Abstract
Cochaperones are essential for Hsp70/Hsc70-mediated folding of proteins and include nucleotide exchange factors (NEF) that assist protein folding by accelerating ADP/ATP exchange on Hsp70. The cochaperone Bag2 binds misfolded Hsp70 clients and also acts as a NEF, but the molecular basis of its functions is unclear. We show that, rather than being a member of the Bag domain family, Bag2 contains a new type of Hsp70 NEF domain, which we call the “Brand New Bag” (BNB) domain. Free and Hsc70-bound crystal structures of Bag2-BNB show its dimeric structure in which a flanking linker helix and loop bind to Hsc70 to promote nucleotide exchange. NMR analysis demonstrates that the client-binding sites and Hsc70 interaction sites of Bag2-BNB overlap, and that Hsc70 can displace clients from Bag2-BNB, indicating a distinct mechanism for the regulation of Hsp-70-mediated protein folding by Bag2.
The stress-induced chaperone Hsp70 and its highly homologous, constitutively expressed counterpart Hsc70 exemplify a family of ATP-driven chaperones that play indispensable roles in intracellular protein folding and cellular stress response1,2. Hsp70 family members promote correct folding of nascent substrate proteins and act as chaperones for mature substrates or “clients”. They also prevent aggregation of misfolded substrates and promote their refolding. Hsp70/Hsc70 contain nucleotide binding domains and substrate binding domains that are conformationally and functionally coupled. In the ATP-bound state, Hsp70/Hsc70 exhibits low substrate affinity and high substrate on- and off-rates, whereas the ADP-bound state has high affinity for substrates. Cycling between the two states leads to repeated substrate binding and release, and mediates substrate folding1,2.
Several types of cochaperones maintain the high rates of Hsp70 cycling necessary for proper in vivo chaperone function. J-domain-containing cochaperones such as Hsp40 stimulate ATP hydrolysis3, while Hsp70 nucleotide exchange factors (NEFs) accelerate ADP dissociation from the Hsp70 nucleotide binding domain (NBD)4. Diverse classes of J-domain proteins and NEFs differ in their intracellular compartmentalization and possess different accessory properties, lending additional variability to the regulation of Hsp70-family chaperones5. At least four structurally and functionally distinct classes of Hsp70-NEFs have been characterized: the bacterial and mitochondrial GrpE/Mge1p family, and the eukaryotic Bag-domain, HspBP1/Fes1 and Hsp110/Hsp170 families4.
The cochaperone Bag2 is a NEF that also has intrinsic chaperone client-binding activity. Bag2 has been classified as a member of the Bag-domain family of Hsp70-NEFs6, and promotes nucleotide exchange through its C-terminal domain, putatively a three-helix bundle Bag domain7. Bag domains promote nucleotide exchange by binding to subdomains Ib and IIb of the Hsp70-NBD. This causes a rotation of subdomain IIb and disrupts the nucleotide binding site, which lies at the interface between these subdomains8. Although it is classified as a Bag domain, however, the C-terminal domain of Bag2 has relatively low homology to Bag domains of other Bag family members compared to the similarity of the Bag domains to each other. For example, key Bag domain motifs that contact the Hsp70-NBD are poorly conserved or mis-positioned in the Bag2 C-terminal domain. Unlike other Bag family members, Bag2 can also bind directly to misfolded chaperone substrates and suppresses their aggregation9. These characteristics of Bag2 raise the question as to whether the generally accepted model of Bag domain function is applicable to Bag2.
We have now determined the crystal structures of the C-terminal domain of murine Bag2, both in the free form and in complex with the Hsc70-NBD. Unexpectedly, we found that the Bag2 C-terminal domain adopts a novel dimeric structure and Hsc70 binding mode that differ greatly from those of Bag domains as well as those of other Hsp70-NEF proteins. In addition, we used NMR to map the client binding activity of Bag2 onto two C-terminal domain surfaces that overlap with the Hsc70-binding surface, suggesting that Hsc70 and chaperone clients compete for binding to Bag2. We suggest that the Bag2 C-terminal domain should be considered a unique Hsp70/Hsc70 nucleotide exchange factor domain and propose that it be called the BNB (“Brand New Bag”) domain. Our results establish a new type of Hsp70-NEF, define the molecular basis of its NEF and substrate binding activities, and provide insights into coordination between Hsp70 and Bag2 during the chaperoning process.
RESULTS
Novel dimeric structure of Bag2-BNB domain
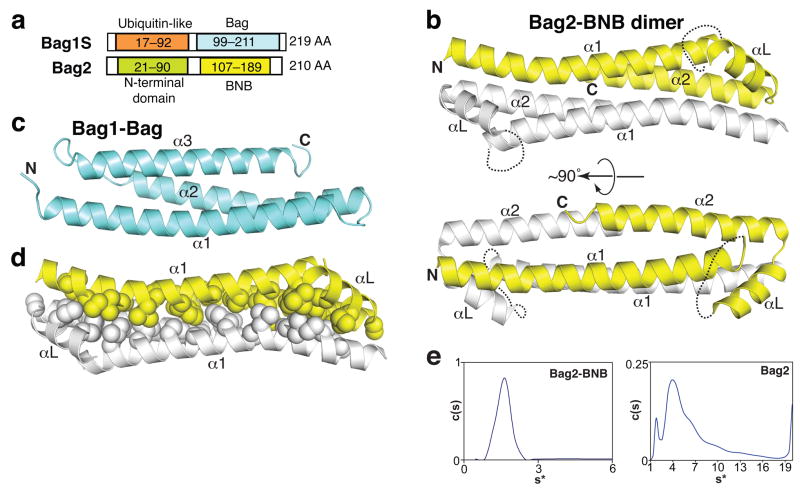
Full-length human, murine or zebrafish Bag2 proteins form oligomeric species of varying size, as gauged by gel filtration chromatography (data not shown) and analytical ultracentrifugation experiments (see below). To obtain a monodisperse protein suitable for crystallization, we tested numerous constructs of the C-terminal half of Bag2, which contains the NEF activity of Bag29,10. We successfully crystallized residues 107–189 of murine Bag2 (Fig. 1a; sequence in Supplementary Fig. 1 online) and solved the corresponding structure to 2.55 Å resolution. The domain crystallized as antiparallel dimers (Fig. 1b). Unexpectedly, given previous annotations of the Bag2 sequence6,7, the structure differs substantially from that of canonical Bag domains, such as the Bag domain from Bag1, which are monomeric 3-helix bundles (Fig. 1c)8,11. We therefore call this domain the BNB (“Brand New Bag”) domain.
Figure 1.
Structure of the Bag2-BNB domain. (a) Comparison between domain organizations of Bag2 and Bag1S, the shortest isoform of Bag1. The C-terminal domain of each protein binds to Hsp70 and has nucleotide exchange factor activity. (b) Cartoon representations of the Bag2-BNB dimer, related by an approximately 90° rotation. Dotted lines indicate the disordered linker loops of each monomer. (c) Structure of the Bag1-Bag domain8 (RCSB ID: 1HX1). (d) Packing between protomers of the Bag2-BNB dimer. Sidechains of nonpolar residues are shown as spheres. (e) Sedimentation velocity analysis of Bag2-BNB (left) and Bag2 (right) at 48,000 rpm and 20 °C. The sedimentation coefficient distribution c(s) of Bag2-BNB shows a single species corresponding to a dimer, in agreement with the crystal structure. The differential sedimentation coefficient distribution of Bag2 shows multiple sedimenting species. Mass estimates suggest that the large peak corresponds to a tetramer (Supplementary Table 1 online), followed by higher-order oligomeric species.
The BNB domain contains two long antiparallel helices (α1 and α2) connected by an orthogonal linker region consisting of a 9-residue disordered linker loop and a short helix (αL). The electron density for the linker loop is very poor, suggesting that the loop is mobile in the free form of the BNB domain. The asymmetric unit of the crystal contains four BNB molecules arranged as two nearly identical, 2-fold-symmetric homodimers. The structures of the BNB protomers are very similar (Rmsd = 0.44 Å). The long helices (α1 and α2) of two BNB protomers pack together in a 4-helix bundle, burying an interface of 1100 Å2 (per protomer). More than 70% of the buried interface is hydrophobic as defined by the POPSCOMP program12. The extensive hydrophobic packing between the BNB domains (Fig. 1d) suggests that the BNB domains may form bona fide dimers in solution, and may also dimerize in the context of full-length Bag2.
We therefore investigated the solution state of the BNB domain using analytical ultracentrifugation. In sedimentation velocity experiments, Bag2-BNB sedimented as a single species with an estimated molecular mass of 19 kDa, corresponding to a BNB dimer (Fig. 1e; Supplementary Table 1 online). Sedimentation equilibrium experiments show that Bag2-BNB undergoes a reversible monomer-dimer equilibrium with a KD of 4.5±2 μM (Supplementary Fig. 2a online). In contrast, near full-length murine Bag2 (Bag221–210) sedimented as multiple species corresponding to a complex mixture of oligomerization states (Fig. 1e). Mass estimates of individual peaks suggest the presence of tetramer along with many poorly resolved higher-order species (Supplementary Table 1 online). The N-terminal domain of Bag2 (Bag2-NTD) has itself been shown to homodimerize with high affinity in a yeast 2-hybrid assay9, and we also observed Bag2-NTD dimerization using gel filtration chromatography (Supplementary Fig. 2b online). Rather than dimerization, however, the combination of the NTD and BNB domains appears to promote a higher-order oligomerization of full-length Bag2. This may occur by a given Bag2 molecule making an NTD:NTD contact with one Bag2 partner and a BNB:BNB contact with another Bag2 partner (as shown schematically in Supplementary Fig. 2c online).
Bag2-BNB binds to Hsc70 through the linker loop and helix αL
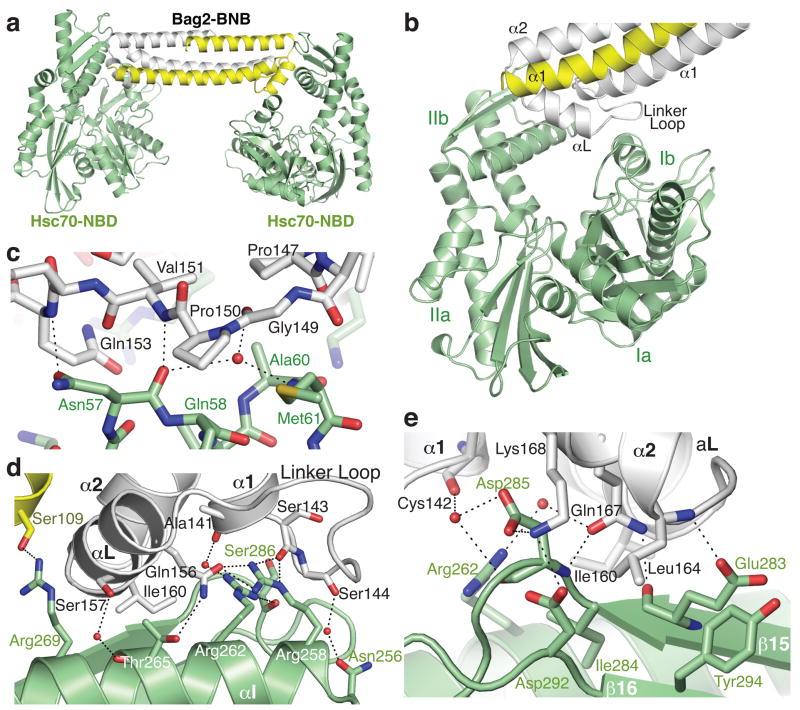
To understand how Bag2 promotes nucleotide exchange on Hsp70/Hsc70, we cocrystallized the BNB domain with the Hsc70 nucleotide binding domain (Hsc701–381, termed Hsc70-NBD) and solved the structure of the complex to 2.3 Å resolution. The BNB domain retains its dimeric state in the Hsc70-bound form. The asymmetric unit of the crystal contains a 2:2 complex, in which each protomer of a BNB dimer contacts an Hsc70-NBD (Fig. 2a). Hsc70:Bag2 contacts are formed by the linker loops of the BNB domains, the linker helices (αL), and the N-terminal ends of the α2 helices. The BNB linker loops become ordered upon Hsc70 binding, with well-resolved electron density. The BNB domains contact subdomains Ib and IIb of the Hsc70-NBDs (Fig. 2b). The distal tip of each BNB linker loop and the N-terminal end of αL make contact with subdomain Ib, while the N-terminal part of the linker loop, the C-terminal half of αL and the N-terminal portion of α2 make extensive contacts with subdomain IIb.
Figure 2.
Structure of the Hsc70-NBD:Bag2-BNB domain complex. (a) Asymmetric unit of the crystal, containing two Hsc70-NBD molecules and a Bag2-BNB dimer. (b) Closer view of the interface between the Bag2-BNB dimer and one of the Hsc70-NBD molecules. The BNB dimer contacts Hsc70-NBD subdomains Ib and IIb, largely through the Bag2 linker loop and αL helix. (c) Closeup of contacts between the Bag2-BNB linker loop and residues on subdomain Ib of the Hsc70-NBD. Dotted lines represent likely hydrogen bonds or salt bridges. (d) Closeup of contacts between the Bag2-BNB dimer and subdomain IIb of the Hsc70-NBD. (e) Closeup of additional contacts between Bag2-BNB and subdomain IIb of the Hsc70-NBD. This view is related to panel (d) by an approximate 180° rotation about the vertical axis.
Details of the Hsc70-NBD:Bag2-BNB contact interface are shown in Fig. 2c-e. The interface contains extensive electrostatic and hydrogen-bonding interactions, including several mediated through solvent molecules. Prominent hydrophobic interactions are also present. The tip of the BNB linker loop, containing the sequence 146VPPGPV151, makes both hydrophobic and polar contacts with an exposed helical turn on Hsc70 subdomain Ib. This turn consists of residues Hsc70-Asn57 to Met61 (Fig. 2c) and connects Hsc70 helices αA and αB. A series of basic side-chains (Hsc70-Arg258, Arg262, Arg269), located on helix αI of subdomain IIb, interact with a shallow groove bounded by the linker loop and the N-terminal half of Bag2-αL. Hsc70-R269 interacts with the other side of αL and with the sidechain of S109 on the other protomer of the BNB dimer (Fig. 2d); this is the only contact formed with the other protomer. Several acidic side chains from the 2-stranded β-sheet of subdomain IIb (β15-β16) interact with residues at the N-terminal end of Bag2-α2 (Fig. 2e). In addition, several nonpolar residues that connect Bag2-αL and α2, including Bag2-I160, make hydrophobic contacts with αI and β15 on subdomain IIb of the Hsc70-NBD.
Bag2 promotes nucleotide exchange by rotating NBD domain II
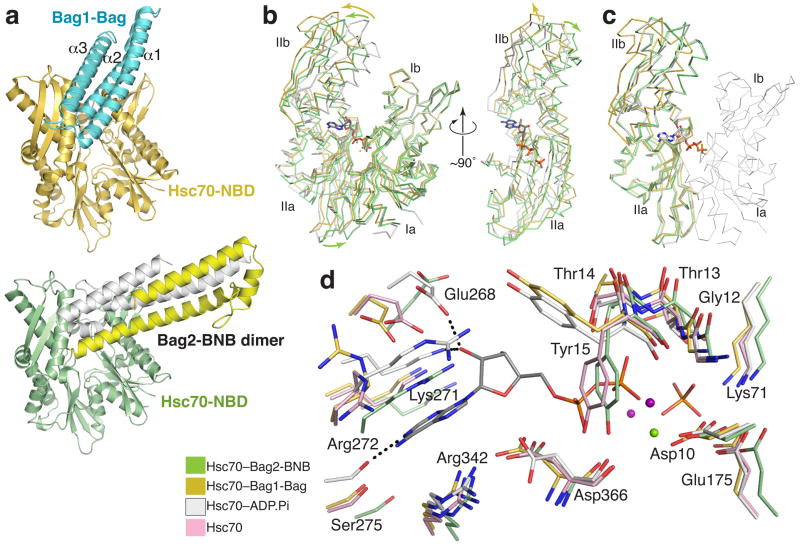
The Bag2-BNB dimer binds to the Hsc70-NBD in an end-on fashion. In contrast, Bag domains bind in a more parallel fashion, along the cleft between subdomains Ib and IIb, as shown by the complex between the Bag domain of Bag1 and the Hsc70-NBD8 (Fig. 3a). Several key Hsc70 surface residues, including Asn57, Met61, Arg258, Arg262, Thr265, Glu283 and Asp292, are involved in binding to both Bag2 and Bag1. In contrast, the Hsc70-binding regions on the Bag2-BNB and Bag1-Bag domains differ greatly in structure and orientation. In addition, while the Bag2-BNB a and Bag domain binding surfaces overlap, Bag2-BNB induces a different conformational change in the Hsc70-NBD.
Figure 3.
Mechanism of Bag2-BNB-mediated nucleotide exhange. (a) Comparison between overall Hsc70-NBD binding modes of Bag2-BNB and Bag1-Bag domain8; RCSB ID: 1HX1. (b) Overall conformational changes in Hsc70-NBD induced by Bag2-BNB or Bag1-Bag domain binding. Structural alignment of the Bag2-BNB-bound (green), Bag1-Bag-bound (gold), or ADP.Pi-bound13 (white; RCSB ID: 3HSC) forms of Hsc70-NBD based on superimposition of domain I, emphasizing changes in the orientations of subdomains IIa and IIb relative to domain I. The bound nucleotide in the ADP-Pi complex is shown in stick representation. In the right panel, Hsc70-NBD domain I has been removed and the view is looking at the nucleotide binding site on domain II. (c) Structural alignment of domain II, showing that subdomains IIa and IIb are in the same relative orientation in the ADP.Pi- and Bag2-BNB-bound forms of Hsc70-NBD, but not in the Bag1-Bag domain-bound form. Domain I from the ADP.Pi-bound form is shown as a thin Cα ribbon representation. (d) Closeup of the nucleotide-binding site in the nucleotide-free14 (pink), ADP-Pi, Bag2-BNB and Bag1-Bag-bound Hsc70-NBDs. The structural alignment is as shown in panel (b). Dotted lines represent key polar interactions between the nucleotide and Hsc70-NBD in the ADP.Pi-bound form. The green and purple spheres represent Mg+2 and Na+ ions respectively.
The Bag1-Bag domain promotes nucleotide exchange by causing a marked (~14°) rotation of subdomain IIb about a hinge connecting subdomains IIb and IIa8, relative to ADP.Pi-bound Hsc70-NBD13. This results in local perturbations at the nucleotide binding site. The separation between residues that interact with the adenine and residues that interact with the phosphate groups increases, so that the nucleotide cannot simultaneously interact with both sets of residues. In contrast, the Bag2-BNB domain effectively displaces subdomain IIb away from subdomain Ib (Fig. 3b) without altering the relative orientation between subdomains IIa and IIb (Fig. 3c). Instead, the entirety of domain II pivots ~10° as a single unit. When viewed from the nucleotide binding cleft, domain II also undergoes a distinct clockwise rotation of ~10° parallel to the plane separating domains I and II (Fig. 3b). Displacement of domain II from domain I as a single unit was also observed in the nucleotide-free structure of the NBD14, raising the possibility that the conformational change in our structure is due to absence of nucleotide rather than directly due to Bag2 binding. Comparison of the nucleotide-free and BNB-bound structure of Hsc70-NBD, however, shows that the Bag2-bound structure is also distinct from that of nucleotide-free Hsc70-NBD (Supplementary Fig. 3 online).
As in the case of Bag1, binding of the Bag2-BNB dislocates several nucleotide-interacting sidechains (Fig. 3d). Hsc70-Glu268 and Lys271, which interact with the ribose, and Ser275, Arg272 and Arg342, which interact with the adenine moiety, are displaced “downward” relative to residues that interact with the phosphate groups. However, several of these residues shift in position so that they clash sterically with the nucleotide. Notably, Arg272, which stacks against the adenine in the ADP-bound Hsc70, is displaced into the position of the adenine. The sidechain of Tyr15 adopts a rotamer that overlaps with the position of the β-phosphate and is likely stabilized through hydrogen bonding between its hydroxyl and the mainchain carbonyl of Asp366. Such steric clashes are not observed in the structure of the Hsc70-NBD complexed with the Bag1-Bag domain8 or with other Hsc70-NEF domains15-18. While Tyr15 adopts a similar rotamer in the nucleotide-free form of Hsc70-NBD14, the Tyr15 sidechain is located 1 Å further away from the Asp366 carbonyl and has less optimal hydrogen bonding geometry, suggesting that this rotamer is more stable in Bag2-bound Hsc70-NBD. The Bag2-induced conformational change thus promotes nucleotide exchange by disrupting nucleotide-binding interactions and inducing several direct sidechain clashes with the nucleotide.
Mutational studies of the Hsc70-binding interface of Bag2
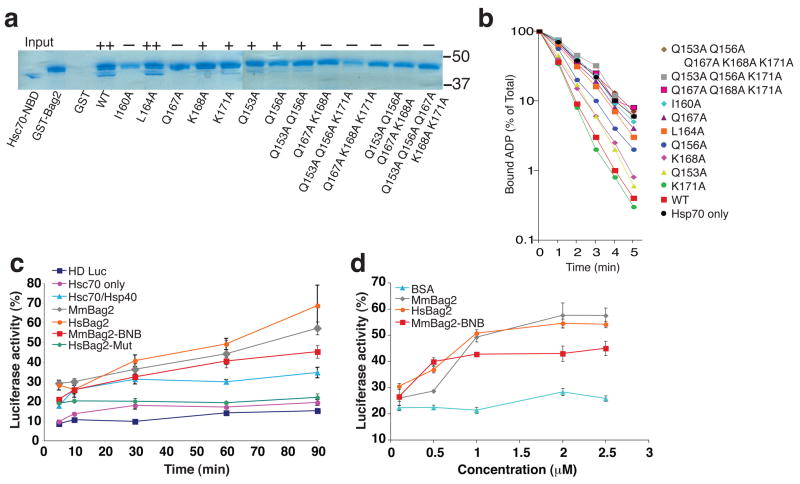
To further investigate the Hsc70-NBD:Bag2-BNB interface, we mutated selected interface residues in GST-fused full-length human Bag2 and examined the ability of the mutants to interact with Hsc70-NBD (Fig. 4a). Mutation of Bag2-Ile160, which is mostly buried by packing against subdomain IIb, and Bag2-Gln167, which makes hydrogen bonds with several backbone groups on subdomain IIb, resulted in complete loss of Bag2:Hsc70 interaction. Individual mutations of other interfacial residues resulted in partial loss of binding and, when combined, in complete loss of binding. We also assessed the effects of the same mutations in single-turnover nucleotide exchange assays on Hsc70 (Fig. 4b). As previously observed10, Bag2 greatly accelerated the release of ADP from Hsc70. ADP release rates were reduced for the Bag2 mutants in agreement with their reduced binding to Hsc70-NBD. These data verify that the Bag2-BNB:Hsc70-NBD interfaces observed in our crystal structure mediate functionally relevant interactions between Hsc70 and Bag2.
Figure 4.
Effects of Bag2 mutations on Hsc70 binding, nucleotide exchange and substrate refolding activity. (a) GST-pulldown assays using wild-type or mutant GST-tagged human Bag2. Hsc70-NBD was incubated with GST-Bag2 prebound to GST-minicolumns, which were subsequently washed and eluted with 10 mM glutathione. Eluted proteins were resolved using SDS-PAGE and stained with Coomassie blue. Symbols above lanes qualitatively designate the amount of Hsc70-NBD bound to and co-eluting with GST-Bag2 (++: strong binding, +: weak binding; −: no binding). In lane “GST”, the GST protein is not within the portion of the gel shown in the figure. Positions of 50 and 37 kD molecular weight markers are shown at right. (b) Single-turnover nucleotide exchange measurements on Hsc70. Time course of α-32P-ATP release from Hsc70 in the absence (black circles) or presence of wild-type Bag2 (red squares) or Bag2 mutants. Data shown are the means of three independent experiments. (c) Effect of Bag2 on refolding of heat-denatured luciferase by Hsc70/Hsp40. “HD Luc” designates background luminescence signal from heat-denatured luciferase alone, without any chaperones present. “Hsc70 only” designates signal from Hsc70 without Hsp40 present. “HsBag2-Mut” corresponds to the human Bag2-Q153A/Q156A/Q167A/K168A/K171A mutant. Hs and Mm designate human and murine Bag2 constructs respectively. All of the experiments containing Bag2 constructs also contained 1 μM Hsc70 and 1 μM Hsp40. Signal from non-denatured luciferase incubated for equivalent amounts of time in refolding buffer was set to 100%. (d) Concentration dependence of promotion of heat-denatured luciferase refolding by Bag2. All experiments also contained 1 μM Hsc70 and 1 μM Hsp40. Luciferase signal was recorded 1 hour after initiation of refolding reaction. Bovine serum albumin (BSA) was used as a negative control. Signal from non-denatured luciferase incubated for equivalent amounts of time in refolding buffer was set to 100%.
We further examined whether Bag2-BNB could promote Hsc70-mediated protein folding in a functional protein refolding assay, using heat-denatured luciferase as the chaperone substrate19. Inclusion of Bag2-BNB in luciferase refolding reactions containing Hsc70 and the J-domain cochaperone Hsp40 noticeably enhanced refolding of heat-denatured luciferase (Fig. 4c,d). The Bag2 mutant Q153A/Q156A/Q167A/K168A/K171A, which does not bind Hsc70-NBD (Fig. 4a), did not enhance luciferase refolding, and in fact exhibited a slight suppressive effect (Fig. 4c). The enhancement of luciferase refolding by Bag2-BNB was somewhat less than that induced by full-length Bag2, but nevertheless indicates the substantial cochaperone activity intrinsic to the Bag2-BNB domains.
Overlapping client- and Hsc70- binding sites on Bag2-BNB
In addition to its function as a nucleotide exchange factor, Bag2 exhibits intrinsic chaperone client binding activity, through which it can suppress aggregation of misfolded substrate proteins. Interestingly, the substrate binding preferences of Bag2 are similar to those of the Hsc70 substrate binding domain, in that both proteins bind to hydrophobic peptide sequences flanked by basic residues9. To determine whether the Bag2 client binding sites are located on the BNB domain, we carried out NMR resonance assignment of 15N/13C-labeled Bag2-BNB followed by HSQC-NMR titrations of 15N-labeled Bag2-BNB with two client-derived peptides. The peptides were previously shown to bind to Bag2 and correspond to sequences from nucleotide binding domain-1 (NBD1) of CFTR9, which is chaperoned by Hsc70 during its biogenesis20,21.
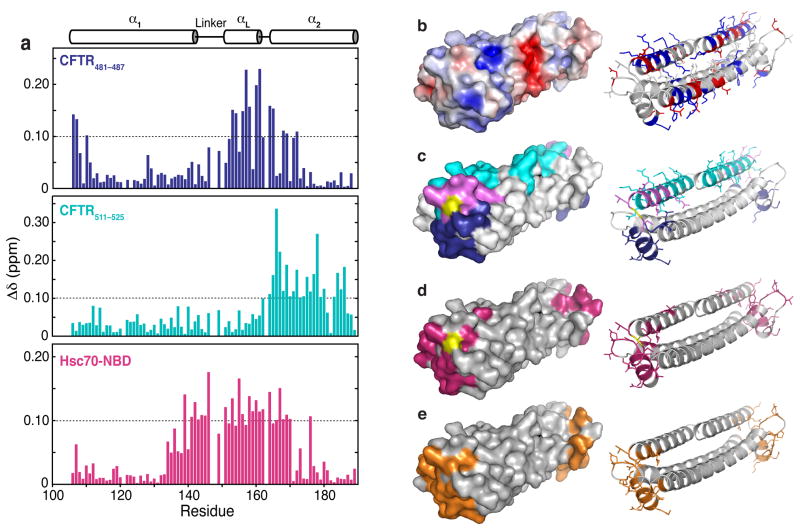
Addition of the peptides CFTR481–487 (KIKHSGR), and CFTR511–525 (SYDEYRYR-SVIKACQ) caused wide-ranging chemical shift perturbations in the spectrum of 15N Bag2-BNB (Fig. 5; full spectra are shown in Supplemental Fig. 4a,c online). The peptides perturb distinct but overlapping sets of chemical shifts, corresponding to two overlapping surfaces on Bag2-BNB. The central surface perturbed by both peptides is comprised of the αL-α2 connector and the N-terminus of α2 (cyan surface in Fig. 5c). This may be the binding site for the core basic-hydrophobic-basic motifs (Lys-Ile-Lys and Arg-Tyr-Arg) of the peptides, while the remaining portions of the peptides may bind on opposite sides of this central surface. The CFTR511–525 binding site extends along α2, while CFTR481–487 binds near αL (Fig. 5d). From the extent of chemical shift perturbations caused by similar peptide:BNB ratios, CFTR511–525 binds more strongly than CFTR481–487, likely because the longer peptide has more interactions with the Bag2-BNB surface. Both peptides cause modest chemical shift perturbations even at high peptide:BNB ratios, in a manner characteristic of fast-exchange binding processes. These results suggest that the peptides have modest (micromolar) binding affinities for the BNB domain.
Figure 5.
Mapping of chaperone client binding sites on Bag2-BNB domains. (a) Chemical shift perturbations induced by addition of CFTR481–487 (KIKHSGR), CFTR511–525 (SYDEYRY-RSVIKACQ) or Hsc70-NBD to 15N-Bag2-BNB. The secondary structure of Bag2-BNB is shown above the chemical shift perturbation histograms. (b) Surface and cartoon representations of the Bag2-BNB dimer. The surface is colored according to a surface potential range of +5 kT (dark blue) to −5 kT (dark red). In the cartoon representation, basic and acidic sidechains are shown as sticks and coloured blue and red, respectively. (c) Surface and cartoon representations of the CFTR481–487 and CFTR511–525 peptide binding sites. Residues on the surface that undergo chemical shift perturbations greater than 0.1 ppm are coloured blue (CFTR481–487), cyan (CFTR511–525) or purple (both peptides). Residues without chemical shift assignments are shown in yellow. The residues are similarly coloured and their sidechains are shown as sticks in the cartoon representation. (d) Surface and cartoon representation of the Hsc70-NBD binding site derived from HSQC titration data. Residues that undergo chemical shift perturbations greater than 0.1 ppm are shown in magenta in both representations, and their sidechains are shown as sticks in the cartoon representation. (e) Surface and cartoon representation of the Hsc70-NBD binding site derived from the Hsc70-NBD:Bag2-BNB crystal structure, shown for comparison with panel (d).
Surprisingly, the peptide binding sites overlap with the Hsc70-NBD binding interface of the BNB domain. We measured HSQC spectra of 15N Bag2-BNB in the presence of unlabeled Hsc70-NBD (Fig. 5a, d; Supplemental Figure 4b, c online). The Hsc70-NBD interaction surface deduced from these experiments is extremely similar to the NBD contact surface in the NBD:BNB crystal structure (Fig. 5e). These data provide a structural explanation for earlier findings9 showing that the Hsc70-NBD competes with chaperone substrates for binding to full-length Bag2.
To determine whether Hsc70-NBD can displace bound clients from the BNB domain, we preincubated 15N-Bag2-BNB with CFTR511–525 and titrated the BNB:peptide complex with Hsc70-NBD. Chemical shift perturbations induced by CFTR511–525 were reversed by addition of Hsc70-NBD, with the final spectrum showing peaks characteristic of the NBD:BNB complex (Supplemental Figure 4d online). Peaks corresponding to residues outside the Hsc70-binding interface but within the CFTR511–525 binding surface reverted to positions characteristic of free Bag2-BNB, showing that the CFTR511–525 peptide was displaced and ruling out the formation of a NBD:BNB:peptide ternary complex. The overlap between the Hsc70- and substrate-interacting surfaces on Bag2 and the known similarity between the substrate sequence specificities of Hsc70 and Bag2 suggest that Bag2 may bind to the Hsc70-NBD while coordinately transferring a bound client onto the substrate binding domain of Hsc70.
DISCUSSION
We have determined that the Bag2-BNB domain is a new type of Hsp70-NEF domain, differing from both canonical Bag domains and other known Hsp70-NEFs in structure, oligomerization state, and Hsp70/Hsc70-binding mode. We conclude that Bag2 was improperly classified as a member of the Bag family6, which (in mammals) otherwise consists of Bag1, Bag3, Bag4, Bag5 and Bag6, although the overall helical folds and limited sequence similarity of Bag and BNB domains suggest possible common ancestry. We have also structurally characterized the dual roles of the Bag2-BNB domain in nucleotide exchange and chaperone client binding. The fact that these roles are mediated by common structural elements of the Bag2-BNB suggests how Bag2 and Hsp70 may cooperate in chaperoning select Hsp70 clients.
Bag2 supports protein folding by Hsp70 in several ways. Most obviously, the Bag2-BNB domain promotes nucleotide exchange on Hsp70/Hsc70 and accelerates the cycle of chaperone client binding and release, along with the substrate remodeling action that accompanies this cycle. Bag domains or the bacterial Hsp70-NEF GrpE both cause a ~14° rotation in subdomain IIb8,15, while Hsp110 causes a more extensive (~27°) rotation16,18 (Supplementary Fig. 5a,b online). In both cases, however, subdomain IIb rotates about a hinge connecting subdomains IIb and IIa and swivels away from subdomain Ib. Bag2, however, causes domain II to rotate as a rigid unit, about the linkages connecting subdomains Ia and IIa. In contrast to GrpE, Bag1 or Hsp110, HspBP1/Fes1 family members clash strongly with domain I and effectively dislocate it from domain II, sensitizing the interdomain linkages to proteolysis17 (Supplementary Fig. 5c online). Bag2 thus causes a conformational change in the Hsp70-NBD that is intermediate between the changes induced by GrpE, Bag1 or Hsp110 and the extensive dislocations caused by HspBp1 (shown schematically in Supplementary Fig. 5d online). Rigid displacement of domain II (relative to the nucleotide-bound form) also occurs in nucleotide-free Hsc7014, but Bag2 induces a more extensive rotation and also pivots the domain in the plane separating domain I and II (Supplementary Fig. 3 online). Compared to other Hsp70-NEFs and to nucleotide-free Hsc70, Bag2 also causes somewhat different localized changes at the nucleotide binding site (Fig. 3d). We conclude that the observed structural changes are specific consequences of Bag2, rather than merely reflecting the absence of nucleotide in our crystallization conditions. The displacement of nucleotide-interacting residues to positions that are unfavorable for interactions or that clash directly with the nucleotide provides the impetus for ADP dissociation and nucleotide exchange.
We have shown how BNB domains integrate nucleotide exchange with a second pro-folding role. BNB domains interact with partial hydrophobic epitopes on chaperone substrates to stabilize the substrates, prevent their aggregation and deliver them to Hsp70. We identified two contiguous binding sites for two peptides derived from the Hsp70 client CFTR (Fig. 5). While the presence of two overlapping sites is unexpected, we suggest that the surface common to both sites selects for core basic/hydrophobic motifs of the peptides. Similar core motifs are present in other CFTR peptides that also bind to Bag2-BNB9. As these core motifs are roughly symmetric, the preferred orientation of each of the peptides may be determined by whether residues outside the core motifs interact more favorably with αL or with α2. A longer peptide of appropriate sequence could potentially engage the combined surface covered by both sites with higher affinity than the individual peptides, thereby wrapping around the end of the BNB dimer. Client binding domains must have defined but limited selectivity, to be able to engage many different incorrectly exposed epitopes on misfolded proteins. Binding a core epitope motif through a selective central patch and accommodating flanking residues with less selectivity is one way to fulfill these criteria.
Several other Hsp70 cochaperones also bind to client proteins. The Hsp40 family of J-domain proteins have elongated β-sheet client binding domains that dimerize in a pincer-like configuration and may help to deliver clients to Hsp7022,23. Among Hsp70-NEFs, the Hsp110 family are structurally similar to Hsp70, and contain β-sandwich client-binding domains equivalent to the substrate binding domain of Hsp7024. These may bind and stabilize extended client segments during client remodeling by Hsp70:Hsp110 complexes16. The substrate binding sites in the Bag2-BNB differ, however, from those in Hsp40 or Hsp110 by their α-helical structure, and instead resemble α-helical protrusions in such distinct chaperones as prefoldin25 or trigger factor26. Prefoldin and trigger factor have multiple such client-interacting protuberances in clamp or pincer configurations, and likely engage several client epitopes simultaneously. The dimeric BNB domains may similarly bind physically separated epitopes on a single client, and Bag2 oligomerization (see below) may further juxtapose multiple BNB dimers for more efficient chaperoning.
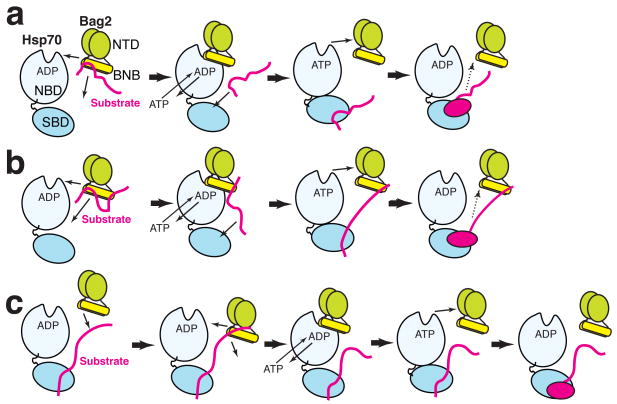
Client peptides and the Hsp70-NBD were previously shown to compete with each other for binding to Bag29. Our data show that this is simply because the client binding sites and the Hsp70-NBD interacting determinants of Bag2-BNB overlap (Fig. 5). This further distinguishes the Bag2 client-binding sites from those of Hsp40 and Hsp110 (in which the client- and Hsp70-interacting sites are located in different domains) and points to a straightforward mechanism by which substrates may be transferred from Bag2 onto Hsp70. We showed evidence that the Hsc70-NBD can displace a client peptide from the Bag2-BNB domain (Supplemental Fig. 4d online). The similar binding preferences of Bag2-BNB and the Hsp70 substrate binding domain suggest that, under such circumstances, the Hsp70 substrate binding domain is well positioned to accept the displaced client segment. Bag2 could thus “hand off” a bound client epitope to the substrate binding domain of Hsp70, while coordinately binding to the Hsp70-NBD to accelerate a mechanistic refolding step, as shown schematically in Fig. 6. In the context of the BNB dimer, one segment of the client could be handed off to Hsp70 while another client epitope remains associated with the binding site at the other end of the dimer (Fig. 6b). A similar “sharing” of a client between chaperone and cochaperone has been proposed for the Hsp70:Hsp110 complex16.
Figure 6.
Models for Hsp70, Bag2 and chaperone client interaction. For simplicity, Bag2 is shown as a dimer in both panels. “SBD” designates the substrate binding domain of Hsp70. The change in Hsp70 as shown is not meant to accurately represent a specific conformational transition, but rather to emphasize remodeling of the bound substrate. (a) Bag2 binds to a chaperone substrate (red) through the BNB domains. Binding of Hsp70-NBD to the BNB domains displaces the substrate. Bag2 promotes nucleotide exchange on Hsp70, while the substrate epitope now binds to the Hsp70-SBD. Hsp70 remodels the substrate and hydrolyzes ATP, aided by a J-domain protein such as Hsp40 (not shown for clarity). The refolded substrate dissociates (dotted arrow), possibly after another round of nucleotide exchange promoted by Bag2. The substrate may reassociate with Bag2 or with Hsp70 to undergo further rounds of refolding. (b) Similar to panel (a), except that a second epitope on the substrate remains associated with the second set of substrate binding sites on the Bag2-BNB dimer. An equivalent model would apply to chaperoning by Bag2 oligomers, which would present multiple substrate and Hsp70 binding sites. (c) Alternately, Bag2 may be recruited by a substrate already bound to Hsp70, and then promote nucleotide exchange and a substrate remodeling step.
Both sedimentation velocity analysis (Fig. 1e) and analytical gel filtration data (not shown) show that, unlike the BNB domains alone, full-length Bag2 forms tetramers and higher order oligomers. In HeLa cell extracts9, Bag2 was found in high molecular weight fractions, primarily complexes too large to be rationalized as Hsp70-bound or substrate-bound Bag2. The modest dimerization constant (~4 μM) of the BNB domain suggests that, in vivo, Bag2 dimerization would be mediated primarily by the Bag2-NTD, while the BNB would mediate further oligomerization. Oligomerization may be promoted through avidity effects, as multiple interactions form between Bag2, Hsc70 and chaperone clients. While our NMR data suggest that the substrate binding sites on the BNB domains have modest affinity individually, oligomeric Bag2 could bind to multiple epitopes on an extended chaperone client with higher avidity. In addition, oligomeric Bag2 would stay engaged with the substrate even as individual substrate epitopes are replaced by binding of Hsp70, and could recruit and coordinate multiple Hsp70 molecules on a single client. These factors may underlie the higher luciferase refolding activity of Bag2 compared to the Bag2-BNB domains alone (Fig. 4c,d). Bag2 may oligomerize to sequester a misfolded substrate, and then interact with Hsp70 molecules through the BNB domains for refolding of the substrate. The oligomerization of full-length Bag2 is reminiscent of such small heat shock proteins as Hsp25 or α-crystallin, which also form oligomers of variable stoichiometry to protect misfolded substrates from aggregation27,28. This model, however, does not rule out, and may coexist with, simpler mechanisms such as recruitment of Bag2 by Hsc70:client complexes (Fig. 6c). In this mechanism, the client-binding function of Bag2 is not utilized for its chaperoning properties but rather to help recruit Bag2 as a NEF.
The unanticipated but comprehensive differences between the Bag2-BNB and Bag domains suggest that the Bag2-BNB should be categorized as a novel type of Hsp70 nucleotide exchange factor domain. Given the diversity of NEFs for Hsp70, what specific role might Bag2 play in cells? Recently, Bag2 was shown to inhibit the ubiquitin ligase CHIP, which interacts with Hsp70/Hsc70 and ubiquitinates Hsp70-bound client proteins in situ29,30. The Bag2 N-terminal domain (Bag2-NTD) directly inhibits CHIP, possibly by disrupting interactions between CHIP and ubiquitin conjugating enzymes, but cannot do so without localizing to the Hsp70-CHIP chaperone complexes through the Bag2-BNB:Hsp70-NBD association9,10. Interestingly, Bag2 homologues are present in the genomes of organisms that have CHIP (Supplementary Fig. 1 online) but not in fungi, which also lack CHIP. Bag2 colocalizes with CHIP, especially at the endoplasmic reticulum10, and is part of chaperone complexes that interrogate the folding of cytoplasmic domains of large slow-folding membrane proteins, including the ion channels CFTR9 and HERG31. Bag2 may have a specific role in promoting the correct folding of such proteins while preventing their premature CHIP-induced ubiquitination and degradation. The architectures of the CHIP- and Bag2-containing Hsp70 complexes that monitor such clients remain relatively poorly defined, and additional studies on the Bag2-NTD and full-length Bag2 will be necessary to better understand their mechanisms and intracellular functions.
METHODS
Expression and crystallization of Bag2-BNB and Bag2-BNB:Hsc70-NBD complex
Mouse Bag2-BNB (murine Bag2107–189) and Hsc70-NBD (human Hsc701–381) were cloned into pHis||2 vector32 and expressed as His-tagged fusions in E. coli Rosetta2(DE3) cells (Novagen) at 20°C. The proteins were purified by nickel affinity chromatography in 20–500 mM imidazole, 50 mM Sodium Phosphate pH 7.8, 300 mM NaCl. His-tags were removed after overnight cleavage with Tobacco Etch Virus (TEV) protease. Additional purification was performed on a Superdex 75-16/60 gel filtration column (GE Healthcare) in 50 mM Tris pH 7.6, 150 mM NaCl. Selenomethionylated Bag2-BNB was expressed in E. coli strain B834(DE3) in defined media supplemented with 40 mg l−1 Seleno-L-methione (Sigma) and purified as described above. Bag2-BNB in 50mM NaCl, 20 mM Tris-HCl (pH 7.4), 2 mM DTT was concentrated to 20–30 mg ml−1 for hanging-drop crystallization trials at 20°C. Native crystals were obtained against 35–42% (v/v) PEG 400, 0.1 M Bis-Tris, pH 6.2–6.5. SeMet Bag2-BNB crystals were grown in 36% (v/v) PEG 400, 0.1 M Bis-Tris pH 6.6, 3% (v/v) ethanol. Crystals were cryoprotected directly with reservoir solution.
Bag2-BNB:Hsc70-NBD domain complexes were mixed at a 1.2:1 ratio and concentrated to 24 mg ml−1. The complex crystallized at 20°C by hanging-drop vapor-diffusion against 120 mM NaSCN (pH6.9), 12% (w/v) PEG3350. Crystals were cryoprotected in reservoir solution supplemented with 20% (v/v) glycerol.
X-ray crystallographic data collection and structure solution
All diffraction data were collected at Advanced Light Source beamline 4.2.2, Lawrence Berkeley National Laboratory from crystals cryocooled to 100°K, and processed using d*TREK33. For free Bag2-BNB, SAD data sets were collected from SeMet crystals at the selenium K-edge (0.9741 Å wavelength). All Bag2-BNB crystals were in spacegroup R32. A SeMet SAD data set with 2.8 Å resolution was used for heavy atom searches and phasing. Phasing and density modification were carried out in PHENIX34. While initial phases were poor (figure-of-merit 0.48) extensive use of iterative density modification and automated partial model building and refinement in PHENIX resulted in an interpretable electron density map. Further model building and rebuilding was carried out in COOT35. The final model was refined against a 2.55 Å native data set (collected at 0.9951 Å) using CNS36; the final working and free R factors are 25.3% and 28.4% respectively and the model has no Ramachandran violations.
The Bag2-BNB:Hsc70-NBD complex crystallized in space group P212121. Native data from the Bag2-BNB:Hsc70-NBD crystals were collected at ALS4.2.2. (at 1.07 Å wavelength). The structure of the complex was solved by molecular replacement in Phaser37 using the Hsc70 NBD domain structure8 (RCSB code:1HX1) and the BNB domain structure as search models. Refinement was carried out in CNS against a 2.3 Å native data set to generate a final model with an R-factor of 22.2% and a free R-factor of 25.6%. The refined structure has no Ramachandran violations.
Crystallographic data collection and refinement statistics are summarized in Table 1. Molecular graphics were generated and rendered using PyMol v.0.99 (www.pymol.org). Surface electrostatics were calculated using APBS38.
Table 1.
Data collection, phasing and refinement statistics
| Bag2-BNB (Native) | Bag2-BNB (SeMet SAD) | Hsc70-ATPase: Bag2-BNB | |
|---|---|---|---|
| Data collection | |||
| Space group | R32 | R32 | P212121 |
| Cell dimensions | |||
| a, b, c (Å) | 104.7, 104.7, 164.3 | 104.3, 104.3, 168.0 | 52.9, 105.8, 210.7 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 | 90, 90, 90 |
| Resolution (Å) | 52.34–2.55 (2.64–2.55)* | 29.99–2.80 (2.90–2.80) | 43.13–2.30 (2.38–2.30) |
| Rsym or Rmerge | 5.9 (24.2) | 9.2 (22.1) | 5.4 (17.9) |
| I/σI | 14.5 (5.7) | 32.3 (9.0) | 18.1 (8.0) |
| Completeness (%) | 98.8 (99.9) | 96.3 (80.2) | 100.0 (100.0) |
| Redundancy | 5.9 (5.9) | 19.2 (15.9) | 6.9 (7.1) |
| Refinement | |||
| Resolution (Å) | 39.69–2.55 | 43.13–2.30 | |
| No. reflections | 11295 | 53515 | |
| Rwork/Rfree | .253/.284 | .222/.256 | |
| No. atoms | |||
| Protein | 2430 | 7126 | |
| Ligand/ion | 19 | ||
| Water | 53 | 240 | |
| B-factors | |||
| Protein | 54.5 | 34.0 | |
| Ligand/ion | 56.9 | ||
| Water | 57.9 | 33.7 | |
| R.m.s deviations | |||
| Bond lengths (Å) | .007 | 0.006 | |
| Bond angles (°) | 1.20 | 1.15 | |
One crystal was used for each of the datasets.
Values in parentheses are for highest-resolution shell.
NMR spectroscopy
Uniformly 15N- and 15N/13C-labeled Bag2-BNB were expressed in Rosetta2(DE3) cells in minimal media with 1g l−1 15N-NH4Cl and 2g l−1 13C-glucose (Isotec) and purified as described above. NMR samples contained approximately 0.2 mM Bag2-BNB in 20 mM sodium phosphate (pH 6.4), 20 mM sodium chloride, 10% (v/v) D2O and 0.5 mM sodium azide. Spectra were acquired at 25°C on a cryoprobe-equipped Bruker 600MHz Avance ICE spectrometer. Datasets were processed using NMRPipe39 and visualized with Sparky (www.cgl.ucsf.edu/home/sparky/). Backbone chemical shift assignments for the free form of Bag2-BNB utilized 1H/13C/15N-HNCA, -HNCO, and -CBCA(CO)NH experiments. Assignments were made for 97.7% of backbone 1HN, 15NH atoms and 91.1% of backbone 13Cα, 13Cβ and 13C’ atoms. Assignments were made for 2 or more backbone atoms for all residues except Ala163 for which no 1HN or 15NH resonances were observed.
For HSQC titrations, Hsc70-NBD was prepared at 1.8 mM in the same buffer as Bag2-BNB. CFTR peptides were synthesized by standard methods by Lerner Research Institute core services. CFTR481–487 (KIKHSGR) was prepared at 127 mM in H2O and CFTR511–525 (SYDEYRYRSVIKACQ) was prepared at 24.5 mM in DMSO. Two-dimensional 1H/15N-HSQC spectra were acquired using a spin-state selective gradient-enhanced HSQC pulse sequence40. BNB:CFTR peptide and BNB:Hsc701–381 ratios varied from 1:0.2 to 1:93 for CFTR481–487, 1:0.2 to 1:10 for CFTR511–525 and 1:0.2 to 1:9 for Hsc70-NBD. In the HSQC competition experiment, spectra were collected from 15N-Bag2-BNB before and after the addition of 1:5 CFTR511–525. Hsc70-NBD was subsequently added to the sample at 1:2 and 1:5 ratios. Normalized chemical shift perturbations (Δ δ) were calculated and plotted according to the following equation41: Δ δ = [(Δ1HN)2 + (Δ1NH/5)2]1/2.
Expression and purification of constructs for in vitro assays
Expression and purification of Bag2, Hsc70 and Hsp40 constructs used for GST-minicolumn-binding experiments, nucleotide exchange assays, analytical ultracentrifugation, and luciferase refolding assays are described in the Supplemental Methods online.
GST-pulldown (minicolumn binding) assays
GST-minicolumn-binding experiments were carried out as described in the Supplemental Methods online.
Analytical ultracentrifugation
Sedimentation velocity and equilibrium experiments were carried out in a Beckman XL-I ProteomeLab analytical ultracentrifuge with absorbance optics. Details of these experiments are described in the Supplemental Methods online.
Nucleotide exchange and Luciferase refolding assays
Single-turnover nucleotide exchange assays and Luciferase refolding assays were performed as described previously10,19, and are detailed further in the Supplemental Methods online.
Accession codes
Coordinates and structure factors have been deposited in the RCSB Protein Data Bank with accession codes 3D0T and 3CQX for the Bag2-BNB domain and the Hsc70-NBD:Bag2-BNB complex structures respectively.
Supplementary Material
Acknowledgments
This work was funded by grants from the U.S. National Institutes of Health (R01-GM61728 to C.P. and RO1-GM080271 to S.M.), the American Heart Association (SDG 0735313N to S.M.) and by funds from the State of Ohio Eminent Scholar Program (to A.B.H.). E.K. was supported by funds from the Ralph Wilson Medical Research Foundation. The Advanced Light Source is supported by the U.S. DOE under contract number DE-AC03-76SF00098 at Lawrence Berkeley National Laboratory.
References
- 1.Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62:670–84. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morano KA. New tricks for an old dog: the evolving world of Hsp70. Ann NY Acad Sci. 2007;1113:1–14. doi: 10.1196/annals.1391.018. [DOI] [PubMed] [Google Scholar]
- 3.Qiu XB, Shao YM, Miao S, Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol Life Sci. 2006;63:2560–70. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cyr DM. Swapping nucleotides, tuning Hsp70. Cell. 2008;133:945–7. doi: 10.1016/j.cell.2008.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Young JC, Barral JM, Ulrich Hartl F. More than folding: localized functions of cytosolic chaperones. Trends Biochem Sci. 2003;28:541–7. doi: 10.1016/j.tibs.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 6.Takayama S, Reed JC. Molecular chaperone targeting and regulation by BAG family proteins. Nat Cell Biol. 2001;3:E237–41. doi: 10.1038/ncb1001-e237. [DOI] [PubMed] [Google Scholar]
- 7.Takayama S, Xie Z, Reed JC. An evolutionarily conserved family of Hsp70/Hsc70 molecular chaperone regulators. J Biol Chem. 1999;274:781–6. doi: 10.1074/jbc.274.2.781. [DOI] [PubMed] [Google Scholar]
- 8.Sondermann H, et al. Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science. 2001;291:1553–7. doi: 10.1126/science.1057268. [DOI] [PubMed] [Google Scholar]
- 9.Arndt V, Daniel C, Nastainczyk W, Alberti S, Hohfeld J. BAG-2 acts as an inhibitor of the chaperone-associated ubiquitin ligase CHIP. Mol Biol Cell. 2005;16:5891–900. doi: 10.1091/mbc.E05-07-0660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai Q, et al. Regulation of the cytoplasmic quality control protein degradation pathway by BAG2. J Biol Chem. 2005;280:38673–81. doi: 10.1074/jbc.M507986200. [DOI] [PubMed] [Google Scholar]
- 11.Briknarova K, et al. Structural analysis of BAG1 cochaperone and its interactions with Hsc70 heat shock protein. Nat Struct Biol. 2001;8:349–52. doi: 10.1038/86236. [DOI] [PubMed] [Google Scholar]
- 12.Kleinjung J, Fraternali F. POPSCOMP: an automated interaction analysis of biomolecular complexes. Nucleic Acids Res. 2005;33:W342–6. doi: 10.1093/nar/gki369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flaherty KM, DeLuca-Flaherty C, McKay DB. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature. 1990;346:623–8. doi: 10.1038/346623a0. [DOI] [PubMed] [Google Scholar]
- 14.Jiang J, Prasad K, Lafer EM, Sousa R. Structural basis of interdomain communication in the Hsc70 chaperone. Mol Cell. 2005;20:513–24. doi: 10.1016/j.molcel.2005.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrison CJ, Hayer-Hartl M, Di Liberto M, Hartl F, Kuriyan J. Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science. 1997;276:431–5. doi: 10.1126/science.276.5311.431. [DOI] [PubMed] [Google Scholar]
- 16.Polier S, Dragovic Z, Hartl FU, Bracher A. Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding. Cell. 2008;133:1068–79. doi: 10.1016/j.cell.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 17.Shomura Y, et al. Regulation of Hsp70 function by HspBP1: structural analysis reveals an alternate mechanism for Hsp70 nucleotide exchange. Mol Cell. 2005;17:367–79. doi: 10.1016/j.molcel.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 18.Schuermann JP, et al. Structure of the Hsp110:Hsc70 nucleotide exchange machine. Mol Cell. 2008;31:232–43. doi: 10.1016/j.molcel.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gassler CS, Wiederkehr T, Brehmer D, Bukau B, Mayer MP. Bag-1M accelerates nucleotide release for human Hsc70 and Hsp70 and can act concentration-dependent as positive and negative cofactor. J Biol Chem. 2001;276:32538–44. doi: 10.1074/jbc.M105328200. [DOI] [PubMed] [Google Scholar]
- 20.Meacham GC, Patterson C, Zhang W, Younger JM, Cyr DM. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat Cell Biol. 2001;3:100–5. doi: 10.1038/35050509. [DOI] [PubMed] [Google Scholar]
- 21.Younger JM, et al. Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell. 2006;126:571–82. doi: 10.1016/j.cell.2006.06.041. [DOI] [PubMed] [Google Scholar]
- 22.Li J, Qian X, Sha B. The crystal structure of the yeast Hsp40 Ydj1 complexed with its peptide substrate. Structure. 2003;11:1475–83. doi: 10.1016/j.str.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 23.Hu J, et al. The crystal structure of the putative peptide-binding fragment from the human Hsp40 protein Hdj1. BMC Struct Biol. 2008;8:3. doi: 10.1186/1472-6807-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Q, Hendrickson WA. Insights into Hsp70 chaperone activity from a crystal structure of the yeast Hsp110 Sse1. Cell. 2007;131:106–20. doi: 10.1016/j.cell.2007.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siegert R, Leroux MR, Scheufler C, Hartl FU, Moarefi I. Structure of the molecular chaperone prefoldin: unique interaction of multiple coiled coil tentacles with unfolded proteins. Cell. 2000;103:621–32. doi: 10.1016/s0092-8674(00)00165-3. [DOI] [PubMed] [Google Scholar]
- 26.Ludlam AV, Moore BA, Xu Z. The crystal structure of ribosomal chaperone trigger factor from Vibrio cholerae. Proc Natl Acad Sci USA. 2004;101:13436–41. doi: 10.1073/pnas.0405868101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ehrnsperger M, Lilie H, Gaestel M, Buchner J. The dynamics of Hsp25 quaternary structure. Structure and function of different oligomeric species. J Biol Chem. 1999;274:14867–74. doi: 10.1074/jbc.274.21.14867. [DOI] [PubMed] [Google Scholar]
- 28.Haley DA, Horwitz J, Stewart PL. The small heat-shock protein, alphaB-crystallin, has a variable quaternary structure. J Mol Biol. 1998;277:27–35. doi: 10.1006/jmbi.1997.1611. [DOI] [PubMed] [Google Scholar]
- 29.McDonough H, Patterson C. CHIP: a link between the chaperone and proteasome systems. Cell Stress Chaperones. 2003;8:303–8. doi: 10.1379/1466-1268(2003)008<0303:calbtc>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murata S, Chiba T, Tanaka K. CHIP: a quality-control E3 ligase collaborating with molecular chaperones. Int J Biochem Cell Biol. 2003;35:572–8. doi: 10.1016/s1357-2725(02)00394-1. [DOI] [PubMed] [Google Scholar]
- 31.Walker VE, Atanasiu R, Lam H, Shrier A. Co-chaperone FKBP38 promotes HERG trafficking. J Biol Chem. 2007;282:23509–16. doi: 10.1074/jbc.M701006200. [DOI] [PubMed] [Google Scholar]
- 32.Sheffield P, Garrard S, Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of "parallel" expression vectors. Protein Expr Purif. 1999;15:34–9. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- 33.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr D. 1999;55:1718–25. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 34.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D. 2002;58:1948–54. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 35.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 36.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 37.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–41. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 40.Schleucher J, et al. A general enhancement scheme in heteronuclear multidimensional NMR employing pulsed field gradients. J Biomol NMR. 1994;4:301–6. doi: 10.1007/BF00175254. [DOI] [PubMed] [Google Scholar]
- 41.Grzesiek S, Stahl SJ, Wingfield PT, Bax A. The CD4 determinant for downregulation by HIV-1 Nef directly binds to Nef. Mapping of the Nef binding surface by NMR. Biochemistry. 1996;35:10256–61. doi: 10.1021/bi9611164. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.