Abstract
Cytosolic sulfotransferases (SULTs) are a family of Phase II drug-metabolizing enzymes that catalyze the transfer of a sulfonate group from 3′-phosphoadenosine 5′-phosphosulfate to endogenous and xenobiotic compounds. Several SULT isoform messages have been detected in the human brain; however, protein expression patterns have not been characterized. Immunoblot analysis of the SULT1A1 and 1A3 isoforms was carried out with cytosolic fractions isolated from superior temporal gyrus, hippocampus, cerebellum, occipital pole, frontal pole, and temporal pole regions of normal adult human brains. SULT1A1 expression was highest in cytosolic fractions isolated from cerebellum, occipital, and frontal lobes, whereas, SULT1A3 expression was highest in cytosol from superior temporal gyrus, hippocampus, and temporal lobe. SULT1A1 and SULT1A3 immunoreactivities were found in both neurons and glial cells by immunohistochemical analysis in all brain regions studied. SULT1A1 is known to catalyze the metabolism of small phenols, whereas SULT1A3 sulfates catecholamine neurotransmitters. Because SULT1A1 and 1A3 have distinct substrate specificities, the differences in expression pattern and cellular localization of the SULT1A isoforms are probably associated with the distribution and function of their selective substrates in the different brain regions.
Cytosolic sulfotransferases (SULTs) are a family of Phase II drug-metabolizing enzymes that catalyze the transfer of a sulfonate moiety from the universal sulfate donor 3-phosphoadenosine 5-phosphosulfate to the hydroxyl, sulfhydryl, amino, or N-oxide groups of a variety of endogenous and xenobiotic substrates (Falany, 1991). The cytosolic SULTs have an important role in the metabolism of many endogenous compounds including steroids, bile acids, thyroid hormones, and monoamine neurotransmitters (MNTs) (Weinshilboum et al., 1997). These enzymes are widely distributed throughout the body and serve to inactivate and increase water solubility of xenobiotics and therapeutic drugs (Falany et al., 2006). The cytosolic SULTs are also known to bioactivate certain prodrugs as well as to initiate chemical carcinogenesis (Glatt 2000).
Human cytosolic SULTs are divided into two major subfamilies, the phenol SULTs (SULT1) and the hydroxysteroid SULTs (SULT2), based upon their specific substrate reactivities (Blanchard et al., 2004). The SULT isoforms have a widespread tissue distribution and are expressed in many tissues including liver, lung, brain, skin, platelets, breast, kidney, and gastrointestinal tissue (Nowell and Falany, 2006) while the SULT1A isoforms have been studied extensively in platelets (Hart et al., 1979), liver (Young et al., 1988), breast (Falany et al., 1993), and gastrointestinal tract (Teubner et al., 2007); little is known concerning SULT expression in the brain. The expression of the SULT1A isoforms have been reported in whole human brain preparations (Hart et al., 1979; Whittemore et al., 1986; Zou et al., 1990; Vietri et al., 2003; Yasuda et al., 2007); however, protein expression in distinct brain regions has not been extensively characterized. Other investigators have reported global expression of SULT1 isoforms in human fetal brain (Richard et al., 2001; Stanley et al., 2005) as well as in rodent models (Falany et al., 2000; Alnouti and Klaassen, 2006). The normal human adult brain SULT expression pattern cannot be compared with that of the fetal or rodent models, because the isoforms are dynamic throughout ontogeny and phylogeny.
The SULT1, or phenol SULT family, consists of at least eight isoforms involved in the conjugation of small phenolic compounds, estrogens, catecholamines, and xenobiotics, including many therapeutic drugs (Meloche 2003). SULT1A isoforms are the classic drug-metabolizing subfamily and have a variety of substrates including small phenols, aromatic amines, hydroxylamines, estrogens, and MNTs (Falany et al., 1993; Hempel et al., 2005, 2007). The best characterized members of the SULT1A family are SULTs 1A1 and 1A3. These isoforms have been purified from whole human brain and demonstrated to sulfonate many endogenous compounds found in the brain, including dopamine and epinephrine, as well as many therapeutic drugs (Richard et al., 2001; Vietri et al., 2003; Yasuda et al., 2007). The SULT1A isoforms are hypothesized to be involved in catecholamine neurotransmitter metabolism (SULT1A3), clearance of drugs and toxins, and generation of mutagens and teratogens (SULT1A1) in the brain.
Understanding the expression patterns of the SULT1A isoforms is clinically important because they metabolize many therapeutic drugs. Polymorphisms have been reported in the SULT1A family and have been shown to result in altered enzyme activity, which may have an effect on the processing of xenobiotics (therapeutic drugs) or brain physiology (MNTs) (Bardakci et al., 2008; Gjerde et al., 2008). In therapeutic studies, different SULT1A1 allozymes have been shown to exhibit substantial differences in catalysis of xenobiotic conjugation (Raftogianis, 1997; Bardakci et al., 2008). In particular, these polymorphisms have been implicated in altered metabolism of 4-hydroxytamoxifen (Gjerde et al., 2008) and changes in bioactivation of pathological electrophiles (Falany, 2004). Differences in tamoxifen metabolism may have considerable clinical consequences because they are associated with therapeutic outcome and should be considered when choosing a treatment regimen (Nowell and Falany, 2006).
Alterations in SULT1A expression may also affect normal brain physiology. More recently, genetic linkage of the SULT1A locus and a predisposition to autism spectrum disorders (ASD) has been reported (Dooley et al., 1993; Kumar et al., 2008; Weiss et al., 2008). Furthermore, individuals with ASD have decreased expression of SULT1A isoforms in platelets and in the gastrointestinal tract (Waring et al., 1996; Waring and Klovrza, 2000; Whiteley and Shattock, 2002; Kern et al., 2004). These decreases in SULT1A catalytic activity are also associated with increases in circulating levels of putative substrates such as neurotransmitters and catechols (Whiteley and Shattock, 2002). Currently, the levels of SULT1A isoforms in the brains of individuals with ASD is unknown. Identifying the normal expression profiles for the SULT1A isoforms in human brain will assist in clarifying the functions of the SULTs in normal human brain as well as lead to progress in understanding the pathological outcomes when these isoforms are aberrantly expressed.
Materials and Methods
Immunohistochemistry. Paraffin blocks of human brain regions were obtained from the Tissue Procurement Service of the Comprehensive Cancer Center of the University of Alabama at Birmingham (UAB) (Birmingham, Alabama). The blocks were cut into 5-μm serial slices, mounted to microscope slides coated with poly-l-lysine, and oven-dried by the UAB Comparative Histology Laboratory. Slides were deparaffinized in xylenes and rehydrated in alcohol baths (absolute, 95%, 70%) then rinsed in dH20 and Tris wash buffer (0.5 M Tris, 0.15 M NaCl, 0.01% Triton X-100, pH 7.6). Optimal staining was achieved without the use of antigen retrieval techniques. Endogenous peroxidase activity was quenched with 3% H2O2 for 10 min then slides were blocked in 1% goat serum for 1 h in humidity chambers. Primary antibody, either rabbit anti-human SULT1A1 IgG or a polyclonal rabbit anti-human SULT1A3 peptide, was diluted in PBE (phosphate-buffered saline with 500 mM EDTA, 1% bovine serum albumin, pH 7.6) at a ratio of 1:100 and incubated on slides for 1 h in humidity chambers. The rabbit anti-SULT1A1 antibody was raised against pure SULT1A1 and due to the high sequence identity between the SULT1A isoforms reacts with both isoforms (Falany, 2004). A selective rabbit polyclonal anti-human SULT1A3 antibody was generated using a peptide with the sequence EVNDPGEPSGLETLK (83–97) as the immunogen (Open Biosystems, Huntsville, AL). Monoclonal antibodies against neuronal nuclei and glial fibrillary acid protein were purchased from Millipore Bioscience Research Reagents (Temecula, CA) and diluted in PBE according to the manufacturer's specifications. Negative control slides were incubated in preimmune or 1% goat serum instead of primary antibody. Slides were washed two times in Tris wash buffer for 5 min before incubation for 10 min with anti-rabbit biotin conjugate (Jackson ImmunoResearch Laboratories Inc., West Grove, PA) diluted 1:500 in PBE. Slides were washed twice in Tris wash buffer for 5 min then incubated for 5 min with streptavidin-labeled conjugate (Jackson ImmunoResearch Laboratories Inc.), diluted 1:500 in PBE. Slides were washed twice in Tris wash buffer for 5 min, and the liquid 3,3′-diaminobenzidine (DAB kit; Biogenex, San Ramon, CA) was used to develop the slides according to manufacturer's instructions. Slides were rinsed in dH2O then counterstained in hematoxylin for 45 s. After rinsing in H2O for 4 min, slides were dehydrated in graded alcohols and xylenes and permanently coverslipped. Stained slides were imaged using a Zeiss Axiostar microscope with camera (Carl Zeiss Microimaging, Inc., Thornwood, NJ) and corresponding AxioVision software (Carl Zeiss Microimaging, Inc.). Tissues stained for immunohistochemical analysis using the SULT1A1 Ig antibody were compared with serial sections of the same tissue stained with the SULT1A3 peptide antibody to determine which isoforms were expressed in distinct cell types.
Immunoblot Analysis. Frozen sections of different regions of normal adult human brains were obtained from the UAB Alzheimer's Disease Research Center. Tissue from normal superior temporal gyrus, hippocampus, cerebellum, occipital pole, frontal pole, and temporal pole was used to prepare cytosolic fractions. Approximately 200 mg of tissue was homogenized in 1 ml of ice-cold phosphate buffer (10 mM KH2PO4, 1 M dithiothreitol, 10% glycerol, pH 7.4) then centrifuged at 100,000g for 1 h. Supernatant fractions were aliquoted and stored at –80°C. Total cytosolic protein was determined by the Bradford assay (Bio-Rad, Hercules, CA) using gamma globulin as a standard. For immunoblots, 100 to 200 μg of cytosolic protein was heated to 95°C for 5 min in sample loading buffer containing SDS and β-mercaptoethanol. Bacterially expressed purified human SULTs were used as positive controls, to generate standard curves for isoform quantitation and to standardize immunoreactivity (data not shown). Samples were loaded onto a 10% SDS/polyacrylamide gel and electrophoresed at 200 V for 45 min. Proteins resolved in polyacrylamide gels were transferred to nitrocellulose membranes via semidry transfer (Bio-Rad) at 12 V for 20 min in semidry transfer buffer (48 mM Tris, 39 mM glycine, 20% methanol, pH 9.2). Membranes were blocked from 4 h to overnight at 4°C in absolute SEA Block (Thermo Fisher Scientific, Waltham, MA). After blocking, membranes were washed three times in Tris-buffered saline in Tween 20 (TBST) before overnight incubation at 4°C with the primary antibody, diluted 1:1000 in 0.1% milk in TBST. Membranes were washed twice for 5 min in TBST at room temperature then incubated in goat anti-rabbit horseradish peroxidase diluted 1:50,000 in 0.1% milk in TBST for 1 h at room temperature. Immunoblots were developed in West Pico or Fempto SuperSignal (Thermo Fisher Scientific) and exposed to autoradiograph film. Relative quantitation of bands was performed using Un-Scan-IT (version 6.1) software (Silk Scientific Inc., Orem, UT) for densitometry and normalizing data to the ratio of SULT1A1 to SULT1A3 immunoreactivity. The ratios were corrected to represent actual protein expression of the isoforms because SULT1A1 is more immunoreactive than SULT1A3 for the same amount of protein using the SULT1A1 IgG antibody.
Results and Discussion
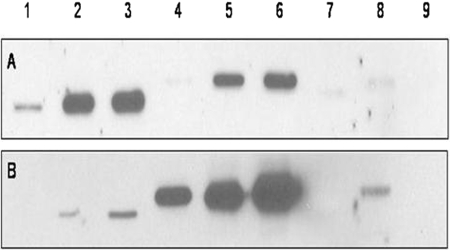
Specific Immunoreactivity. SULT1A1 and SULT1A3 protein expression was detected predominantly with rabbit anti-human SULT1A1 IgG (Fig. 1). The rabbit anti-SULT1A3 peptide antibody showed only slight reactivity to SULT1A1-expressed human protein (Fig. 1). Only negligible cross-reactivity to other SULT1 isoforms was shown with either antibody, except for SULT1C2 (Fig. 1). Although SULT1C2 did show cross-reactivity for the SULT1A3 peptide antibody, the isoforms are distinguished easily by their electrophoretic migration patterns. Despite the cross-reactivity, SULT1C2 was not detected in the brain sections and the SULT1A3 antibody was not used in the brain immunoblot experiments. The anti-SULT1A1 IgG detects both SULT1A1 and SULT1A3 proteins with similar affinity due to the high degree of sequence identity (93%) (Falany, 2004) between these isoforms. Because the anti-SULT1A1 antibody detects both SULT1A proteins and shows no significant cross-reactivity, it was used to quantify the relative amount of the proteins expressed in cytosol isolated from normal human brain sections.
Fig. 1.
Immunoblot of standardized expressed human enzymes. Human expressed protein purified from bacteria was resolved by 10% SDS/polyacrylamide gel electrophoresis, semidry transferred to nitrocellulose, blocked in SEA Block, and incubated with anti-SULT1A1 IgG (1:1000) overnight for blot A and with anti-SULT1A3 peptide (1:1000) overnight for blot B. The immunoblots were developed in Pico West (Thermo Fisher Scientific) and exposed to film 20 m: Lane 1, SULT1A1 0.1 μg; Lane 2, 2 μg; Lane 3, 4 μg; Lane 4, SULT1A3 0.25 μg; Lane 5, 2 μg; Lane 6, 4 μg; Lane 7, SULT1B1 4 μg; Lane 8, SULT 1C2 0.75 μg; Lane 9, SULT 1E1 4 μg.
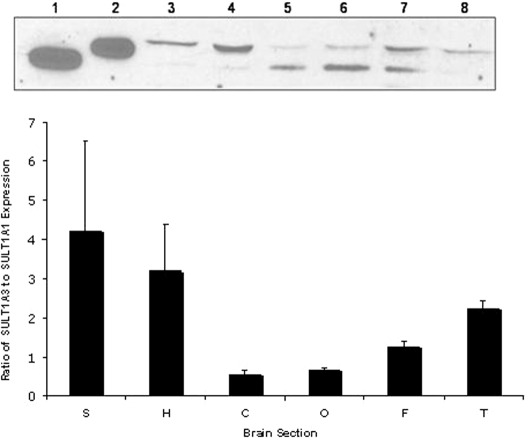
Expression Patterns of SULTs 1A1 and 1A3. Immunoreactive SULT1A1 and SULT1A3 were detected in all regions of the normal human brains analyzed (Fig. 2). However, the expression of these two isoforms varies between brain sections and among brains. Figure 2 depicts an inverse expression pattern between SULT1A1 and SULT1A3 in select brain regions. In general, in sections where SULT1A1 showed higher protein expression levels, SULT1A3 showed lower protein expression, and where SULT1A3 expression was higher, SULT1A1 expression was lower. This was apparent in all regions except the frontal lobe, which showed similar immunoreactivity for SULT1A1 and SULT1A3. In general, SULT1A1 was most highly expressed in the cerebellum and occipital lobe, whereas SULT1A3 demonstrated highest expression in the superior temporal gyrus, hippocampus, and temporal lobe. In the frontal lobe, both isoforms were highly expressed.
Fig. 2.
Expression and distribution of SULT1A isoforms in human brain. Top, cytosol from brain sections (200 μg of total protein) of a 64-year-old white man was blotted against anti-human 1A1 IgG diluted 1:1000: Lane 1, expressed human SULT1A1; Lane 2, expressed human SULT1A3; Lane 3, superior temporal gyrus; Lane 4, hippocampus; Lane 5, cerebellum; Lane 6, occipital lobe; Lane 7, frontal lobe; Lane 8, temporal lobe. Bottom, relative protein expression of SULT1A1 compared with SULT1A3 by densitometric analysis and normalization to standard curves; n ≥ 3, error bars represent mean ± S.E.M.
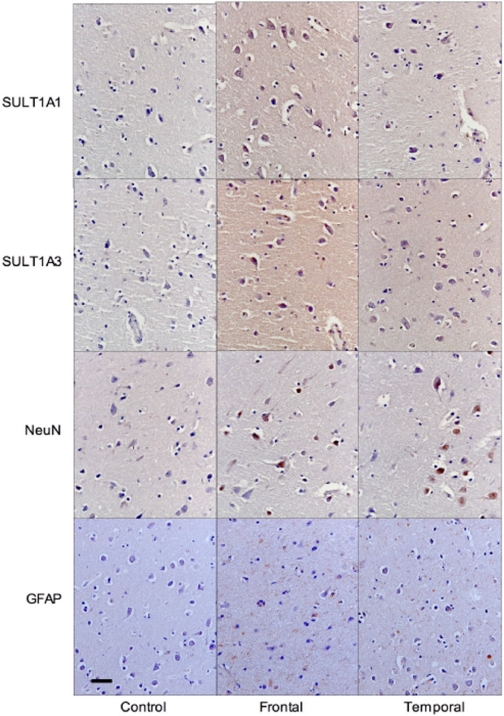
Cellular Localization of SULTs 1A1 and 1A3. SULT1A1 and SULT1A3 are expressed in multiple cell types in the human brain (Fig. 3). Immunohistochemical analysis of normal frontal and temporal lobes shows the expression of these isoforms in both neurons and in glial cells. The antibodies to neuronal nuclei and glial fibrillary acid protein were used as markers for these cell types, respectively. However, the SULT1A isoforms were not expressed uniformly in all neurons and glial cells. As shown in Fig. 3, only select neurons are immunoreactive for SULT1A in frontal and temporal lobes. Figure 3 also shows that SULT1A staining is observed in microglial and oligodendrocyte cells. Immunoreactivity for the SULT1A isoforms is localized to the cytosol in the subcellular compartments of these cell types. This is relevant because some of the human cytosolic SULT isoforms (SULT2B1b) have been reported to have inducible nuclear localization in non-neural tissues (Falany et al., 2006). Neuronal localization of the phenol SULT isoforms was first reported by Zou et al. (1990) using a nonspecific anti-SULT1A antibody. These investigators did not report the differential expression of the two isoforms. Whereas the SULT1A isoforms are expressed in neurons, this report demonstrates that SULT1A expression is not limited to neurons, and expression can be detected in multiple regions of the human brain. In the temporal lobe, SULT1A3 is expressed predominantly in microglia and oligodendrocytes, and this may account for the increased immunoreactivity of SULT1A3 in this brain region. Differential immunoreactivity may be associated with epitope availability, substrate distribution, neuronal subtype, and synaptic status. Western blot analysis of cultured human SH-SY5Y neurons and CCF-STTG1 astrocytes was performed to further examine cell type-specific expression of the SULT1A isoforms. SULT1A1 and SULT1A3 protein expression was not detected in these cultured cells (data not shown), implying that the SULT1A isoforms are most likely expressed in specific neural subtypes rather than constitutively in all cells of the human brain.
Fig. 3.
Immunohistochemical analysis of SULT1A1 and SULT1A3 in normal human brain. Normal frontal lobe from a 70-year-old African American female and normal temporal lobe from a 44-year-old white female were stained for 1 h in appropriate dilution of antibody after blocking in goat serum. Control slides incubated in goat serum with no antibody. Slides counterstained in hematoxylin after biotin-streptavidin labeling. Slides imaged at 40× magnification, bar represents 50 μm. NeuN, neuronal nuclei; GFAP, glial fibrillary acid protein.
This report demonstrates that SULT1A1 and SULT1A3 are differentially expressed in multiple regions of normal adult human brain. Furthermore, it has been shown that the SULT1A isoforms are located in the cytosol of multiple cell types in regions of normal human brain. A varying pattern of expression between brain regions is probably due to the different physiological functions of the isoforms. SULT1A1 is a xenobiotic-conjugating enzyme with a broad substrate range, whereas SULT1A3 is responsible for the regulation of the rapidly fluctuating levels of neurotransmitters. A better understanding of the specific cell types that express the SULT1A isoforms needs to be elucidated. By exploring the role of the SULT1A isoforms in the normal human brain, the underlying pathology in neurodevelopmental diseases, such as ASD, can be better understood. Future investigation will include characterization of the SULT isoforms in cell culture models, multiple regions of normal fetal brain, as well as comparison of expression patterns of the SULT isoforms in regions of non-normal brains, e.g., Alzheimer's disease and schizophrenia.
This work was supported in part by the National Institutes of Health National Institute of General Medical Sciences [Grant GM38953]; and the National Institutes of Health National Cancer Institute [Grant CA128897].
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
doi:10.1124/dmd.108.025767.
ABBREVIATIONS: SULT, cytosolic sulfotransferase; MNTs, monoamine neurotransmitters; ASD, autism spectrum disorders; PBE, phosphate-buffered saline with EDTA; TBST, Tris-buffered saline in Tween 20.
References
- Alnouti Y and Klaassen CD (2006) Tissue distribution and ontogeny of sulfotransferase enzymes in mice. Toxicol Sci 93 242–255. [DOI] [PubMed] [Google Scholar]
- Bardakci F, Arslan S, Bardakci S, Binatli AO, and Budak M (2008) Sulfotransferase 1A1 (SULT1A1) polymorphism and susceptibility to primary brain tumors. J Cancer Res Clin Oncol 134 109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard RL, Freimuth RR, Buck J, Weinshilboum RM, and Coughtrie MW (2004) A proposed nomenclature system for the cytosolic sulfotransferase (SULT) superfamily. Pharmacogenetics 14 199–211. [DOI] [PubMed] [Google Scholar]
- Dooley TP, Obermoeller RD, Leiter EH, Chapman HD, Falany CN, Deng Z, and Siciliano MJ (1993) Mapping of the phenol sulfotransferase gene (STP) to human chromosome 16p12.1-p11.2. Genomics 18 440–443. [DOI] [PubMed] [Google Scholar]
- Falany CN (1991) Molecular enzymology of human liver cytosolic sulfotransferases. Trends Pharmacol Sci 12 255–259. [DOI] [PubMed] [Google Scholar]
- Falany JL, Lawing L, and Falany CN (1993) Identification and characterization of cytosolic sulfotransferase activities in MCF-7 human breast carcinoma cells. J Steroid Biochem Mol Biol 46 481–487. [DOI] [PubMed] [Google Scholar]
- Falany CN, He D, Dumas N, Frost AR, and Falany JL (2006) Human cytosolic sulfotransferase 2B1: isoform expression, tissue specificity and subcellular localization. J Steroid Biochem Mol Biol 102 214–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falany C (2004). Human cytosolic sulfotransferases: properties, physiological functions, and toxicology, in Methods in Pharmacology and Toxicology, Drug Metabolism and Transport: Molecular Methods and Mechanism (Lash LH ed) pp 341–378, Humana Press, Totowa.
- Falany CN, Xie X, Wang J, Ferrer J, and Falany JL (2000) Molecular cloning and expression of novel sulfotransferase-like cDNAs from human and rat brain. Biochem J 346 857–864. [PMC free article] [PubMed] [Google Scholar]
- Gjerde J, Hauglid M, Breilid H, Lundgren S, Varhaug JE, Kisanga ER, Mellgren G, Steen VM, and Lien EA (2008) Effects of CYP2D6 and SULT1A1 genotypes including SULT1A1 copy number on tamoxifen metabolism. Ann Oncol 19 56–61. [DOI] [PubMed] [Google Scholar]
- Glatt HR (2000) An overview of bioactivation and chemical carcinogens. Biochem Soc Trans 28 1–6. [DOI] [PubMed] [Google Scholar]
- Hart RF, Renskers KJ, Nelson EB, and Roth JA (1979) Localization and characterization of phenol sulfotransferase in human platelets. Life Sci 24 125–130. [DOI] [PubMed] [Google Scholar]
- Hempel N, Gamage N, Martin JL, and McManus ME (2007) Human cytosolic sulfotransferase SULT1A1. Int J Biochem Cell Biol 39 685–689. [DOI] [PubMed] [Google Scholar]
- Hempel N, Negishi M, and McManus ME (2005) Human SULT1A genes: cloning and activity assays of the SULT1A promoters. Meth Enzymol 400 147–165. [DOI] [PubMed] [Google Scholar]
- Kern J, Grannemann BD, Trivedi MH, Waring RH, Ramsden DB, and Garver CR (2004) Abnormal sulfation chemistry in autism, in Trends in Autism Research (Ryaskin OT ed) pp 197–219, Nova Science Publishers, Hauppauge.
- Kumar RM, KaraMohamed S, Sudi J, Conrad DF, Brune C, Badner JA, Gilliam TC, Nowak NJ, Cook EH Jr, Dobyns WB, et al. (2008) Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet 17 628–638. [DOI] [PubMed] [Google Scholar]
- Meloche C (2003). Activity and Expression of the SULT2B1 Isoforms in Human Prostate, Placenta, Breast and Skin. Doctoral dissertation, University of Alabama at Birmingham, Birmingham, AL.
- Nowell S and Falany CN (2006) Pharmacogenetics and human cytosolic sulfotransferases. Oncogene 25 1673–1678. [DOI] [PubMed] [Google Scholar]
- Raftogianis RB, Wood TC, Otterness DM, Van Loon JA, and Weinshilboum RM (1997) Phenol sulfotransferase pharmacogenetics in humans: association of common SULT1A1 alleles with TS PST phenotype. Biochem Biophys Res Commun 239 298–304. [DOI] [PubMed] [Google Scholar]
- Richard K, Hume R, Kaptein E, Stanley EL, Visser TJ, and Coughtrie MW (2001) Sulfation of thyroid hormone and dopamine during human development: ontogeny of phenol sulfotransferases and arylsulfatase in liver, lung, and brain. J Clin Endocrinol Metab 86 2734–2742. [DOI] [PubMed] [Google Scholar]
- Stanley EL, Hume R, and Coughtrie MW (2005) Expression profiling of human fetal cytosolic sulfotransferases involved in steroid and thyroid hormone metabolism and detoxification. Mol Cell Endocrinol 240 32–42. [DOI] [PubMed] [Google Scholar]
- Teubner W, Meinl W, Florian S, Kretzschmar M, and Glatt H (2007) Identification and localization of soluble sulfotransferases in the human gastrointestinal tract. Biochem J 404 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietri M, Vaglini F, Cantini R, and Pacifici GM (2003) Quercetin inhibits the sulfation of r(-)-apomorphine in human brain. Int J Clin Pharmacol Ther 41 30–35. [PubMed] [Google Scholar]
- Waring R and Klovrza LV (2000) Sulphur metabolism in autism. J Nutri Environ Med 10 25–32. [Google Scholar]
- Waring R, Ngong JM, Klovrza L, Green S, and Sharp H (1997) Biochemical parameters in autistic children. Devel Brain Dysfunct 10 40–43. [Google Scholar]
- Weinshilboum RM, Otterness DM, Aksoy IA, Wood TC, Her C, and Raftogianis RB (1997) Sulfation and sulfotransferases 1: sulfotransferase molecular biology: cDNAs and genes. FASEB J 11 3–14. [PubMed] [Google Scholar]
- Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemundsen E, Stefansson H, Ferreira MAR, Green T, et al. (2008) Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med 358 667–675. [DOI] [PubMed] [Google Scholar]
- Whiteley P and Shattock P (2002) Biochemical aspects in autism spectrum disorders: updating the opioid-excess theory and presenting new opportunities for intervention. Expert Opin Ther Targets 6 175–183. [DOI] [PubMed] [Google Scholar]
- Whittemore RM, Pearce LB, and Roth JA (1986) Purification and kinetic characterization of pheno-sulfating form of phenol sulfotransferase from human brain. Arch Biochem Biophys 249 464–471. [DOI] [PubMed] [Google Scholar]
- Yasuda S, Liu MY, Suiko M, Sakakibara Y, and Liu MC (2007). Hydroxylated serotonin and dopamine as substrates and inhibitors for human cytosolic SULT1A3. J Neurochem 103 2679–2689. [DOI] [PubMed] [Google Scholar]
- Young WF Jr, Gorman CA, and Weinshilboum RM (1988) Triiodothyronine: a substrate for the thermostable and thermolabile forms of human phenol sulfotransferase. Endocrinology 122 1816–1824. [DOI] [PubMed] [Google Scholar]
- Zou JY, Petney R, and Roth JA (1990) Immunohistochemical detection of phenol sulfotransferase-containing neurons in human brain. J Neurochem 55 1154–1158. [DOI] [PubMed] [Google Scholar]