tandem mass spectrometry; ESI, electrospray ionization.
References
- Born SL, Rodriguez PA, Eddy CL, and Lehman-McKeeman LD (1997) Synthesis and reactivity of coumarin 3,4-epoxide. Drug Metab Dispos 25 1318–1324. [PubMed] [Google Scholar]
- Dicke KE, Skrlin SM, and Murphy SE (2005) Nicotine and 4-(methylnitrosamino)-1-(3-pyridyl)-butanone (NNK) metabolism by P450 2B6. Drug Metab Dispos 33 1760–1764. [DOI] [PubMed] [Google Scholar]
- Hanna IH, Teiber JF, Kokones KL, and Hollenberg PF (1998) Role of the alanine at position 363 of cytochrome P450 2B2 in influencing the NADPH- and hydroperoxide-supported activities. Arch Biochem Biophys 350 324–332. [DOI] [PubMed] [Google Scholar]
- Hecht SS (1998) Biochemistry, biology, and carcinogenicity of tobacco-specific N-nitrosamines. Chem Res Toxicol 11 559–603. [DOI] [PubMed] [Google Scholar]
- Honkakoski P and Negishi M (1997) The structure, function, and regulation of cytochrome P450 2A enzymes. Drug Metab Rev 29 977–996. [DOI] [PubMed] [Google Scholar]
- International Agency for Research on Cancer (2007) Smokeless tobacco and tobacco-specific nitrosamines, in IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, vol 89, pp 564, International Agency for Research on Cancer, Lyon, France. [Google Scholar]
- Jalas JR, Ding X, and Murphy SE (2003) Comparative metabolism of the tobacco-specific nitrosamines 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL) by rat cytochrome P450 2A3 and human cytochrome P450 2A13. Drug Metab Dispos 31 1199–1202. [DOI] [PubMed] [Google Scholar]
- Jalas JR, Hecht SS, and Murphy SE (2005) Cytochrome P450 enzymes as catalysts of metabolism of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), a tobacco-specific carcinogen. Chem Res Toxicol 18 95–110. [DOI] [PubMed] [Google Scholar]
- Kim D, Wu ZL, and Guengerich FP (2005) Analysis of coumarin 7-hydroxylation activity of cytochrome P450 2A6 using random mutagenesis. J Biol Chem 280 40319–40327. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Martin MV, and Guengerich FP (2001) Random mutagenesis of human cytochrome p450 2A6 and screening with indole oxidation products. Arch Biochem Biophys 395 25–31. [DOI] [PubMed] [Google Scholar]
- Omura T and Sato R (1964) The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J Biol Chem 239 2370–2378. [PubMed] [Google Scholar]
- Sansen S, Hsu MH, Stout CD, and Johnson EF (2007) Structural insight into the altered substrate specificity of human cytochrome P450 2A6 mutants. Arch Biochem Biophys 464 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlicht KE, Michno N, Smith BD, Scott EE, and Murphy SE (2007) Functional characterization of CYP2A13 polymorphisms. Xenobiotica 37 1439–1449. [DOI] [PubMed] [Google Scholar]
- Shultz MA, Choudary PV, and Buckpitt AR (1999) Role of murine cytochrome P-4502F2 in metabolic activation of naphthalene and metabolism of other xenobiotics. J Pharmacol Exp Ther 290 281–288. [PubMed] [Google Scholar]
- Smith BD, Sanders JL, Porubsky PR, Lushington GH, Stout CD, and Scott EE (2007) Structure of the human lung cytochrome P450 2A13. J Biol Chem 282 17306–17313. [DOI] [PubMed] [Google Scholar]
- Soucek P (1999) Expression of cytochrome P450 2A6 in Escherichia coli: purification, spectral and catalytic characterization, and preparation of polyclonal antibodies. Arch Biochem Biophys 370 190–200. [DOI] [PubMed] [Google Scholar]
- Su T, Bao Z, Zhang QY, Smith TJ, Hong JY, and Ding X (2000) Human cytochrome P 450 CYP2A13: Predominant expression in the respiratory tract and in high efficiency metabolic activation of a tobacco-specific carcinogen, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Cancer Res 60 5074–5079. [PubMed] [Google Scholar]
- Vassallo JD, Hicks SM, Born SL, and Daston GP (2004) Roles for epoxidation and detoxification of coumarin in determining species differences in Clara cell toxicity. Toxicol Sci 82 26–33. [DOI] [PubMed] [Google Scholar]
- Vassallo JD, Morrall SW, Fliter KL, Curry SM, Daston GP, and Lehman-McKeeman LD (2003) Liquid chromatographic determination of the glutathione conjugate and ring-opened metabolites formed from coumarin epoxidation. J Chromatogr B Analyt Technol Biomed Life Sci 794 257–271. [DOI] [PubMed] [Google Scholar]
- von Weymarn LB and Murphy SE (2003) CYP 2A13-catalyzed coumarin metabolism, comparison to CYP2A5 and CYP2A6. Xenobiotica 33 73–81. [DOI] [PubMed] [Google Scholar]
- Von Weymarn LB, Sridar C, and Hollenberg PF (2004) Identification of amino acid residues involved in the inactivation of cytochrome P450 2B1 by two acetylenic compounds: the role of three residues in non-substrate recognition sites. J Pharmacol Exp Ther 311 71–79. [DOI] [PubMed] [Google Scholar]
- von Weymarn LB, Zhang QY, Ding X, and Hollenberg PF (2005) Effects of 8-methoxypsoralen on cytochrome P450 2A13. Carcinogenesis 26 621–629. [DOI] [PubMed] [Google Scholar]
- Wong HL, Murphy SE, and Hecht SS (2005a) Cytochrome P450 2A-catalyzed metabolic activation of structurally similar carcinogenic nitrosamines: N′-nitrosonornicotine enantiomers, N-nitrosopiperidine, and N-nitrosopyrrolidine. Chem Res Toxicol 18 61–69. [DOI] [PubMed] [Google Scholar]
- Wong HL, Zhang X, Zhang QY, Gu J, Ding X, Hecht SS, and Murphy SE (2005b) Metabolic activation of the tobacco carcinogen 4-(methylnitrosamino)-(3-pyridyl)-1-butanone by cytochrome P450 2A13 in human fetal nasal microsomes. Chem Res Toxicol 18 913–918. [DOI] [PubMed] [Google Scholar]
- Yano JK, Hsu MH, Griffin KJ, Stout CD, and Johnson EF (2005) Structures of human microsomal cytochrome P450 2A6 complexed with coumarin and methoxsalen. Nat Struct Mol Biol 12 822–823. [DOI] [PubMed] [Google Scholar]
- Zhu LR, Thomas PE, Lu G, Reuhl KR, Yang GY, Wang LD, Wang SL, Yang CS, He XY, and Hong JY (2006) CYP2A13 in human respiratory tissues and lung cancers: an immunohistochemical study with a new peptide-specific antibody. Drug Metab Dispos 34 1672–1676. [DOI] [PubMed] [Google Scholar]