Abstract
The pregnane X receptor (PXR) plays crucial roles in multiple physiological processes. However, the signaling mechanisms responsible are not well defined; it is most likely that multiple functions of PXR are modulated by its phosphorylation. Therefore, we sought to determine whether mutation at a highly conserved Thr57 affects human PXR (hPXR) function. Site-directed mutagenesis was performed to generate phosphorylation-deficient (hPXRT57A) and phosphomimetic (hPXRT57D) mutants. Gene reporter, Western blotting, immunocytochemistry, mammalian two-hybrid, and electrophoretic mobility shift assays were used to study cytochrome P450 3A4 (CYP3A4) promoter activation, protein levels, localization, cofactor interaction, and CYP3A4 promoter binding of the hPXR mutants, respectively. hPXRT57D, but not hPXRT57A, lost its transcriptional activity. Neither mutation altered hPXR's protein levels and interaction with steroid receptor coactivator-1. hPXR and hPXRT57A exhibited a homogenous nuclear distribution, whereas hPXRT57D exhibited a distinctive punctate nuclear localization pattern similar to that of hPXR mutants with impaired function that colocalize with silencing mediator of retinoid and thyroid receptors (SMRT), although silencing of SMRT did not rescue the altered function of hPXRT57D. However, hPXRT57D, but not hPXRT57A, impaired hPXR's ability to bind to the CYP3A4 promoter, consistent with the mutant's transactivation function. Furthermore, the 70-kDa form of ribosomal protein S6 kinase (p70 S6K) phosphorylated hPXR in vitro and inhibited its transcriptional activity, whereas hPXRT57A partially resisted the inhibitory effect of p70 S6K. Our studies identify a functionally significant phosphomimetic mutant (hPXRT57D) and show p70 S6K phosphorylation and regulation of hPXR transactivation to support the notion that phosphorylation plays important roles in regulating hPXR function.
Pregnane X receptor (PXR) is a member of the nuclear receptor (NR) family of ligand-activated transcription factors and is activated by a huge variety of endobiotics and xenobiotics, including many clinical drugs, due to its promiscuous ligand-binding nature (Kliewer et al., 1998; Lehmann et al., 1998; Harmsen et al., 2007). PXR has a key role not only as a xenosensor in the regulation of both drug metabolism and elimination (Kliewer et al., 1998; Lehmann et al., 1998; Harmsen et al., 2007) but also as a physiological sensor in the homeostasis of bile acid and cholesterol metabolism (Xie et al., 2001). In addition, growing evidence points to its role or implications in adverse drug-drug interactions (Lehmann et al., 1998; Harmsen et al., 2007), hepatic glucose and lipid metabolism (Bhalla et al., 2004; Kodama et al., 2004), bone homeostasis (Tabb et al., 2003; Pascussi et al., 2005; Igarashi et al., 2007), endocrine homeostasis (Zhai et al., 2007; Lim and Huang, 2008), inflammatory bowel disease and inflammation (Gu et al., 2006; Zhou et al., 2006; Shah et al., 2007), cancer cell growth and drug resistance (Chen et al., 2007; Mensah-Osman et al., 2007; Zhou et al., 2008), uterine contractility and vascular tone (Mitchell et al., 2005; Hagedorn et al., 2007), blood-brain barrier permeability (Bauer et al., 2006), and neuroprotection (Langmade et al., 2006). Nevertheless, the signaling mechanisms responsible for its diverse cellular responses are not well defined, although analogous to other NRs, it is most likely that multiple functions of PXR are modulated and integrated by its phosphorylation.
Phosphorylation is a dynamic regulatory mechanism that not only affects the function of a protein but also enables both specificity and cross-talk among diverse signaling pathways (Cohen, 2000, 2001). Phosphorylation has been shown to modulate the activity of many NRs, and it provides an important mechanism for cross-talk between signaling pathways (Shao and Lazar, 1999; Weigel and Moore, 2007). All the aspects of NR function are known to be regulated when specific sites are phosphorylated, including expression, stability, subcellular localization, dimerization, ligand and DNA binding, coregulator interaction, and transcriptional activity (Ortí et al., 1992; Shao and Lazar, 1999; Rochette-Egly, 2003; Sun et al., 2007; Weigel and Moore, 2007). Furthermore, it is known that NR phosphorylation plays a crucial role in the development of diseases such as breast, ovarian, and prostate cancers (Rochette-Egly, 2003). In contrast to other NRs, our current understanding on phosphorylation of PXR is extremely poor despite substantial evidence for its considerable role as a xenobiotic sensor with a promiscuous ligand-binding nature resulting in adverse drug-drug interactions (Kliewer et al., 1998; Lehmann et al., 1998; Harmsen et al., 2007) and as a physiological sensor and signal cross-talker in diverse cellular responses. Therefore, an understanding of phosphorylation-dependent events in PXR signaling is crucial for effective drug design and clinical therapeutic strategies.
Similar to steroid receptors, the N-terminal part of the PXR protein contains a highly conserved DNA binding domain (DBD) (Fig. 1A), and the C-terminal part of the protein contains a ligand binding domain (LBD) with an additional ligand-inducible transactivation function 2 (AF-2). The DBD and LBD are separated by a hinge region (Fig. 1A). It can be argued that, in contrast to most NRs, PXR does not harbor ligand-independent AF-1 (Fig. 1A), which is the most variable region in NRs in terms of length and sequence similarities and is believed to be regulated by growth factor signaling. NRs are phosphorylated on all the domains at specific sites depending on the cellular context, and such site-specific phosphorylation can lead to either modulation or termination of the activity of the NRs (Rochette-Egly, 2003; Weigel and Moore, 2007). Although it is known that protein kinase A (PKA) (Ding and Staudinger, 2005a), protein kinase C (PKC) (Ding and Staudinger, 2005b), and cyclin-dependent kinase 2 (CDK2) (Lin et al., 2008) are involved in the regulation of PXR activity, and that PKA and CDK2 can phosphorylate the PXR in vitro (Ding and Staudinger, 2005a; Lin et al., 2008), very little is known about the specific site(s) of phosphorylation. Therefore, we examined one of the highly conserved putative phosphorylation sites by performing a phosphomimetic mutation at Thr57 in the DBD and by testing the transcriptional activity, promoter-binding activity, expression, cofactor interaction, and localization of the mutant. Furthermore, results from phosphorylation site prediction and rapamycin-induced PXR modulation (see under Results) led us to test whether the 70-kDa form of ribosomal protein S6 kinase (p70 S6K) regulates the transcriptional activity of human pregnane X receptor (hPXR) through phosphorylation of Thr57.
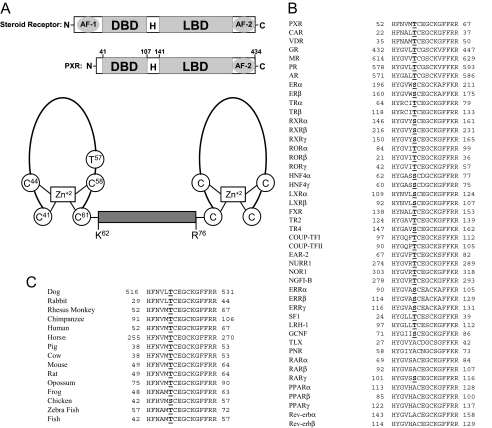
Fig. 1.
Thr57 in hPXR is conserved in other NRs. A, top panel showing the schematic comparison of the domain structures of a steroid receptor and hPXR. H, hinge region. Bottom panel shows the schematic representation of the two zinc finger motifs in the DBD of hPXR. The two zinc finger motifs are separated by a linker (from Lys62 to Arg76). The diagram was not drawn as per the scale in terms of number of residues. Note that Thr57 is located within the first zinc finger motif. B, Thr57 in hPXR is highly conserved among other human NRs. Of the 46 human NRs with DBD, 37 receptors (including all the classic steroid receptors) have a conserved T/S in the DBD. Nine receptors are exceptions, and they have an A instead of Thr/Ser at this position. Expansion of the abbreviated nomenclature for the receptors is as follows: CAR, constitutive androstane receptor; VDR, vitamin D receptor; GR, glucocorticoid receptor; MR, mineralocorticoid receptor; PR, progesterone receptor; AR, androgen receptor; ER, estrogen receptor; TR, thyroid hormone receptor; ROR, retinoid-related orphan receptor; LXR, liver X receptor; FXR, farnesoid X receptor; TR2, testicular receptor 2; TR4, testicular receptor 4; COUP-TF, chicken ovalbumin upstream promoter transcription factor; EAR-2, v-erbA-related; NURR1, NR-related 1; NOR1, neuron-derived orphan receptor 1; NGFI-B, nerve growth factor-induced clone B; ERR, estrogen receptor-related; SF-1, steroidogenic factor 1; LRH-1, liver receptor homolog-1; GCNF, germ cell nuclear factor; TLX, human homolog of the Drosophila tailless gene; PNR, photoreceptor cell-specific NR. C, Thr57 in hPXR is conserved in all the vertebrate species with a fully known PXR sequence. Chicken PXR is the only exception and has an S instead of T at that position.
In this report, we identify a phosphomimetic mutant of hPXR that loses its transactivation function and displays a punctate nuclear distribution. Our results indicate that a phosphomimetic mutation at Thr57 (hPXRT57D) impairs the ability of hPXR to bind to the promoter of cytochrome P450 3A4 (CYP3A4), an important hPXR target gene. This is consistent with the loss of function of hPXRT57D. In addition, we show that p70 S6K phosphorylates hPXR in vitro and inhibits its transactivating function, and that phosphorylation-deficient mutation at Thr57 (hPXRT57A) confers significant resistance to the inhibitory effect of p70 S6K. Taken together, our observations suggest that Thr57 might be regulated by phosphorylation, which in turn regulates the function of hPXR. These observations might apply to other NRs due to the highly conserved nature of this residue.
Materials and Methods
Cell Culture, Plasmids, and Transient Transfections. HepG2 human liver carcinoma and COS7 African green monkey kidney fibroblast cell lines were obtained from the American Type Culture Collection (Manassas, VA). The cells were grown in Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS; HyClone, Logan, UT), 100 U/ml penicillin, 100 μg/ml streptomycin (Invitrogen), 2 mM l-glutamine (Invitrogen), and 1 mM sodium pyruvate (Invitrogen). The cells were cultured in an incubator with a humidified atmosphere maintained at 5% CO2 and 95% air at 37°C.
The pcDNA3-hPXR, pcDNA3-FLAG-hPXR, and CYP3A4 luciferase reporter (pGL3-CYP3A4-luc) plasmids were generated as described previously (Lin et al., 2008). Site-directed mutagenesis was performed to generate alanine (hPXRT57A) and aspartate (hPXRT57D) mutant hPXR plasmids. Polymerase chain reaction (PCR) was performed using pcDNA3-hPXR as the template and specific mutant primers, with forward primers containing a mutation corresponding to either A or D and a PPUMI restriction enzyme cutting site and a reverse primer containing a BsgI restriction enzyme cutting site. The PCR product was cloned into a TOPO vector (Zero Blunt TOPO PCR Cloning Kit; Invitrogen), followed by cleaving the sequence-verified TOPO-PCR product for the desired mutations with PPUMI (New England Biolabs, Ipswich, MA) and BsgI (New England Biolabs), and ligating the resulting fragment into PPUMI- and BsgI-cleaved pcDNA3-hPXR or pcDNA3-FLAG-hPXR. Only the open reading frame of hPXR, but not pcDNA3 or the FLAG tag, had cleavage sites for PPUMI and BsgI. Sequence-verified pcDNA3-hPXRT57A, pcDNA3-hPXRT57D, pcDNA3-FLAG-hPXRT57A, and pcDNA3-FLAG-hPXRT57D were used for transient transfections. pACT contains the transcriptional activation domain of VP16, a herpes simplex virus protein (Promega, Madison, WI). pBIND contains the DBD of the yeast transcription factor Gal4 (Promega). Full-length wild-type hPXR, hPXRT57A, and hPXRT57D were fused to VP16 in pACT to generate VP16-hPXR, VP16-hPXRT57A, and VP16-hPXRT57D, respectively. DNA encoding the NR interaction domain of steroid receptor coactivator (SRC)-1 (amino acids 621–765) (Oñate et al., 1995) was fused to Gal4 in pBIND to generate Gal4-SRC-1. pG5-luc contains the luciferase reporter gene controlled by a promoter containing Gal4 DNA binding sites (Promega). A constitutively active p70 S6K plasmid, Myc-p70S6K D3E-E389, was a gift from Dr. George Thomas (The Genome Research Institute of The University of Cincinnati, Cincinnati, OH).
Transient transfections were performed using FuGENE 6 (Roche Diagnostics, Indianapolis, IN), unless otherwise mentioned, following the manufacturer's protocol. Twenty-four hours post-transfection, the cells were grown in DMEM supplemented with 10% charcoal/dextran-treated FBS (HyClone) and above-mentioned antibiotics. They were then treated with 0.1% dimethyl sulfoxide (DMSO; Sigma-Aldrich, St. Louis, MO) or 10 μM compounds such as rifampicin (Sigma-Aldrich), SR12813 (Sigma-Aldrich), and ketoconazole (Sigma-Aldrich) for an additional 24 h (unless otherwise indicated) before performing luciferase assays for transactivation analyses, harvesting of cell lysates for Western blotting analyses for protein expression, and fixing of cells for immunolocalization studies.
For the rapamycin (Sigma-Aldrich) experiments, the final DMSO concentration was 1%. In p70 S6K overexpression experiments, hPXR-transactivating function was analyzed using 1 μM rifampicin with 0.01% final concentration of DMSO. In mammalian two-hybrid assays, 5 μM rifampicin with 0.1% final DMSO concentration was used.
Transfections were performed in six-well culture plates (Corning Inc., Corning, NY). Each well has approximately 10-cm2 surface for cell culture growth. Cells in each well were transfected with 2 μg of total plasmid DNA. To test the transactivating activity of wild-type and mutant hPXRs, the cells were transfected with 50 ng of pcDNA3, wild-type or mutant hPXR, and 1950 ng of pGL3-CYP3A4-luc plasmids. To examine the protein expression and localization, the cells were transfected with 2 μg of pcDNA3 and wild-type or mutant hPXR plasmids. For the rapamycin experiments, the cells were transfected with 50 ng of pcDNA3 or hPXR and 1950 ng of pGL3-CYP3A4-luc plasmids. In p70 S6K overexpression experiments, the cells were transfected with 100 ng of FLAG-pcDNA3, pcDNA3-FLAG-hPXR, or pcDNA3-FLAG-hPXRT57A; 500 ng of constitutively active p70 S6K; and 1000 ng of pGL3-CYP3A4-luc and 100 ng of pGL4-hRluc; FLAG-pcDNA3 was used to make up the total plasmid DNA to 2 μg in each transfection. In mammalian two-hybrid assays, the cells were transfected with 500 ng of pACT (empty vector control for VP16-hPXR), VP16-hPXR, VP16-hPXRT57A, or VP16-hPXRT57D; 500 ng of Gal4-SRC-1; 500 ng of pG5-luc; and 500 ng of pcDNA3 to make up the total plasmid DNA to 2 μg in each transfection.
Western Blotting Analysis. HepG2 and COS7 cells were lysed with radioimmunoprecipitation assay buffer (Thermo Fisher Scientific, Waltham, MA) containing protease inhibitor mixture (Thermo Fisher Scientific) and phosphatase inhibitor mixture (Thermo Fisher Scientific). Whole cell lysates containing approximately 25 μg of total protein were solubilized at 95°C for 4 min in Laemmli sample buffer (Bio-Rad, Hercules, CA) containing β-mercaptoethanol and were resolved using electrophoresis on Nupage 4 to 12% bis-Tris gels (Invitrogen) with Nupage MES SDS running buffer (Invitrogen). For immunoblotting, the proteins were transferred from unstained gels to nitrocellulose membranes using the iBlot gel transfer system (Invitrogen) and iBlot gel transfer stacks (Invitrogen). After blocking with Odyssey blocking buffer (LI-COR Biosciences, Lincoln, NE) for 1 h, membranes were incubated for 1 h with primary antibodies against FLAG-tag (mouse monoclonal anti-FLAG M2 antibody; Sigma-Aldrich) at 1:1000 dilution or silencing mediator of retinoid and thyroid receptor (SMRT) (mouse monoclonal anti-SMRT antibody; GeneTex, Irvine, CA) at 1:500 dilution. In p70 S6K overexpression experiments, hPXR was probed using rabbit polyclonal anti-hPXR serum (Saradhi et al., 2005). Infrared dye-conjugated secondary goat anti-rabbit (LI-COR Biosciences) or goat anti-mouse (LI-COR Biosciences) antibodies were diluted to 1:20,000 and then used to incubate the membrane for 1 h. The sites of antibody-antigen reaction were visualized using an Odyssey infrared imager (LI-COR Biosciences). The same membranes that were probed for anti-FLAG, anti-hPXR, or anti-SMRT were stripped using Restore PLUS Western blot stripping buffer (Thermo Fisher Scientific) and then reprobed with anti-actin antibody (mouse monoclonal anti-β-actin; Sigma-Aldrich) at 1:5000 dilution to confirm equal loading of total protein in each lane. All of the steps were conducted at room temperature.
Immunocytochemistry. Twenty-four hours post-transfection, approximately 5000 live cells were seeded in each well of a 96-well culture plate (PerkinElmer Life and Analytical Sciences, Waltham, MA) containing either DMSO or rifampicin and grown for an additional 24 h. After aspirating the media, the cells were washed with phosphate-buffered saline (PBS) and fixed in 3.7% formaldehyde for 20 min, followed by four 5-min washes with PBS containing 0.1% Triton X-100 to remove the fixative and to permeabilize the fixed cells. The cells were blocked in Odyssey blocking buffer (LI-COR Biosciences) for 2 h and then incubated with primary antibody in the blocking buffer for 2 h, followed by four 5-min washes with PBS containing 0.1% Tween 20. Cy3-conjugated anti-FLAG M2 mouse monoclonal antibody (Sigma-Aldrich), rabbit polyclonal anti-hPXR serum (Saradhi et al., 2005), and mouse monoclonal anti-SMRT antibody (GeneTex) were used at 1:200, 1:500, and 1:100 dilutions, respectively. After incubating with appropriate secondary antibody in the blocking buffer for 1 h, the cells were washed with PBS containing 0.1% Tween 20 for a total of four washes with 5 min/wash. Cy3-conjugated goat anti-mouse IgG (Jackson ImmunoResearch Laboratories Inc., West Grove, PA) and fluorescein isothiocyanate-conjugated donkey anti-rabbit IgG (Jackson ImmunoResearch Laboratories Inc.) were used at a dilution of 1:200. Finally, the cells were incubated with Hoechst dye (AppliChem, Cheshire, CT) for 10 min to stain the nuclear DNA at a final concentration of 0.5 μg/1 ml in PBS, followed by three PBS washes, before acquiring the fluorescence images using an immunofluorescence microscope (Olympus America, Center Valley, PA). All of the steps were performed at room temperature while gentle shaking of the plate; during cell fixation, there was no shaking of the plate.
Electrophoretic Mobility Shift Assay. Double-stranded 32P-labeled oligonucleotides representing the PXR DNA binding sequence within the CYP3A4 promoter (an everted repeat with a 6-base pair spacer; hereafter referred to as CYP3A4-ER6), 5′-GATCAATATGAACTCAAAGGAGGTCAGTG-3′, or a mutant CYP3A4-ER6 5′-GATCAATATGCCATCAAAGGAATACAGTG-3′ (bolded bases disrupt the PXR binding half-site) was incubated with in vitro-transcribed and -translated protein and binding buffer. Labeled oligonucleotide (600,000 cpm) was added to each reaction along with FLAG-hPXR, FLAG-hPXRT57A, FLAG-hPXRT57D, and human retinoid X receptor (RXR)-α proteins, which were synthesized in vitro using the TNT rabbit reticulocyte lysate system (Promega) following the manufacturer's protocol. Equal amount of the in vitro-translated protein was added in each reaction. Oligonucleotide competition was performed using a 500-fold molar excess of unlabeled oligonucleotides. To validate the specific binding between FLAG-hPXR and hot oligonucleotide, 2 μg of either mouse monoclonal anti-FLAG M2 antibody (Sigma-Aldrich) or rabbit polyclonal anti-hPXR serum (Saradhi et al., 2005) was added to some reactions. To verify the specific binding between FLAG-hPXR and the antibodies, 2 μg of either normal mouse IgG (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) or normal rabbit IgG (Santa Cruz Biotechnology, Inc.) was added to some reactions as negative controls. Binding buffer contained 10 mM Tris, pH 8.0, 40 mM KCl, 0.05% nonidet P-40, 6% glycerol, 1 mM DTT, 0.2 μg of poly(dI-dC), 10 μM zinc chloride, and 2 μl of in vitro-translated protein in a total of 20-μl reaction. Oligonucleotides and synthesized proteins were added to the inside wall of the microcentrifuge tube, mixed via vortexing, and incubated on ice for 30 min. Complexes were resolved using electrophoresis through a nondenaturing 4% polyacrylamide gel and analyzed with a Storm 860 PhosphoImager (GE Healthcare, Little Chalfont, Buckinghamshire, UK).
Silencing of SMRT Using Small Interfering RNA. The cells were transfected with Accell nontargeting SMARTpool control small interfering RNA (siRNA) (Thermo Fisher Scientific) or Accell SMARTpool human SMRT siRNA (Thermo Fisher Scientific) using Accell siRNA delivery media (Thermo Fisher Scientific) for 72 h according to the manufacturer's instructions. Very briefly, when the cell growth reached approximately 75% confluence, growth medium was aspirated and replaced with Accell delivery medium containing a final concentration of 1 μM Accell siRNA. Protein expression of SMRT was analyzed from whole cell lysates collected 72 h after transfection with siRNA. For functional studies, the cells were first transfected with 100 ng of pcDNA3-FLAG-PXRT57D, 100 ng of pGL4-hRluc (Promega), and 3800 ng of pGL3-CYP3A4-luc using Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol. Five hours after transfection, growth medium was aspirated and replaced with Accell delivery medium containing a final concentration of 1 μM Accell siRNA. Compounds were added 48 h after transfection with siRNA, and luciferase activity was measured 24 h after compound treatments. Renilla luciferase (pGL4-hRluc) was used as a control for transfection efficiency. Firefly luciferase activity followed by Renilla luciferase activity was measured using the Dual-Glo Luciferase Assay System (Promega). Firefly luciferase activity was first normalized with Renilla luciferase activity before normalizing the data for rifampicin with DMSO, as described below for the PXR transactivation assay.
PXR Transactivation Assay. The cells were transfected with pcDNA3, hPXR, p70 S6K, pGL4-hRluc, and CYP3A4-luc plasmids using FuGENE 6 or Lipofectamine. Twenty-four hours after transfection in growth media, approximately 10,000 live cells were plated in each well of a 96-well culture plate (PerkinElmer Life and Analytical Sciences) and grown for an additional 24 h in phenol red-free DMEM (Invitrogen) supplemented with 5% charcoal/dextran-treated FBS (HyClone) and other additives, as described in the cell culture section. Forty-eight hours after transfection, luciferase assay was performed to measure luminescence using the Steady Lite luciferase substrate (PerkinElmer Life and Analytical Sciences) and PHERAstar microplate reader (BMG Labtech, Durham, NC). CYP3A4 promoter activity induced by hPXR after incubation with rifampicin, SR12813, or ketoconazole was shown and described as relative luminescence units (RLUs), a normalized ratio between the luminescence observed in the presence of compounds and the DMSO vehicle. In p70 S6K and hPXR cotransfection studies, Renilla luciferase (pGL4-hRluc) was used as an internal control for transfection efficiency. Firefly luciferase activity followed by Renilla luciferase activity was measured using the Dual-Glo Luciferase Assay System (Promega). Firefly luciferase activity was normalized with Renilla luciferase to determine relative luminescence. Relative luminescence is shown as mean values with S.D. from four to six independent observations. The Student's t test was used to determine statistical significance of unpaired samples. Differences were considered significant for p < 0.05 (*), 0.01 (**), or 0.001 (***) and nonsignificant (N.S.) for p > 0.05.
For the rapamycin experiments, HepG2 cells were transfected with either pcDNA3 or hPXR and pGL3-CYP3A4-luc. Twenty-four hours after transfection, cells were treated with either DMSO or rapamycin, and firefly luciferase activity was measured 24 h after rapamycin treatment. Luminescence, measured from the samples transfected with hPXR, was normalized with pcDNA3-transfected samples to determine RLU. The Student's t test was used to determine statistical significance of four unpaired samples by comparing the RLU in the presence of rapamycin to DMSO. Rapamycin treatment did not exert significant cytotoxic effect in HepG2 cells between 10 nM and 10 μM concentrations (data not shown).
In Vitro p70 S6K Kinase Assay. The in vitro kinase assays were performed as described previously (Lin et al., 2008). In brief, kinase assays were performed at 30°C for 30 min in 25-μl reactions with approximately 1 μgof substrate protein, 25 ng of p70 S6K (Assay Designs, Ann Arbor, MI), 0.5 μM cold ATP, and 5 μCi of [γ-32P]ATP (PerkinElmer Life and Analytical Sciences; 6000 Ci/mmol). The reactions were loaded to SDS-polyacrylamide gel electrophoresis, and the amount of the substrate samples was visualized by staining with SimplyBlue SafeStain (LC6060; Invitrogen) followed by desiccation of the stained gel using the LABCONCO gel dryer (Labconco, Kansas City, MO). The dried gel was then subjected to overnight exposure in the Storage Phosphor Screen (GE Healthcare). Phosphoimages were obtained using the Storm scanner (GE Healthcare). Glutathione S-transferase (GST) was expressed and purified using pGex-4T-1 (GE Healthcare) in Escherichia coli BL21. Purified hPXR protein was purchased from Origene Technologies, Inc. (Rockville, MD). p70 S6K positive control substrate peptide was purchased from Assay Designs.
Mammalian Two-Hybrid Assay. The mammalian two-hybrid system (Promega) consists of VP16-hPXR (wild-type or mutant), Gal4-SRC-1, and a luciferase reporter pG5-luc cotransfected into HepG2 cells. The Gal4 vector (pBIND) also constitutively expresses Renilla luciferase, which was used as an internal transfection control. Dual-Glo Luciferase Assay (Promega) was used to measure luciferase activity, which is an indicator of protein-protein interactions. Expression of hPXR and SRC-1 was confirmed by Western analysis (data not shown). The relative luciferase activity for pG5-luc was determined by normalizing firefly luciferase activity with Renilla luciferase activity. The values represent the means of eight independent experiments, and the bars denote the S.D. The Student's t test was used to determine statistical significance of unpaired samples. Differences were considered significant for p < 0.05 (*), 0.01 (**), or 0.001 (***) and nonsignificant (N.S.) for p > 0.05.
Results
Thr57 in the First Zinc Finger Motif of the DBD of hPXR Is Highly Conserved among NRs. Sequence alignment of all the human NRs showed that there is a highly conserved threonine (Thr) or serine (Ser) in the DBD one residue N-terminal to a highly conserved cysteine (Cys) and five residues C-terminal to a highly conserved histidine (His) (Fig. 1B). Of the 46 human NRs with DBD, all but nine receptors have a Thr or Ser at that position (Fig. 1B). The exceptions—retinoic acid receptor (RAR)-α and RARβ, peroxisome proliferator-activated receptor (PPAR)-α, PPARβ, and PPARγ, human homolog of the Drosophila tailless gene, photoreceptor cell-specific NR, Rev-erbα, and Rev-erbβ—have an alanine (Ala) at that position (Fig. 1B). Of the 15 vertebrate species with a fully known PXR sequence, all except chicken have a Thr at that position; chicken PXR has an Ser at that position (Fig. 1C).
Phosphorylation of a conserved Thr/Ser in this DBD region, particularly the zinc finger motifs and the linker connecting the motifs (Fig. 1A), has been implicated in regulating NR function. As an example, phosphorylation of a highly conserved Ser78 in the DBD of hepatocyte nuclear factor (HNF)4α has been shown to regulate the function of HNF4α, such as its intracellular localization, promoter-binding activity, and transcriptional activity (Sun et al., 2007).
It is interesting to note that the Group-Based Phosphorylation Scoring Method (GPS) (Zhou et al., 2004; Xue et al., 2005), which is one of the bioinformatics tools widely used to identify putative phosphorylation sites and corresponding kinases for a given protein, predicted that Thr57 in hPXR might be phosphorylated by S6K (ribosomal S6 kinase) with a significant GPS score of 24 (the cutoff score is 5.8). S6K, a member of the S/T kinase family, exists in p70 and p90 forms. The p70 form of S6K (p70 S6K), which is known to be activated after phosphorylation by mammalian target of rapamycin (mTOR) kinase, is a downstream kinase in the phosphatidylinositol 3-kinase (PI3K)-Akt pathway, whereas the p90 form is a downstream kinase in the mitogen-activated protein kinase pathway.
mTOR is activated by Akt. Activated mTOR in turn phosphorylates and activates p70 S6K. Rapamycin is a known mTOR inhibitor. Inhibition of mTOR activity by rapamycin results in subsequent inhibition of p70 S6K. It is interesting to note that when tested in cell-based reporter gene assays, rapamycin treatment for 24 h significantly enhanced the transcriptional activity of transiently transfected hPXR in HepG2 cells by approximately 30 and 40% respectively, at 5 and 10 μM compared with DMSO control (RLUs; DMSO 4.21 ± 0.81, 5 μM rapamycin 6.04 ± 0.52, and 10 μM rapamycin 6.96 ± 0.72). Multiple mechanisms may be responsible for this rapamycin effect. However, it is conceivable that signaling mechanisms initiated by rapamycin, which alter hPXR activity via mTOR or p70 S6K, are most likely responsible for the rapamycin effect.
The results described above led us to speculate that Thr57 is an important residue that might be phosphorylated and regulated by p70 S6K signaling. Therefore, we sought to determine whether a phosphomimetic mutation at Thr57 in hPXR would affect hPXR function, and whether p70 S6K would phosphorylate hPXR at Thr57 and regulate the transcriptional activity of hPXR.
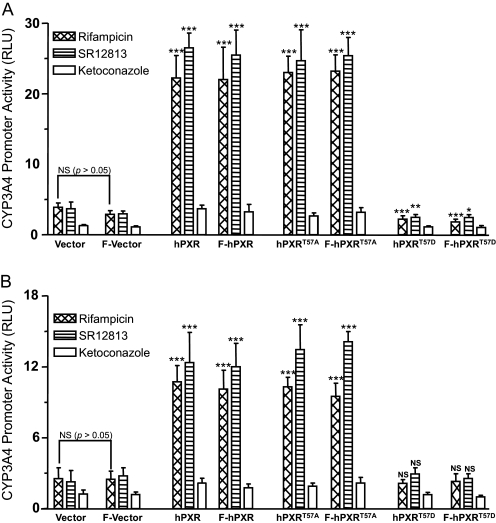
Phosphomimetic Mutation at Thr57 Abolishes hPXR Transcriptional Activity. A mutation of a Thr/Ser to a negatively charged aspartic acid (Asp) is often used to mimic phosphorylation. We tested whether such a phosphomimetic mutation at Thr57 affects hPXR transactivation of the CYP3A4 promoter. The test was performed in HepG2 (Fig. 2A) and COS7 (Fig. 2B) cells transiently transfected with a vector or hPXR plasmid and CYP3A4-luc, in which the expression of luciferase is controlled using an hPXR-regulated CYP3A4 promoter. When transfected with the vector alone, basal promoter activity was observed after treatment with the hPXR agonists rifampicin (10 μM, 24 h) and SR12813 (10 μM, 24 h), suggesting activation of the CYP3A4 promoter by endogenous hPXR (Fig. 2). As expected, wild-type hPXR significantly enhanced the CYP3A4 promoter activity in the presence of its agonists (Fig. 2). As a negative control, a nonspecific PXR inhibitor, ketoconazole, did not enhance the hPXR activity (Fig. 2). It is interesting to note that a mutation of Thr to Ala (hPXRT57A), which places a hydrophobic side chain at position 57, did not affect the rifampicin- or SR12813-mediated hPXR transactivation (Fig. 2). In contrast, a mutation to Asp (hPXRT57D), which mimics phosphorylation, abolished the activation of the CYP3A4 promoter by hPXR (Fig. 2), suggesting that phosphorylation on Thr57 could confer a tremendous functional impact by negatively regulating hPXR activity. Multiple mechanisms might contribute to the impaired transactivation function of hPXRT57D, including protein stability and expression level, nuclear localization, cofactor interaction, and target promoter binding. Therefore, we sought to elucidate the responsible mechanisms. Fusion of a FLAG-tag to the N-terminal wild-type hPXR (F-hPXR) or mutant hPXR (F-hPXRT57A or F-hPXRT57D) did not alter the transcriptional activity of the corresponding hPXR (Fig. 2). Therefore, both FLAG-tagged and untagged hPXR constructs were used interchangeably in this study.
Fig. 2.
Phosphomimetic mutation at Thr57 abolishes hPXR transactivation activity. HepG2 (A) and COS7 (B) cells were cotransfected with either pcDNA3 or hPXR using CYP3A4-luc. F denotes for FLAG-tag. Twenty-four hours post-transfection, the cells were treated with vehicle DMSO (0.1%) or 10 μM compounds: rifampicin, SR12813, and ketoconazole. Luciferase activity was measured 24 h after compound treatments. CYP3A4 promoter activity induced by hPXR after treatment with the compounds was shown as RLUs. RLUs were determined by normalizing the luminescence observed in the presence of one of the compounds with the luminescence observed in the presence of DMSO. Data are shown as mean values from six independent experiments with bars indicating the S.D. The Student's t test was used to determine statistical significance of unpaired samples by comparing the RLU obtained from the samples transfected with wild-type or mutant hPXR with the samples transfected with the vector. Likewise, statistical significance was determined from the samples transfected with FLAG-tagged wild-type or mutant hPXR to the samples transfected with the FLAG vector. Furthermore, statistical significance was ascertained similarly by comparing the RLU obtained from the samples transfected with FLAG-tagged plasmids with the samples transfected with corresponding untagged plasmids, and statistics were shown only for one comparison, i.e., between the vector and FLAG vector after rifampicin treatment. No statistical significance was observed between the samples transfected with the FLAG-tagged plasmids and the untagged plasmids (statistics were not shown for all the sample comparisons). Differences were considered significant for p < 0.05 (*), 0.01 (**), or 0.001 (***), and nonsignificant (N.S.) for p > 0.05.
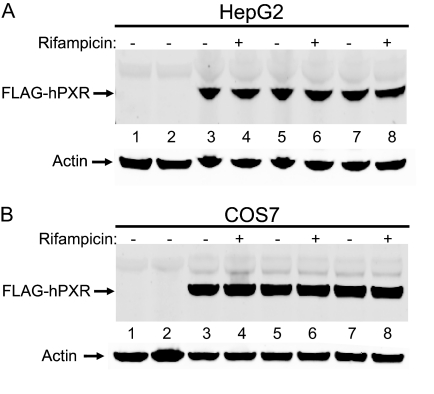
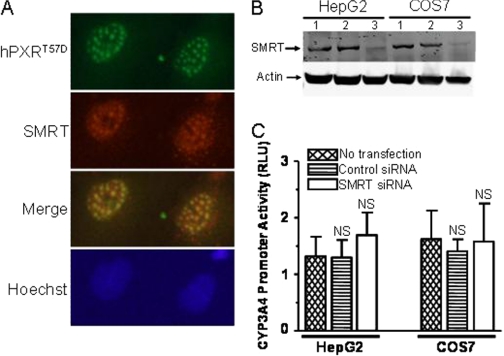
Mutation at Thr57 Does Not Affect hPXR Protein Levels. It is possible that mutation in a protein could lead to reduced protein stability or expression level. Therefore, we tested whether the remarkable reduced activity of hPXRT57D was caused by reduced mutant protein levels in HepG2 (Fig. 3A) and COS7 (Fig. 3B) cells. Western blotting results showed that there was no significant change in protein levels of hPXR after mutation to either Ala or Asp at Thr57 (Fig. 3). Furthermore, rifampicin treatment did not alter the levels of mutant protein (Fig. 3), suggesting that the impaired function of the phosphomimetic mutant was not caused by its reduced expression or stability. Equal amounts of actin protein expression were observed in each lane, verifying that equal amounts of total protein were loaded in each lane (Fig. 3).
Fig. 3.
Mutation at Thr57 does not affect hPXR protein expression levels. HepG2 (A) and COS7 (B) cells were transfected with either wild-type or mutant FLAG-hPXR plasmids. Twenty-four hours post-transfection, the cells were treated with either 0.1% DMSO (indicated by a “–” symbol) or 10 μM rifampicin (indicated by a“+” symbol). Whole cell lysates were collected 24 h after treatment with DMSO or rifampicin and subjected to Western blotting analysis using anti-FLAG and anti-actin antibodies as described under Materials and Methods. pcDNA3-FLAG vector transfection (lane 1) and no transfection (lane 2) are “negative” controls. Wild-type FLAG-hPXR protein expression was shown in lanes 3 and 4. Protein expression for the alanine mutant (FLAG-hPXRT57A) was shown in lanes 5 and 6 and for the aspartate (FLAG-hPXRT57D) in lanes 7 and 8. Actin protein expression was analyzed as a mean of loading control. Data shown are from a representative experiment.
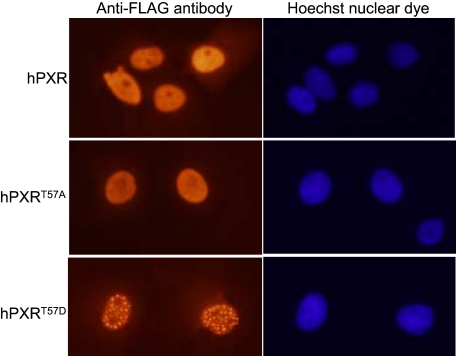
Phosphomimetic Mutation at Thr57 Alters hPXR Nuclear Localization Pattern. hPXR is a transcriptional factor that induces a transactivation response in the nucleus. Therefore, we asked whether a phosphomimetic mutation alters the localization of hPXR in HepG2 and COS7 cells and thereby contributes to impaired transcriptional activity. Immunocytochemistry experiments revealed that both hPXRT57A and hPXRT57D, like wild-type, were localized to the nucleus in COS7 cells (Fig. 4). Similar to wild-type hPXR, hPXRT57A exhibited a homogenous distribution (Fig. 4), whereas hPXRT57D predominantly (in approximately 70% of the cells expressing hPXRT57D) exhibited a distinct punctate pattern, suggesting that phosphorylation of hPXR on Thr57 could dramatically alter nuclear localization pattern. Rifampicin treatment did not affect the localization pattern of either wild-type or mutant hPXR proteins (data not shown). A similar localization phenotype for wild-type and mutant hPXR was also observed in HepG2 cells (data not shown). hPXR mutants with impaired function have been shown to colocalize with an NR corepressor (SMRT) to the nucleus with a punctate pattern similar to that of hPXRT57D (Johnson et al., 2006), suggesting that the altered nuclear localization pattern of hPXRT57D might contribute to its impaired function, and SMRT might be involved.
Fig. 4.
Phosphomimetic mutation at Thr57 alters hPXR nuclear localization pattern. COS7 cells were transfected with FLAG-hPXR, FLAG-hPXRT57A, or FLAG-hPXRT57D plasmids. The cells were treated with vehicle 0.1% DMSO or 10 μM rifampicin 24 h post-transfection. The cells were processed for immunocytochemistry 24 h after treatment with DMSO or rifampicin as described under Materials and Methods. Wild-type and mutant hPXR were probed with Cy3-conjugated anti-FLAG M2 mouse monoclonal antibody. Nuclear DNA was stained with Hoechst dye. Data shown are from a representative experiment.
Silencing of SMRT Cannot Rescue the Impaired Function of hPXRT57D. Johnson et al. (2006) showed in COS7 cells that transiently transfected PXR deletion mutants with partial or complete loss of function colocalize with SMRT to the nucleus with a punctate pattern at discrete nuclear foci. Similar colocalization results were observed for the loss-of-function phosphomimetic mutant, i.e., hPXRT57D colocalizes with SMRT to the nucleus at discrete nuclear foci (Fig. 5A). It has also been shown that SMRT physically interacts with the LBD of the hPXR (Johnson et al., 2006). Activation of an NR by its agonist usually causes a release of corepressors and allows the recruitment of coactivators. Activation of hPXR by its agonist rifampicin after binding to the LBD causes an exchange of the corepressor SMRT with the NR coactivators such as SRC-1 and SRC-3 (Kliewer et al., 1998; Ding and Staudinger, 2005a,b; Johnson et al., 2006). It is possible that the impaired function of hPXRT57D is caused by its sustained association with SMRT. Therefore, we asked whether the silencing of SMRT can rescue the function of hPXRT57D. This question was answered by knocking down the endogenous SMRT protein levels using SMRT SMARTpool Accell siRNA (Thermo Fisher Scientific) as described under Materials and Methods, in HepG2 and COS7 cells transiently transfected with hPXRT57D. Western blotting results showed that SMRT siRNA, but not control nonsilencing siRNA, selectively knocked down the protein expression levels of SMRT (Fig. 5B). Equal actin expression indicates that an equal amount of total protein was loaded into each lane (Fig. 5B). However, silencing of SMRT did not significantly rescue the impaired rifampicin-mediated transactivation function of hPXRT57D (Fig. 5C), suggesting that SMRT is not responsible for the impaired transactivation function.
Fig. 5.
Silencing of SMRT cannot rescue the impaired function of hPXRT57D. A, hPXRT57D colocalizes with SMRT to the nucleus at discrete nuclear foci. COS7 cells were transfected with FLAG-hPXRT57D. Twenty-four hours post-transfection, the cells were treated with vehicle 0.1% DMSO or 10 μM rifampicin. The cells were processed for immunocytochemistry 24 h after treatment with DMSO or rifampicin. FLAG-hPXRT57D was probed using rabbit anti-hPXR polyclonal antibody; SMRT was probed using anti-SMRT mouse monoclonal antibody; and nuclear DNA was stained with Hoechst dye. Data shown are from a representative experiment. B, Accell SMARTpool human SMRT siRNA knocks down the protein expression of SMRT. HepG2 and COS7 cells were transfected with 1 μM Accell nontargeting pool control siRNA (lane 2) or Accell SMARTpool human SMRT siRNA (lane 3) in Accell siRNA delivery media. Whole cell lysates were collected 72 h after transfection and subjected to Western blotting analysis using anti-SMRT and anti-actin antibodies. Protein expression for SMRT without transfection was shown in lane 1. Actin protein expression was shown as a loading control. Data shown are from a representative experiment. C, silencing of SMRT cannot rescue the impaired transactivation function of hPXRT57D. HepG2 and COS7 cells were transfected with pcDNA3-FLAG-PXRT57D, pGL4-hRluc, and pGL3-CYP3A4-luc using Lipofectamine. Five hours after transfection with the plasmids, the cells were transfected with 1 μM Accell nontargeting SMARTpool control siRNA or Accell SMARTpool SMRT siRNA in Accell siRNA delivery media for 72 h. Forty-eight hours after transfection with the siRNAs, the cells were treated with vehicle DMSO (0.1%) or 10 μM rifampicin. Firefly and Renilla luciferase activities were measured 24 h after rifampicin treatment. Firefly luciferase activity was first normalized with Renilla luciferase activity before normalizing the data for rifampicin with DMSO. Data are shown as mean values from four independent experiments with bars indicating the S.D. The Student's t test was used to determine statistical significance of unpaired samples by comparing the RLU obtained from the samples transfected with control siRNA or SMRT siRNA with the samples that were not transfected with siRNA. Differences were considered nonsignificant (N.S.) for p > 0.05.
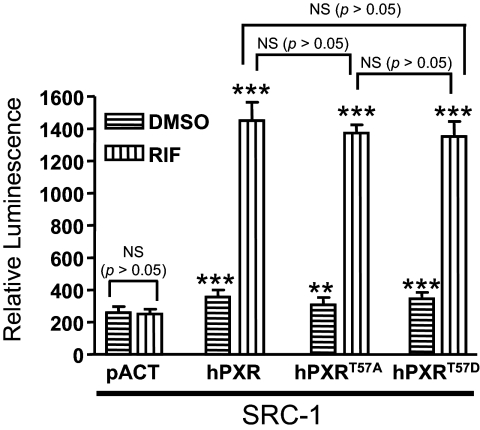
Phosphomimetic Mutation at Thr57 Does Not Affect hPXR and SRC-1 Interaction. Phosphorylation of NRs often regulates their ability to interact with their coregulators (Rochette-Egly, 2003). In the absence of an agonist, hPXR is associated with transcriptional corepressors such as nuclear receptor corepressor 1 and nuclear receptor corepressor 2 (SMRT) (Kliewer et al., 1998; Ding and Staudinger, 2005a,b; Johnson et al., 2006). Agonist binding disrupts hPXR-corepressor association and induces the association between hPXR and transcriptional coactivators such as SRC-1 and SRC-3 (Kliewer et al., 1998; Ding and Staudinger, 2005a,b; Johnson et al., 2006). Thr57 is located in the DBD, whereas PXR interacts with its transcriptional coregulators through its LBD. Although it is a remote possibility that a phosphomimetic mutation at Thr57 in the DBD affects the ability of hPXR to interact with its coregulators, we tested whether the loss of function of the phosphomimetic mutation was caused by a weakened or loss of interaction with its transcriptional coactivator SRC-1. Mammalian two-hybrid assay results showed that rifampicin induced the interaction between hPXR and SRC-1. However, the ability of hPXRT57D to interact with SRC-1 was similar to that of hPXR and hPXRT57A (Fig. 6), suggesting that impaired function of the phosphomimetic mutant was not because of its inability to interact with SRC-1. The result also suggests that rifampicin binds to hPXR, hPXRT57D, and hPXRT57A with similar affinity. The rifampicin-inducible interaction with SRC-1 was not observed in the absence of hPXR, as indicated by the pACT vector control.
Fig. 6.
Phosphomimetic mutation at Thr57 does not affect hPXR interaction with SRC-1. Mammalian two-hybrid assays were performed in HepG2 cells transiently cotransfected with plasmids encoding Gal4-SRC-1 and the reporter gene pG5-luc, along with empty vector pACT, VP16-hPXR, VP16-hPXRT57A, or VP16-hPXRT57D, as indicated. The cells were treated with DMSO or 5 μM rifampicin (RIF) 24 h after transfection. Luciferase assays were performed 24 h after the compound treatment. The relative luminescence for pG5-luc was determined by normalizing firefly luciferase activity with Renilla luciferase activity. The values represent the means of eight independent experiments, and the bars denote the S.D. The Student's t test was used to determine statistical significance of unpaired samples by comparing the relative luminescence obtained between the samples cotransfected with the vector and hPXR plasmids for corresponding DMSO or rifampicin treatments. Comparisons between other samples were noted in brackets. Differences were considered significant for p < 0.05 (*), 0.01 (**), or 0.001 (***), and nonsignificant (N.S.) for p > 0.05. SRC-1, Gal4-SRC-1; hPXR, VP16-hPXR; hPXRT57A, VP16-hPXRT57A; hPXRT57D, VP16-hPXRT57D.
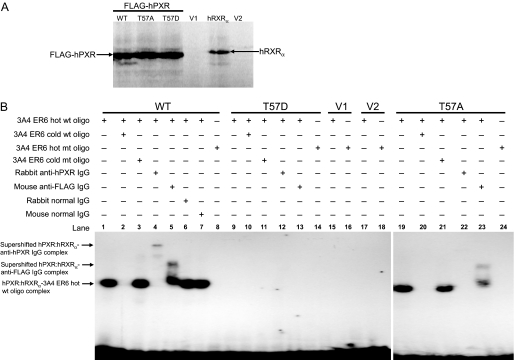
Phosphomimetic Mutation at Thr57 Impairs hPXR Promoter DNA Binding. hPXR transactivation of the CYP3A4 promoter requires direct binding of hPXR to the CYP3A4 promoter. It is known that the first zinc finger motif in the DBD of hPXR is essential for DNA binding (Staudinger et al., 2001). Thr57 is located in the first zinc finger motif (Fig. 1A), and phosphorylation of this residue might alter the DNA-binding activity of the hPXR. Therefore, we investigated whether the loss of transactivation function of hPXRT57D was because of its lack of ability to bind to the CYP3A4 promoter. Electrophoretic mobility shift assay results showed that whereas both wild-type hPXR and hPXRT57A bound to CYP3A4 promoter, hPXRT57D failed to do so (Fig. 7).
Fig. 7.
Phosphomimetic mutation at Thr57 impairs DNA binding of hPXR. A, in vitro synthesis of proteins for wild-type and mutant FLAG-hPXR, as well as for hRXRα, as described under Materials and Methods. Equal amounts of protein were observed from 2-μl in vitro transcription and translation reactions for wild-type (WT), alanine mutant (T57A), and aspartate mutant (T57D) hPXRs. No specific proteins were observed from the negative control reactions using the vectors, pcDNA3-FLAG (V1) and pSG5 (V2), for hPXR and hRXRα, respectively. B, electrophoretic mobility shift assay was performed with in vitro-translated FLAG-hPXR, FLAG-hPXRT57D, or FLAG-hPXRT57A and hRXRα proteins, as well as 32P-labeled CYP3A4 PXR DNA binding sequence (indicated as 3A4ER6) as described under Materials and Methods. Equal amount of hRXRα was added to all the reactions and was not shown in the figure. FLAG-hPXR and FLAG-hPXRT57A bound to and formed a complex with hot wild-type (wt) oligo, and this complex (lanes 1 and 19) was efficiently competed with by cold wt oligo (lanes 2 and 20) but not by cold mutant (mt) oligo (lanes 3 and 21) at 500 M excess. Addition of 2 μg of rabbit anti-hPXR IgG supershifted the FLAG-hPXR-hot wt oligo complex (lane 4) and vanished the FLAG-hPXRT57A-hot wt oligo complex (lane 22) (Saradhi et al., 2005). Addition of 2 μg of mouse anti-FLAG IgG supershifted both the FLAG-hPXR-hot wt oligo complex (lane 5) and the FLAG-hPXRT57A-hot wt oligo complex (lane 23). Addition of 2 μg of rabbit (lane 6) and mouse (lane 7) normal IgG failed to supershift or vanish the FLAG-hPXR-hot wt oligo complex. No complex was detected for FLAG-hPXR (lane 8) and FLAG-hPXRT57A (lane 24) with hot mt oligo. No complex was detected with phosphomimetic mutant FLAG-hPXRT57D (lanes 9–14) and vectors, pcDNA3-FLAG (V1) (lanes 15 and 16), and pSG5 (V2) (lanes 17 and 18). Data shown are from a representative experiment.
As shown in Fig. 7A, equal amounts of hPXR protein were used. Both in vitro-translated wild-type hPXR and hPXRT57A, but not hPXRT57D, bound to radiolabeled (hot) wild-type CYP3A4 promoter oligo (i.e., oligo containing the hPXR response element) (Fig. 7B, lanes 1, 9, and 19). To verify the specificity of the binding of hPXR to its response element, we showed that higher concentrations of unlabeled (cold) wild-type oligo competed with and inhibited hot wild-type oligo to bind to both hPXR and hPXRT57A (Fig. 7B, lanes 2 and 20). In addition, hot mutant oligo (i.e., oligo with the hPXR response element mutated) failed to bind to hPXR and hPXRT57A (Fig. 7B, lanes 8 and 24). Furthermore, rabbit polyclonal anti-hPXR IgG and mouse monoclonal anti-FLAG IgG (Fig. 7B, lanes 4 and 5), but not rabbit normal IgG and mouse normal IgG (Fig. 7B, lanes 6 and 7), supershifted the hPXR-hot wild-type oligo complex from the original position. Similar results were obtained for hPXRT57A: whereas anti-hPXR IgG vanished the hPXRT57A-hot wild-type oligo complex (Fig. 7B, lane 22) (Saradhi et al., 2005), anti-FLAG IgG supershifted the complex (Fig. 7B, lane 23). Taken together, these observations show that hPXRT57D loses its ability to bind to the CYP3A4 promoter and decipher a mechanism for the impaired function of the phosphomimetic hPXR mutant.
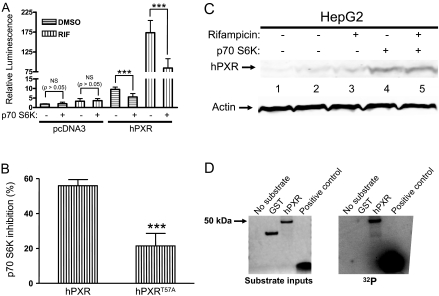
p70 S6K Attenuates the Transactivation Activity of hPXR. Kinases have been shown to alter the function of the proteins through phosphorylation. Both GPS bioinformatics prediction and rapamycin-enhanced hPXR transactivation in HepG2 cells encouraged us to speculate that p70 S6K might be involved in regulating the transcriptional activity of hPXR. Therefore, we tested whether cotransfection of constitutively active p70 S6K alters hPXR transactivation in HepG2 cells. Constitutively active p70 S6K attenuated both basal (DMSO) and rifampicin (1 μM, 24 h)-induced hPXR transactivation (Fig. 8A). It is interesting to note that activation of p70 S6K inhibited the rifampicin-induced hPXR activation by approximately 55% but only inhibited that of the phosphorylation-deficient hPXRT57A by approximately 20% (Fig. 8B), suggesting that Thr57 might be one of the functionally significant target sites on hPXR for phosphorylation by p70 S6K and that hPXR may have more than one functionally important phosphorylation sites for p70 S6K.
Fig. 8.
p70 S6K regulates hPXR function. A, p70 S6K attenuates the transactivating activity of hPXR. HepG2 cells were cotransfected with pGL4-hRluc and pGL3-CYP3A4-luc, along with either FLAG-pcDNA3, pcDNA3-FLAG-hPXR, or pcDNA3-FLAG-hPXRT57A (for B), with or without the constitutively active p70 S6K, as indicated. Twenty-four hours after transfection, the cells were treated with vehicle DMSO (0.01%) or 1 μM rifampicin (RIF). Firefly and Renilla luciferase activities were measured 24 h after rifampicin treatment. CYP3A4 promoter activity was shown as relative luminescence determined by normalizing the firefly luciferase activity with Renilla luciferase activity. Data are shown as mean values from four to six independent experiments, with bars indicating the S.D. The Student's t test was used to determine statistical significance of unpaired samples by comparing the relative luminescence between samples as noted in brackets. Differences were considered significant for p < 0.05 (*), 0.01 (**), or 0.001 (***), and nonsignificant (N.S.) for p > 0.05. pcDNA3, FLAG-pcDNA3; hPXR, pcDNA3-FLAG-hPXR. B, hPXRT57A partially resists the regulation of p70 S6K. Percentage inhibition of rifampicin-inducible hPXR-transactivating activity by p70 S6K was shown. The change (inhibition) in rifampicin-inducible hPXR activity after p70 S6K cotransfection, for the corresponding wild-type or mutant hPXR, was calculated and expressed as percentage inhibition. Data are shown as mean values from four to six independent experiments, with bars indicating the S.D. The Student's t test was used to determine statistical significance of unpaired samples. Differences were considered significant for p < 0.05 (*), 0.01 (**), or 0.001 (***). hPXR, pcDNA3-FLAG-hPXR; hPXRT57A, pcDNA3-FLAG-hPXRT57A. C, p70 S6K cotransfection does not reduce hPXR protein levels. Transient transfections were performed as described in A. Whole cell lysates were collected 24 h after treatment with DMSO (0.01%) or 1 μM rifampicin and subjected to Western blotting analysis using rabbit polyclonal anti-hPXR serum (Saradhi et al., 2005). FLAG-pcDNA3 vector transfection (lane 1) is a negative control. pcDNA3-FLAG-hPXR protein expression without p70 S6K cotransfection was shown in lanes 2 and 3, and with p70 S6K cotransfection was shown in lanes 4 and 5. Actin protein expression was analyzed as a mean of loading control. D, p70 S6K phosphorylates hPXR in vitro. Right, recombinant active p70 S6K (20 ng) was used with 1 μg of substrate as indicated. The kinase assays were performed as described under Materials and Methods. Left, SimplyBlue staining of the proteins resolved on SDS-polyacrylamide gel electrophoresis indicating the amounts of input substrate proteins used in the kinase assay reactions.
We tested whether p70 S6K inhibition of hPXR transcriptional activity was caused by reduced protein levels of hPXR. Western blot results show that cotransfection of constitutively active p70 S6K did not reduce the protein levels of hPXR (Fig. 8C). In contrast, It is interesting to note that higher levels of hPXR were observed with the kinase cotransfection, confirming that inhibition of hPXR by p70 S6K was not caused by reduced protein levels of hPXR (Fig. 8C). Equal amounts of actin protein expression were observed in each lane, verifying that equal amounts of total protein were loaded in each lane (Fig. 8C).
p70 S6K Phosphorylates hPXR in Vitro. The results described in Fig. 8, A through C, suggest that hPXR might be phosphorylated by p70 S6K. In an in vitro kinase assay, we showed that reconstituted complexes of purified p70 S6K directly phosphorylate purified hPXR (Fig. 8D). Purified GST protein was used as a negative substrate control, and a p70 S6K substrate peptide was used as a positive control. Similar amounts of GST and hPXR were used in the kinase assays (Fig. 8D). These results indicate that p70 S6K directly phosphorylates hPXR in vitro and are consistent with the negative regulation of hPXR-transactivating function by p70 S6K.
Discussion
Regulated site-specific phosphorylation provides an important mechanism by which the activity of a protein is regulated (Cohen, 2000, 2001). It is well established for NRs that site-specific phosphorylation occurs on all the domains and plays a vital role to modulate or terminate the activity of NRs (Rochette-Egly, 2003; Weigel and Moore, 2007). It is known that PKA (Ding and Staudinger, 2005a), PKC (Ding and Staudinger, 2005b), and CDK2 (Ding and Staudinger, 2005a; Lin et al., 2008) are involved in the regulation of PXR activity. It is also known that PKA and CDK2 can phosphorylate the PXR in vitro (Ding and Staudinger, 2005a; Lin et al., 2008). However, very little is known about specific phosphorylation of PXR. Therefore, we examined one of the putative phosphorylation sites by performing a phosphomimetic mutation at Thr57—a highly conserved Thr/Ser site in the DBD among the human NRs, including all the classic steroid receptors (Fig. 1B), and among PXR from various species (Fig. 1C)—and by testing the mutant's transactivation function, promoter-binding activity, expression, cofactor interaction, and localization. This is the first report on the identification and characterization of a PXR phosphomimetic mutant that loses its transactivation function, due to impaired promoter-binding activity, and exhibits a punctate nuclear distribution. In addition, this is the first report to identify hPXR as an in vitro substrate for p70 S6K and to show p70 S6K regulation of hPXR transcriptional activity in HepG2 cells. Our observations suggest that Thr57 might be regulated by phosphorylation, which in turn regulates hPXR target promoter binding and transactivation function. These observations might apply to other NRs due to the highly conserved nature of this residue.
The DBD is a highly conserved domain among NRs. Phosphorylation of a conserved Thr/Ser in this DBD region has been implicated in regulating NR function. Several studies have revealed that phosphorylation of a conserved Thr/Ser in the DBD can lead to attenuation and/or termination of promoter binding and transcriptional activity (Hsieh et al., 1993; Chen et al., 1999; Delmotte et al., 1999; Rochette-Egly, 2003; Sun et al., 2007). For example, phosphorylation of a highly conserved Ser78 in the DBD of HNF4α has been shown to regulate the function of HNF4α by altering its intracellular localization and reducing its promoter-binding activity (Sun et al., 2007). Phosphorylation of conserved Thr/Ser residues involved in the recognition of cognate response elements, which results in loss of promoter-binding activity, has also been described for the vitamin D receptor after phosphorylation of Ser51 in the DBD by PKC (Hsieh et al., 1993). Inhibition of the transcriptional activity of other NRs also occurs subsequently to phosphorylation of conserved Thr/Ser residues located within the DBD dimerization surface. For example, estrogen receptor α and RARα lose their transcriptional activity after phosphorylation of Ser236 or Ser157 by PKA (Chen et al., 1999) or PKC (Delmotte et al., 1999), respectively. In agreement with these observations, our results show that the phosphomimetic mutant of hPXR loses its promoter-binding activity and provide a mechanism for the loss of transactivation function.
The fact that the first zinc finger motif in the DBD of hPXR is essential for the DNA binding (Staudinger et al., 2001) leads us to speculate that the phosphorylation of Thr57 in the first zinc finger motif might contribute to the regulation of DNA binding and transactivation activities of the hPXR. Phosphomimetic mutation of this site in other NRs may bring about similar functional effects as a result of the highly conserved nature of this residue.
Wild-type and both hPXR mutants (i.e., hPXRT57A and hPXRT57D) are localized to the nucleus in HepG2 and COS7 cells (Fig. 4), and this observation is in agreement with previously published results (Saradhi et al., 2005; Johnson et al., 2006). Punctate nuclear distribution was observed for the phosphomimetic mutant in approximately 70% of the hPXRT57D-expressing cells. The localization phenotype for both wild-type and mutant hPXR was not affected by treatment with the hPXR agonists rifampicin and SR12813, as well as the nonspecific hPXR inhibitor, ketoconazole (data not shown). Nuclear punctate distribution for some NRs, such as the glucocorticoid receptor (Ogawa et al., 2004) and androgen receptor (Marcelli et al., 2006), has been observed in response to their ligands. It is interesting to note that the nuclear punctate pattern has also been observed for hPXR mutants with impaired function (Johnson et al., 2006), and these mutants have been shown to colocalize with the corepressor SMRT to the nucleus with a punctate pattern. This suggests that mutant hPXR may constitutively associate with SMRT and that altered nuclear localization pattern might contribute to impaired function. Similar results were observed in our study for the phosphomimetic mutant in terms of impaired function and nuclear colocalization with punctate distribution. However, SMRT knockdown did not reverse the functional phenotype of the phosphomimetic mutant, indicating that SMRT is not responsible for the impaired function, although SMRT colocalizes with the phosphomimetic mutant at discrete nuclear foci. At this juncture, we do not have any data to offer a logical speculation for the punctate phenotype, but it would be interesting to define the molecular mechanism(s) responsible for the altered nuclear localization of the phosphomimetic mutant.
The fact that the agonist-mediated transactivation of the endogenous hPXR (i.e., the relative luminescence observed when the cells were transfected with an empty vector) was significantly lower when HepG2 cells were transfected with hPXRT57D (Fig. 2) suggests that hPXRT57D is a properly folded protein and acted as a dominant-negative mutant by competing with one or more of the components involved in the PXR transactivation pathway. The interaction between hPXR and the coactivator SRC-1 is regulated by agonist binding and is critical for hPXR-transactivating function. In the mammalian two-hybrid system, hPXRT57D interacts, similarly to hPXR and hPXRT57A, with SRC-1 in a rifampicin-inducible manner (Fig. 6), showing that the phosphomimetic mutant retains its ability to bind to rifampicin and interact with SRC-1. These data further support the notion that the loss-of-function phosphomimetic mutant is an appropriately folded protein and can act as a dominant-negative protein by competing with wild-type hPXR for interaction with the coactivator SRC-1, which is a crucial protein-protein interaction step in the process of hPXR transactivation pathway. This could partially, if not fully, account for the reduced endogenous hPXR transcriptional activity in HepG2 cells when transfected with hPXRT57D. When a protein is misfolded, it is usually bound to chaperones such as heat shock proteins and subsequently undergoes degradation by proteasomes, and these events occur predominantly in the cytoplasm. The fact that hPXRT57D, translocated to the nucleus and stably expressed similarly to the wild-type and nonphosphomimetic mutant hPXR, argues against the idea that hPXRT57D displays the punctate nuclear pattern because of its association with proteasomes or heat shock proteins.
It has been reported that kinases could phosphorylate nonconsensus Thr/Ser sites depending on the cellular context (Lehtinen et al., 2006). Thr57 in hPXR, along with its flanking residues, does not fulfill the known consensus sequence for known kinases. However, our bioinformatics search results predicted that Thr57 is a putative phosphorylation site for S6K and/or protein kinase B (Akt). PI3K-Akt pathway, a major signaling pathway activated by insulin and other growth factors, is known to be involved in negatively regulating the transcriptional activity of the PXR by affecting the interaction between PXR and its coactivator forkhead in rhabdomyosarcoma (FKHR) (Kodama et al., 2004). Akt possibly accomplishes this negative regulation by phosphorylating and translocating the nuclear FKHR into the cytoplasm for proteasomal degradation (Tang et al., 1999), consequently minimizing the levels of nuclear FKHR available for interacting with and activating PXR. However, it is not known whether Akt phosphorylates PXR and regulates its activity independently of FKHR.
In this report, we show that p70 S6K, a downstream kinase in the PI3K-Akt pathway, phosphorylates and negatively regulates the transcriptional activity of hPXR (Fig. 8). The inhibition of hPXR by p70 S6K was not caused by reduced protein levels of hPXR. In fact, the kinase cotransfection resulted in elevated hPXR protein level (Fig. 8). This particular result might be because of enhanced stability of hPXR by unknown mechanisms. Alanine mutation at Thr57 confers partial but significant resistance to p70 S6K attenuation, hinting that p70 S6K regulates hPXR activity possibly by phosphorylating Thr57 and that p70 S6K may have more than one functionally significant target residues in hPXR. The resistance to p70 S6K attenuation, conferred by the alanine mutation at Thr57, is consistent with the impaired hPXR activity when phosphomimetic mutation was introduced at the same site and supports the suggestion that p70 S6K might regulate hPXR activity via Thr57 phosphorylation. However, the phosphomimetic mutation abolishes, whereas p70 S6K significantly attenuates but not abolishes, the hPXR activity. This discrepancy could be explained in three possible ways. First, it is most likely that all hPXR may not be phosphorylated by p70 S6K in the transient transfections. Second, hPXR may be phosphorylated at multiple sites, resulting in some compensatory effect. Third, the kinase might phosphorylate multiple proteins involved in hPXR transactivation, resulting in some compensatory effect. In summary, our studies identify a functionally significant phosphomimetic mutant of hPXR, show p70 S6K phosphorylation and regulation of hPXR, and support the notion that phosphorylation plays important roles in regulating the multitude of diverse cellular functions of hPXR.
Acknowledgments
We thank Drs. John Obenauer and Jianmin Wang for assistance with bioinformatics tools; Dr. George Thomas for sharing the p70 S6K plasmid; Dr. Hanqing Dong for sharing the reagents and protocols, as well as other members of the Chen group for valuable discussions; Dr. Kip Guy for critical review of the manuscript; and Dr. Donald Samulack for scientific editing.
This work was supported in part by the National Institutes of Health National Institute of General Medical Sciences [Grant GM60346] (to E.G.S.); the National Institutes of Health National Cancer Institute [Grant P30-CA21765]; and the American Lebanese Syrian Associated Charities and St. Jude Children's Research Hospital.
Parts of this work were previously presented as a poster as follows: Pondugula SR, Wu J, Brimer-Cline C, Schuetz EG, and Chen T (2008) Phosphomimetic mutation affects function, localization, and CYP3A4 promoter binding of pregnane X receptor. Nuclear Receptors: Bench to Bedside; 2008 Aug 27–31; Cold Spring Harbor, NY. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
doi:10.1124/dmd.108.024695.
ABBREVIATIONS: PXR, pregnane X receptor; NR, nuclear receptor; DBD, DNA binding domain; LBD, ligand binding domain; AF, activation function; PKA, protein kinase A; PKC, protein kinase C; CDK2, cyclin-dependent kinase 2; p70 S6K, protein S6 kinase; hPXR, human pregnane X receptor; DMEM, Dulbecco's modified Eagle's medium; FBS, fetal bovine serum; PCR, polymerase chain reaction; SRC, steroid receptor coactivator; DMSO, dimethyl sulfoxide; bis-Tris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol (systematic); SMRT, silencing mediator of retinoid and thyroid receptor; PBS, phosphate-buffered saline; RXR, retinoid X receptor; siRNA, small interfering RNA; RLU, relative luminescence unit; GST, glutathione S-transferase; RAR, retinoic acid receptor; PPAR, peroxisome proliferator-activated receptor; HNF, hepatic nuclear factor; GPS, Group-Based Phosphorylation Scoring Method; mTOR, mammalian target of rapamycin; PI3K, phosphatidylinositol 3-kinase; F-hPXR, fusion of a FLAG-tag to the N-terminal wild-type hPXR; Akt, protein kinase B; FKHR, forkhead in rhabdomyosarcoma; SR12813, 3,5-di-tert-butyl-4-hydroxystyrene-β,β-diphosphonic acid tetraethyl ester.
References
- Bauer B, Yang X, Hartz AM, Olson ER, Zhao R, Kalvass JC, Pollack GM, and Miller DS (2006) In vivo activation of human pregnane X receptor tightens the blood-brain barrier to methadone through P-glycoprotein up-regulation. Mol Pharmacol 70 1212–1219. [DOI] [PubMed] [Google Scholar]
- Bhalla S, Ozalp C, Fang S, Xiang L, and Kemper JK (2004) Ligand-activated pregnane X receptor interferes with HNF-4 signaling by targeting a common coactivator PGC-1alpha. Functional implications in hepatic cholesterol and glucose metabolism. J Biol Chem 279 45139–45147. [DOI] [PubMed] [Google Scholar]
- Chen D, Pace PE, Coombes RC, and Ali S (1999) Phosphorylation of human estrogen receptor alpha by protein kinase A regulates dimerization. Mol Cell Biol 19 1002–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Tang Y, Wang MT, Zeng S, and Nie D (2007) Human pregnane X receptor and resistance to chemotherapy in prostate cancer. Cancer Res 67 10361–10367. [DOI] [PubMed] [Google Scholar]
- Cohen P (2000) The regulation of protein function by multisite phosphorylation—a 25 year update. Trends Biochem Sci 25 596–601. [DOI] [PubMed] [Google Scholar]
- Cohen P (2001) The role of protein phosphorylation in human health and disease. The Sir Hans Krebs medal lecture. Eur J Biochem 268 5001–5010. [DOI] [PubMed] [Google Scholar]
- Delmotte MH, Tahayato A, Formstecher P, and Lefebvre P (1999) Serine 157, a retinoic acid receptor alpha residue phosphorylated by protein kinase C in vitro, is involved in RXR.RARalpha heterodimerization and transcriptional activity. J Biol Chem 274 38225–38231. [DOI] [PubMed] [Google Scholar]
- Ding X and Staudinger JL (2005a) Induction of drug metabolism by forskolin: the role of the pregnane X receptor and the protein kinase a signal transduction pathway. J Pharmacol Exp Ther 312 849–856. [DOI] [PubMed] [Google Scholar]
- Ding X and Staudinger JL (2005b) Repression of PXR-mediated induction of hepatic CYP3A gene expression by protein kinase C. Biochem Pharmacol 69 867–873. [DOI] [PubMed] [Google Scholar]
- Gu X, Ke S, Liu D, Sheng T, Thomas PE, Rabson AB, Gallo MA, Xie W, and Tian Y (2006) Role of NF-KappaB in regulation of PXR-mediated gene expression: a mechanism for the suppression of cytochrome P-450 3A4 by proinflammatory agents. J Biol Chem 281 17882–17889. [DOI] [PubMed] [Google Scholar]
- Hagedorn KA, Cooke CL, Falck JR, Mitchell BF, and Davidge ST (2007) Regulation of vascular tone during pregnancy: a novel role for the pregnane X receptor. Hypertension 49 328–333. [DOI] [PubMed] [Google Scholar]
- Harmsen S, Meijerman I, Beijnen JH, and Schellens JH (2007) The role of nuclear receptors in pharmacokinetic drug-drug interactions in oncology. Cancer Treat Rev 33 369–380. [DOI] [PubMed] [Google Scholar]
- Hsieh JC, Jurutka PW, Nakajima S, Galligan MA, Haussler CA, Shimizu Y, Shimizu N, Whitfield GK, and Haussler MR (1993) Phosphorylation of the human vitamin D receptor by protein kinase C. Biochemical and functional evaluation of the serine 51 recognition site. J Biol Chem 268 15118–15126. [PubMed] [Google Scholar]
- Igarashi M, Yogiashi Y, Mihara M, Takada I, Kitagawa H, and Kato S (2007) Vitamin K induces osteoblast differentiation through pregnane X receptor-mediated transcriptional control of the Msx2 gene. Mol Cell Biol 27 7947–7954. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Johnson DR, Li CW, Chen LY, Ghosh JC, and Chen JD (2006) Regulation and binding of pregnane X receptor by nuclear receptor corepressor silencing mediator of retinoid and thyroid hormone receptors (SMRT). Mol Pharmacol 69 99–108. [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Moore JT, Wade L, Staudinger JL, Watson MA, Jones SA, McKee DD, Oliver BB, Willson TM, Zetterström RH, et al. (1998) An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell 92 73–82. [DOI] [PubMed] [Google Scholar]
- Kodama S, Koike C, Negishi M, and Yamamoto Y (2004) Nuclear receptors CAR and PXR cross talk with FOXO1 to regulate genes that encode drug-metabolizing and gluconeogenic enzymes. Mol Cell Biol 24 7931–7940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmade SJ, Gale SE, Frolov A, Mohri I, Suzuki K, Mellon SH, Walkley SU, Covey DF, Schaffer JE, and Ory DS (2006) Pregnane X receptor (PXR) activation: a mechanism for neuroprotection in a mouse model of Niemann-Pick C disease. Proc Natl Acad Sci U S A 103 13807–13812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann JM, McKee DD, Watson MA, Willson TM, Moore JT, and Kliewer SA (1998) The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J Clin Invest 102 1016–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtinen MK, Yuan Z, Boag PR, Yang Y, Villén J, Becker EB, DiBacco S, de la Iglesia N, Gygi S, Blackwell TK, et al. (2006) A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell 125 987–1001. [DOI] [PubMed] [Google Scholar]
- Lim YP and Huang JD (2008) Interplay of pregnane X receptor with other nuclear receptors on gene regulation. Drug Metab Pharmacokinet 23 14–21. [DOI] [PubMed] [Google Scholar]
- Lin W, Wu J, Dong H, Bouck D, Zeng FY, and Chen T (2008) Cyclin-dependent kinase 2 negatively regulates human pregnane X receptor-mediated CYP3A4 gene expression in HepG2 liver carcinoma cells. J Biol Chem 283 30650–30657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcelli M, Stenoien DL, Szafran AT, Simeoni S, Agoulnik IU, Weigel NL, Moran T, Mikic I, Price JH, and Mancini MA (2006) Quantifying effects of ligands on androgen receptor nuclear translocation, intranuclear dynamics, and solubility. J Cell Biochem 98 770–788. [DOI] [PubMed] [Google Scholar]
- Mensah-Osman EJ, Thomas DG, Tabb MM, Larios JM, Hughes DP, Giordano TJ, Lizyness ML, Rae JM, Blumberg B, Hollenberg PF, et al. (2007) Expression levels and activation of a PXR variant are directly related to drug resistance in osteosarcoma cell lines. Cancer 109 957–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell BF, Mitchell JM, Chowdhury J, Tougas M, Engelen SM, Senff N, Heijnen I, Moore JT, Goodwin B, Wong S, et al. (2005) Metabolites of progesterone and the pregnane X receptor: a novel pathway regulating uterine contractility in pregnancy? Am J Obstet Gynecol 192 1304–1313; discussion 1313–1315. [DOI] [PubMed] [Google Scholar]
- Ogawa H, Yu RT, Haraguchi T, Hiraoka Y, Nakatani Y, Morohashi K, and Umesono K (2004) Nuclear structure-associated TIF2 recruits glucocorticoid receptor and its target DNA. Biochem Biophys Res Commun 320 218–225. [DOI] [PubMed] [Google Scholar]
- Oñate SA, Tsai SY, Tsai MJ, and O'Malley BW (1995) Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science 270 1354–1357. [DOI] [PubMed] [Google Scholar]
- Ortí E, Bodwell JE, and Munck A (1992) Phosphorylation of steroid hormone receptors. Endocr Rev 13 105–128. [DOI] [PubMed] [Google Scholar]
- Pascussi JM, Robert A, Nguyen M, Walrant-Debray O, Garabedian M, Martin P, Pineau T, Saric J, Navarro F, Maurel P, et al. (2005) Possible involvement of pregnane X receptor-enhanced CYP24 expression in drug-induced osteomalacia. J Clin Invest 115 177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochette-Egly C (2003) Nuclear receptors: integration of multiple signalling pathways through phosphorylation. Cell Signal 15 355–366. [DOI] [PubMed] [Google Scholar]
- Saradhi M, Krishna B, Mukhopadhyay G, and Tyagi RK (2005) Purification of full-length human pregnane and xenobiotic receptor: polyclonal antibody preparation for immunological characterization. Cell Res 15 785–795. [DOI] [PubMed] [Google Scholar]
- Shah YM, Ma X, Morimura K, Kim I, and Gonzalez FJ (2007) Pregnane X receptor activation ameliorates DSS-induced inflammatory bowel disease via inhibition of NF-KappaB target gene expression. Am J Physiol Gastrointest Liver Physiol 292 G1114–G1122. [DOI] [PubMed] [Google Scholar]
- Shao D and Lazar MA (1999) Modulating nuclear receptor function: may the phos be with you. J Clin Invest 103 1617–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudinger JL, Goodwin B, Jones SA, Hawkins-Brown D, MacKenzie KI, LaTour A, Liu Y, Klaassen CD, Brown KK, Reinhard J, et al. (2001) The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc Natl Acad Sci U S A 98 3369–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K, Montana V, Chellappa K, Brelivet Y, Moras D, Maeda Y, Parpura V, Paschal BM, and Sladek FM (2007) Phosphorylation of a conserved serine in the deoxyribonucleic acid binding domain of nuclear receptors alters intracellular localization. Mol Endocrinol 21 1297–1311. [DOI] [PubMed] [Google Scholar]
- Tabb MM, Sun A, Zhou C, Grün F, Errandi J, Romero K, Pham H, Inoue S, Mallick S, Lin M, et al. (2003) Vitamin K2 regulation of bone homeostasis is mediated by the steroid and xenobiotic receptor SXR. J Biol Chem 278 43919–43927. [DOI] [PubMed] [Google Scholar]
- Tang ED, Nuñez G, Barr FG, and Guan KL (1999) Negative regulation of the forkhead transcription factor FKHR by Akt. J Biol Chem 274 16741–16746. [DOI] [PubMed] [Google Scholar]
- Weigel NL and Moore NL (2007) Steroid receptor phosphorylation: a key modulator of multiple receptor functions. Mol Endocrinol 21 2311–2319. [DOI] [PubMed] [Google Scholar]
- Xie W, Radominska-Pandya A, Shi Y, Simon CM, Nelson MC, Ong ES, Waxman DJ, and Evans RM (2001) An essential role for nuclear receptors SXR/PXR in detoxification of cholestatic bile acids. Proc Natl Acad Sci U S A 98 3375–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Zhou F, Zhu M, Ahmed K, Chen G, and Yao X (2005) GPS: a comprehensive www server for phosphorylation sites prediction. Nucleic Acids Res 33 W184–W187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y, Pai HV, Zhou J, Amico JA, Vollmer RR, and Xie W (2007) Activation of pregnane X receptor disrupts glucocorticoid and mineralocorticoid homeostasis. Mol Endocrinol 21 138–147. [DOI] [PubMed] [Google Scholar]
- Zhou C, Tabb MM, Nelson EL, Grün F, Verma S, Sadatrafiei A, Lin M, Mallick S, Forman BM, Thummel KE, et al. (2006) Mutual repression between steroid and xenobiotic receptor and NF-KappaB signaling pathways links xenobiotic metabolism and inflammation. J Clin Invest 116 2280–2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou FF, Xue Y, Chen GL, and Yao X (2004) GPS: a novel group-based phosphorylation predicting and scoring method. Biochem Biophys Res Commun 325 1443–1448. [DOI] [PubMed] [Google Scholar]
- Zhou J, Liu M, Zhai Y, and Xie W (2008) The antiapoptotic role of pregnane X receptor in human colon cancer cells. Mol Endocrinol 22 868–880. [DOI] [PMC free article] [PubMed] [Google Scholar]