Abstract
Mosquito-borne dengue viruses are maintained in two discrete transmission cycles: a sylvatic cycle between nonhuman primates and sylvatic Aedes mosquitoes, and an endemic cycle between humans and peridomestic Aedes (primarily Ae. aegypti and Ae. albopictus). Most sylvatic strains are genetically distinct from endemic strains, and human infections with sylvatic strains have been detected only rarely. Interestingly, sylvatic strains replicate as well as endemic strains in Ae. aegypti and experimental models of replication in humans, suggesting that adaptive constraints may not explain the limited spillover of sylvatic strains into the endemic cycle. Within-host competition is another mechanism known to decrease emergence of strains into occupied niches. In the current study, we examined the magnitude of competitive suppression between sylvatic and endemic dengue strains of different serotypes in pair-wise mixed infections of cultured Ae. albopictus cells to test whether the ecotype or the initial ratio of the two strains influenced the outcome of competition. Strains isolated from non-human primates were competitively inferior to those isolated from humans. Moreover, competition was density-dependent; the magnitude of suppression increased as the starting density of a strain relative to its competitor decreased. These data suggest that competitive inferiority in endemic vectors coupled with a numerical disadvantage relative to resident endemic strains could restrict reemergence of sylvatic strains into the endemic cycle and contribute to the ecologically correlated genetic divergence between sylvatic and endemic strains.
Key Words: Dengue virus, Competition, Density-dependent, Superinfection, Sylvatic, Aedes
Introduction
The four serotypes of dengue virus (DENV; genus Flavivirus) comprise a genetically and ecologically diverse group of mosquito-borne RNA viruses. They occur in two distinct life cycles: a sylvatic cycle between nonhuman primates and sylvatic Aedes mosquitoes in the forests of Southeast Asia and West Africa (Rudnick 1965, 1978, Saluzzo et al. 1986), and an endemic cycle between humans and peridomestic Aedes spp., occurring globally in the tropics as well as in some temperate regions (Gubler 1998, Mackenzie et al. 2004). Each endemic serotype is composed of multiple major lineages, termed genotypes, which in turn contain multiple distinct strains (Rico-Hesse 2003). Endemic DENVs have shown a dramatic geographic expansion in the last several decades, leading to increased cocirculation of serotypes, genotypes, and strains within particular areas (Gubler 1998, Chareonsook et al. 1999, Endy et al. 2002, De Simone et al. 2004, Fouque et al. 2004, Mackenzie et al. 2004, Gubler 2006) and high levels of mixed infections within individual vectors in some outbreaks (Lorono-Pino et al. 1999, Thavara et al. 2006).
Sylvatic DENVs (serotypes 1, 2, and 4) were isolated from canopy-dwelling sentinel monkeys (Macaca fascicularis and Presbytis obscura) and Aedes spp. almost four decades ago (Rudnick 1965, Rudnick et al. 1965), in a region of Asia where Ae. aegypti was absent and where humans were scarce. A later study showed that DENV infection in humans tended to be highest in populations living adjacent to forest habitat and to be associated with mild disease (Rudnick 1986). From this evidence, it was concluded that sylvatic DENV strains are transmitted in an enzootic cycle, mainly circulating in canopy-dwelling monkeys, with infrequent spillover to human populations via Aedes spp. that feed on both upper and lower canopy primates (Yuwono et al. 1984, Rudnick 1986). Subsequently, phylogenetic analyses revealed that sylvatic strains are genetically distinct from endemic strains of the same serotype (Wang et al. 2000, Shurtleff et al. 2001, Vasilakis et al. 2007a).
It was initially hypothesized that the rarity of human infection with sylvatic DENV was attributable to a lack of adaptation of these viruses to human hosts or peridomestic Aedes. However, recent experiments have demonstrated that sylvatic DENV-2 replicates to the same level as endemic DENV-2 strains in multiple models of human infection (Vasilakis et al. 2007b) as well as in Ae. aegypti (Hanley, unpublished data), indicating that no adaptive barrier exists to the emergence of DENV-2 sylvatic strains. Alternatively, circulation of endemic DENV strains may restrict emergence of sylvatic strains through competitive exclusion. Recent experiments with genetically distinct Plasmodium strains have shown that direct competition of pathogens within hosts can alter invasion success, strain prevalence, and evolutionary rates (de Roode et al. 2005, Wargo et al. 2007). Studies of RNA viruses have found that replication of a given strain may be suppressed during concurrent infection with a different strain or species (Sundin and Beaty 1988, el Hussein et al. 1989, Simon et al. 1990, Karpf et al. 1997, Singh et al. 1997, Alonso et al. 1999, Shinjoh et al. 2000, Geib et al. 2003, Lee et al. 2005, Perales et al. 2007, Tscherne et al. 2007). Across these studies, two general patterns become apparent: (1) when infection of two strains is staggered in time, the strain infecting second is more suppressed than the strain infecting first, and (2) the magnitude of suppression increases as the density of the two competing strains increases. In a previous study, we investigated whether suppression of replication in DENV could also occur by coinfecting cultured Ae. albopictus cells with two endemic DENV serotypes and measuring the titers of each strain over time. We found that the magnitude of suppression depended on the strain and the time interval between infection of the two strains, and that suppression was stronger at later time points during the infection time course (Pepin et al. 2008). We hypothesized that considerable variation in competitive ability may exist among DENV strains and that suppression may be density-dependent.
In the current study, we tested these hypotheses by conducting pair-wise, mixed-strain infections among six DENV strains that differed by serotype, ecotype, and competitor density. Results from this panel of DENV strains revealed that while considerable variation exists among strains in competitive ability, the magnitude of suppression was strongly density-dependent for all strains. Importantly, ecotype was a significant determinant of competitive ability.
Materials and Methods
Cells and viruses
Ae. albopictus epithelial cells (C6/36) (Singh 1967, Igarashi 1978) were maintained in minimal essential media (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS; Invitrogen), 2 mM L-glutamine (Invitrogen), 1X nonessential amino acids (Invitrogen), and 50 μg/mL gentamycin at 32°C, 5% CO2, and 88% RH. The six virus strains used in this study are described in Table 1. Each strain was passaged once in C6/36 cells to produce working pools, which were stored in 1X SPG (218 mM sucrose, 6 mM L-glutamic acid, 3.8 mM potassium phosphate [monobasic], 7.2 mM potassium phosphate [dibasic] [pH 7.2] [final concentrations given]) at −80°C.
Table 1.
Strain Information
| Serotype | Strain | Ecotype | Source | Location | Year | Passage history | Label |
|---|---|---|---|---|---|---|---|
| 1 | P72-1244 | Sylvatic | Sentinel monkey | Malaysia | 1972 | Monkey-1 (passage history unknown), C6/36-2 | 1S |
| 1 | 16007 | Endemic | Human | Thailand | 1964 | BS-C-1-3, LLC-MK2-2, T. amb.-1, C6/36-2 | 1E |
| 2 | 0372 | Endemic | Human | Thailand | 1988 | C6/36-6 | 2E |
| 2 | P8-1407 | Sylvatic | Sentinel monkey | Malaysia | 1970 | SM-3, C6/36-2 | 2S |
| 3 | Sleman/78 | Endemic | Human | Indonesia | 1978 | A. aeg.-1, Vero-5, C6/36-2, Vero-1 | 3E |
| 4 | 052 | Endemic | Human | Thailand | 1985 | C6/36-2 | 4E |
Passage history = the infection model followed by the number of passages that were performed prior to this experiment (SM: suckling mice in vivo; C6/36: Aedes albopictus, epithelial cells; BS-C-1: grivet monkey kidney cells; LLC-MK2: rhesus monkey kidney cells; Vero: African green monkey kidney cells; A. aeg.: Aedes aegyptii in vivo; T. amb.: Toxorhynchites amboinenis in vivo); Label = the designated notation used throughout the manuscript.
Infection and determination of virus titer
Infections were initiated on ∼80% confluent monolayers of C6/36 cells in 6-well tissue culture-treated plates (BD Falcon, Fisher Scientific, Pittsburgh, PA). The multiplicity of infection (MOI) was 0.05, 5, or 10, according to the treatments outlined in Table 2. Prior to infection, cells were washed in 2 mL fresh media. Appropriately diluted virus in total volume of 1 mL was added to cells, which were then incubated for 1 hour and 45 minutes, washed with 2 mL fresh media, replenished with 3 mL fresh media, and returned to incubation in the conditions described above. The protocol for second infections in superinfection treatments was identical except that the initial wash step was omitted. Superinfections were conducted with an 8-hour time lag since the majority of Asian vectors that feed multiply within a gonotropic cycle and tend to do so within the same day (Scott et al. 2000). All treatments were harvested 72 hours after initiation of the experiment, which was based on our previous determination that DENV infected at the specified conditions begins to reach peak titers at this time (data not shown). A 1 mL aliquot of each treatment was frozen in 1X SPG at −80°C for determination of titer.
Table 2.
Description of Treatments
| |
Infection MOI per strainb |
|
|
|
|---|---|---|---|---|
| Infection treatments | 0 hours | 8 hours | Total MOI | X:Y ratioc |
| SINGLE | 0.05 | — | 0.05 | 1:0 |
| X only | — | 0.05 | 0.05 | |
| 5 | — | 5 | ||
| — | 5 | 5 | ||
| 10 | — | 10 | ||
| CO | 0.05 + 0.05 | — | 0.1 | 1:1 |
| X + Y same time | 5 + 5 | — | 10 | |
| SUPER XY | 0.05 | 5 | 5.5 | 1:10 |
| X first | 5 | 5 | 10 | 1:1 |
| Y second | 5 | 0.05 | 5.5 | 10:1 |
| SUPER YX | 0.05 | 5 | 5.5 | 1:10 |
| Y first | 5 | 5 | 10 | 1:1 |
| X second | 5 | 0.05 | 5.5 | 10:1 |
Each single infection was done for all strains listed in Table 1; CO and SUPER infections were done for each of the following six pairs: 1S + 2E, 1S + 3E, 1S + 4E, 2S + 1E, 2S + 3E, 2S + 4E.
Numbers are the MOI at which X and Y were infected; the two columns represent the time of infection from the start of the experiment in hours.
X refers to the strain for which titers were determined, Y is the competitor.
All treatments were sampled at 72 hours postinfection; four independent experiments per treatment were conducted.
Virus titers were determined by infecting ∼80% confluent C6/36 cells in 24-well tissue culture-treated plates (BD Falcon, Fisher Scientific) with a 10-fold serial dilution of the designated sample in duplicate. Plates were incubated with occasional shaking for 2 hours under conditions for maintenance of C6/36 described above and then overlaid with 1 mL/well of 0.8% methylcellulose in Opti-MEM (Invitrogen) supplemented with 2% FBS, 2 mM L-glutamine, and 50 μg/mL gentamycin. Plates were incubated for 5 days and then plaques were detected by antibody and visualized by immunoperoxidase staining as previously described (Durbin et al. 2001, Troyer et al. 2001). Titers were quantified as log10 pfu/mL, where each plaque represents a focus of infection initiated by a single virion and detected by either a monoclonal antibody (Mab) (Henchal et al. 1982) or hyperimmune mouse ascites fluid (HMAF) from mice immunized with the designated serotype. Antibody dilutions were optimized to eliminate nonspecific staining and maximize detection by peroxidase staining, resulting in final dilutions of (serotype specificity: antibody, dilution): 1S: Mab 15F3, 1:1000; 1E: Mab 15F3, 1:300; 2E and 2S: Mab 3H5, 1:1000; 3E: Mab 3D4, 1:400; 4E: HMAF 4, 1:2000.
Experimental design
To test the effect of ecotype on the magnitude of competitive suppression in mixed-strain infections, the two sylvatic strains were each separately competed against three of the endemic strains in the following combinations: 1S + 2E, 1S + 3E, 1S + 4E, 2S + 1E, 2S + 3E, 2S + 4E (Table 2). Competitors always differed in serotype so that individual strains could be distinguished with available antibodies. In mixed infections, competitors were added at two intervals: coinfections, where each strain was added simultaneously, and superinfections, where one strain was added 8 hours before the second strain. Strains in mixed infections were designated X (focal strain) and Y (competitor). Symmetric superinfections were conducted, e.g., XY where X was added before Y, and YX where X was added after Y. To investigate the effects of competitor density in mixed infections, three ratios of X:Y were used in each type of superinfection (1:10, 1:1, and 10:1) for a total of six superinfection treatments per pair of strains. Coinfections were conducted at two multiplicities of infection (MOI = 0.05 and MOI = 5), but the initial ratio was 1:1 in both coinfection treatments. Each experimental block was replicated 4-fold. To determine the replication of each strain in the absence of competition, five single-strain infection treatments that differed in the time of initiation (0 hours versus 8 hours) or MOI (0.05, 5, or 10) were conducted for each of the six strains.
Titer data for each strain in mixed infections were expressed as competitive suppression by subtracting the log titer of a designated strain in single infection from the log titer of that strain in mixed infection. The variability in suppression among experimental strains was tested in the MOI 5 coinfection data using a Kruskal-Wallis test since these data were not normally distributed (JMP, Version 7.0, SAS Institute Inc., Cary, NC). Means from each pair-wise competition under each treatment were calculated and used to test the significance of higher-level effects such as ecotype and competitor ratio. These data were normally distributed and thus analyzed using t-tests and ANCOVA in JMP (Version 7.0, SAS Institute Inc.).
Results
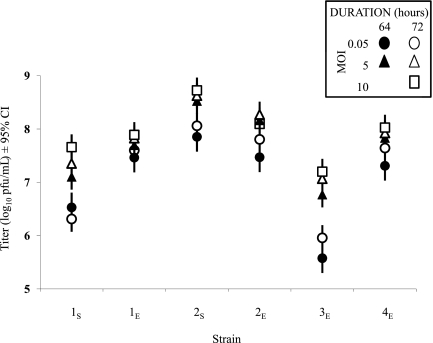
Five single-strain infections were conducted for each strain as controls to assess the combined effects of MOI (0.05, 5, or 10) and duration of infection (64 or 72 hours) on overall titers in the treatments (Fig. 1). An ANOVA with MOI and duration of infection as a combined variable nested within the main effect “strain” showed that: (1) there was significant variation in final titers among strains (main effect: F5,113 = 145.7, p < 0.0001), and (2) final titers of the six strains in single-strain infections depended on these nested variables (nested effect: F24,113 = 12.2, p < 0.0001). The sylvatic strains (1S and 2S) differed in overall final titers, and their titers were comparable to those of endemic strains (Fig. 1). Strain 3E had the lowest titers overall and appeared to be most affected by low MOI. Since MOI and infection duration affected final titers in single-strain infections, data from mixed infection treatments were expressed as “competitive suppression” values: the titer of a given strain in each mixed-strain infection treatment relative to its titer in the corresponding single-strain infection treatment. For example, for 1S superinfected at MOI 0.05, titers for 1S were compared to the single-strain infection of 1S infected at MOI 0.05 with a total duration of 64 hours rather than 72 hours.
FIG. 1.
Final titers of all single-strain infections. Five single-strain infections were conducted for each strain to control for the effects of infection duration and MOI in mixed-strain infection treatments. Data are mean log10 pfu/mL of four replicate infections. Error bars are 95% confidence intervals (CI) from the nested ANOVA model: Titer of X = β0 + β1Strain + β2Treatment + β3Treatment[Strain] + ɛ. Treatment is the combined effects of total infection duration: 64 hours (black) and 72 hours (white), and MOI: 0.05 (circle), 5 (triangle), and 10 (square); e.g., a black circle indicates that infection time was 64 hours and the inoculation MOI was 0.05. Strain labels on the X-axis correspond to those in Table 1.
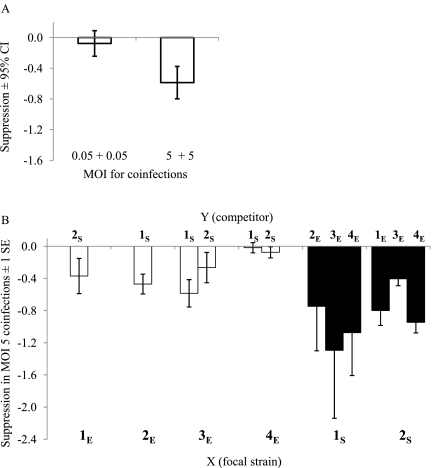
In coinfection treatments, competitive suppression occurred only in the MOI 5 treatments, and was not detected in the MOI 0.05 treatments (t22 = −3.5, p < 0.0012; Fig. 2A: 95% confidence intervals [CI] overlap 0 in MOI 0.05 treatments). Figure 2B shows the mean suppression (± 1 standard error; n = 4 replicates) for experimental strains (indicated along the bottom) with each competitor (indicated along the top) in the MOI 5 coinfections. When grouping suppression data for all replicates by focal strain (i.e., 6 levels: 1S,2S,1E,2E,3E, and 4E), a significant difference in suppression among strains was found (χ2 = 13.7, DF = 5, p < 0.018). Suppression of sylvatic strains was significantly greater than that of endemic strains regardless of competitor (compare white to black columns in Fig. 2B; t9 = 3.8, p < 0.002). Moreover, suppression of 1S was similar to that of 2S (t4 = 1.4, p < 0.11), and each of these strains was significantly different from all other endemic strains (2S versus 1–4E: t3 = 2.3, p < 0.05; 1S versus 1–4E: t3 = 4.1, p < 0.011). Thus, suppression appeared to be stronger in strains isolated from sylvatic habitats.
FIG. 2.
Effects of competitor density and ecotype on suppression in coinfections. Suppression of strain X is: log10 pfu/mL of strain X in single-strain infections minus log10 pfu/mL of strain X in mixed-strain infections with competitor Y (indicated on Y-axis, see Materials and Methods). Zero indicates that titers in mixed infections are similar to those in single infections (no suppression), whereas negative values indicate suppression. (A) Columns are the mean suppression over all mixed-strain coinfections in each MOI treatment. Error bars are 95% confidence intervals (CI) from the t-test: suppression of X at MOI 0.05 versus 5. (B) Mean suppression ± 1 SE of the four replicate infections conducted for each pair of strains for mixed-strain coinfections at MOI 5. Columns for each strain are color-coded by ecotype: endemic (white) and sylvatic (black). Experimental strains (X) are indicated above competitors (Y).
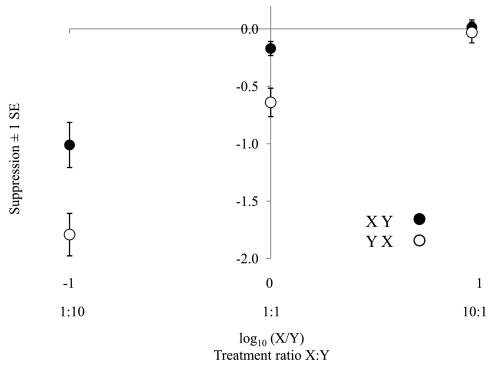
To examine whether the magnitude of suppression depended on the initial ratio of X to its competitor Y in superinfection treatments, an ANCOVA model with initial ratio as the covariate and order of infection (XY versus YX) as the effect was used. Overall, the magnitude of suppression depended on the relative concentration of each strain in mixed infections (F3,71 = 42.1, p < 0.0001), and when X was infected at 10 times the concentration of its competitor, no significant suppression was detected in either treatment (Fig. 3). There was a significant interaction of initial ratio and order of infection (F1,71 = 7.3, p < 0.009); the slope of the relationship was steeper in the treatment where Y was added before X (Fig. 3). Thus, suppression of X depended more strongly on initial density of Y when X was infected 8 hours after Y.
FIG. 3.
Effects of X:Y infection ratio on suppression in superinfections. Suppression of strain X is: log10 pfu/mL of strain X in single-strain infections minus log10 pfu/mL of strain X in mixed-strain infections, as in Figure 2. The X-axis shows the infection ratio of X:Y. The two types of superinfection treatments are differentiated by color: XY (black) and YX (white). Each point is a mean (± 1 SE) of all superinfection treatments at a specific competitor density and order of strain X infection (see Table 2; n = 6 for each point since there were 6 pair-wise strain combinations in each category; each of these 6 data points within each category is a mean of 4 replicates from each pair; see Materials and Methods).
Discussion
The competitive inferiority of sylvatic strains was not due to serotype or intrinsic replicative ability since: (1) the endemic strains used encompassed all four serotypes, and (2) replication rates in single-strain infections showed no trend for lower replication of sylvatic strains (in fact, the 2S strain showed the highest levels of replication). Although both 1S and 2S have been classified as sylvatic strains based on their isolation from sentinel monkeys in a sylvatic habitat, only 2S shows convincing genetic separation from endemic serotype 2 strains. The bootstrap support for the distinction of 1S from endemic DENV-1 strains is weak at best (Wang et al. 2000, Shurtleff et al. 2001, Vasilakis et al. 2007a). This suggests that any potential genetic basis for competitive inferiority in sylvatic-isolated strains would involve relatively few differences in amino acids or in the sequence of the untranslated regions. It has been shown that single amino acid mutations in DENV and chikungunya viruses may have dramatic, specific effects on replication in vectors (Hanley et al. 2003, Tsetsarkin et al. 2007), suggesting that competitive differences among sylvatic and endemic strains could even derive from a single amino acid change. More sampling of Asian sylvatic DENV is needed to understand the underlying genetic differences that determine competitive ability among sylvatic and endemic DENV, and more broadly, to understand the evolutionary relationships of sylvatic and endemic DENV.
For all pairs of strains, coinfection resulted in suppression only when the total MOI was high, a result that is indicative of competition since the frequency of direct interaction among strains and degree of resource depletion increases with MOI. Additionally, when the total MOI was high, DENV strains were only suppressed when they were superinfected at titers equal to or less than those of their competitors. Since competitive suppression was estimated from titers at a single time point (72 hours), and not all strains would be expected to reach peak titer at the same time, the measure of competitive suppression quantifies differences in replication rate rather than effects on peak titers. In other words, it is possible that viruses in treatments that differed in titer at 72 hours postinfection achieved similar peak titers later in the course of infection. However, in addition to peak titer, replication rate is an important determinant of transmission probability since the average life expectancy of vectors can be very short. In order to investigate competitive suppression on replication rate, the sampling time point was determined from growth curve experiments with two serotypes (Pepin et al. 2008), which showed that 72 hours was the earliest that DENVs could reach peak titers. Interestingly, results of this experiment also suggested that a decrease in replication rate translates to decreased peak titer (Pepin et al. 2008).
Competitive suppression among RNA virus strains can occur during virus entry (Steck and Rubin 1966, Bratt and Rubin 1968, Delwart and Panganiban 1989), penetration (Simon et al. 1990, Singh et al. 1997), or replication (Singh et al. 1997, Geib et al. 2003, Lohmann et al. 2003, Lee et al. 2005, Tscherne et al. 2007). However, suppression of DENV is most likely to involve a process downstream of cell entry or penetration since superinfecting strains are still able to begin producing progeny. Suppression in mixed-strain infections could be due to competition for limited viral polymerase and host factors, as has been suggested by Lohmann et al. (2003). Alternatively, viruses may compete for sites of replication (Lee et al. 2005). This may be especially common in flaviviruses, which require attachment to a specific membrane compartment (Westaway 1987), so that sites along the membrane could become saturated once high levels of replication complexes are achieved. The density-dependent patterns observed in DENV are consistent with either of these mechanisms.
Competition within vectors only impacts the epidemiology of DENV when the likelihood of concurrent infections is high. Although the prevalence of DENV in Aedes in Southeast Asia appears low at between 0%–20% (Chow et al. 1998, Romero-Vivas et al. 1998, Chung and Pang 2002, Ritchie et al. 2004, Urdaneta et al. 2005, Thavara et al. 2006, Srisuphanunt et al. 2007), vector susceptibility to DENV is also low and variable at between 0%–62% (Black et al. 2002). Thus, the proportion of infected vectors may not always be an accurate indicator of DENV prevalence, and low numbers of infected vectors could represent infection of a large proportion of susceptible vectors in some populations. In fact, the ratio of vectors with mixed-strain versus single-strain infections may be a better measure than overall proportion of infected vectors of the prevalence of DENV in a given region. Furthermore, documentation of concurrent infections in both hosts and vectors is increasing and levels of concurrent infection are high in some outbreaks (Lorono-Pino et al. 1999, Wang et al. 2003, Thavara et al. 2006, Bharaj et al. 2008), likely reflecting increased sampling efforts and improved detection techniques. More emphasis on screening individual vectors for mixed-strain infections as well as experiments to reveal the outcome of interactions of DENV in mixed infections will provide a clearer understanding of the role of competition in the epidemiology and evolution of DENV.
If replication in cultured cells reflects patterns in mosquitoes, our data suggest that poor competitive ability of sylvatic strains could decrease their emergence to the endemic cycle through density-dependent competition in coinfected cells in the mosquito midgut. Recent studies have shown that DENV replicates in the mosquito midgut for 6 days before dissemination (Linthicum et al. 1996, Salazar et al. 2007). Thus, the virus population in the midgut may become large enough that individual cells are frequently coinfected. Furthermore, a study of Ae. taeniorhynchus mosquitoes fed on high titer Venezuelan equine encephalitis virus showed that: (1) only a small proportion of midgut cells were infected, and (2) greater than 33% of the infected cells were coinfected with two strains of virus carrying different markers, which was significantly higher than expected based on the titer of the inoculum (Smith et al. 2007). Therefore, coinfection of individual cells may be frequent in vectors if only a small proportion of midgut cells can support infection. However, one caveat to generalizing cell culture data in a live mosquito context is that replication dynamics of flaviviruses in cultured cells do not always accurately reflect their replication dynamics in mosquitoes (Ebel et al. 2004, Ciota et al. 2007). This suggests that the ranking of replication rates and competitive abilities may differ during infection in vivo. Nonetheless, our data do show that in coinfected cells DENV strains may affect one another's replication, and that there is variation in competitive ability among strains that can be attributed to ecological differences.
In summary, our data reveal that: (1) competitive suppression among strains of DENV occurs in cultured mosquito cells through a density-dependent mechanism, and (2) sylvatic DENV strains are competitively inferior to endemic strains even though their rates of replication in single-strain infections are comparable to those of endemic strains. Follow-up studies of these patterns in vivo are needed, as are modeling studies to address the epidemiological significance of competitive suppression within vectors. Although the emergence of sylvatic strains does not appear to be limited by adaptation to endemic vectors and hosts (Vasilakis et al. 2007b, Hanley, unpublished data), the present data suggest that competition from endemic strains has the potential to exclude sylvatic strains from the endemic cycle and thereby maintain the genetic distinction of sylvatic DENV.
Acknowledgments
We are grateful to Robert Tesh and the World Reference Center of Emerging Viruses and Arboviruses (UTMB), Stephen S. Whitehead (NIAID, NIH), Aravinda de Silva (UNC), and Duane Gubler (UHI) for providing us with virus isolates. Funding was provided by NIH-NM-INBRE (P20 RR016480-05), NIH-K22 (K22-A164193), and NSF-AD-VANCE (SBE-123690).
References
- Alonso M. Rodriguez S. Perez-Prieto SI. Viral coinfection in salmonids: infectious pancreatic necrosis virus interferes with infectious hematopoietic necrosis virus. Arch Virol. 1999;144:657–673. doi: 10.1007/s007050050534. [DOI] [PubMed] [Google Scholar]
- Bharaj P. Chahar HS. Pandey A. Diddi K, et al. Concurrent infections by all four dengue virus serotypes during an outbreak of 2006 in Delhi, India. Virol J. 2008;5:1. doi: 10.1186/1743-422X-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black WC. Bennett KE. Gorrochotegui-Escalante N. Barillas-Mury CV, et al. Flavivirus susceptibility in Aedes aegypti. Arch Med Res. 2002;33:379–388. doi: 10.1016/s0188-4409(02)00373-9. [DOI] [PubMed] [Google Scholar]
- Bratt MA. Rubin H. Specific interference among strains of Newcastle disease virus. 3. Mechanisms of interference. Virology. 1968;35:395–407. doi: 10.1016/0042-6822(68)90218-3. [DOI] [PubMed] [Google Scholar]
- Chareonsook O. Foy HM. Teeraratkul A. Silarug N. Changing epidemiology of dengue hemorrhagic fever in Thailand. Epidemiol Infect. 1999;122:161–166. doi: 10.1017/s0950268898001617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow VT. Chan YC. Yong R. Lee KM, et al. Monitoring of dengue viruses in field-caught Aedes aegypti and Aedes albopictus mosquitoes by a type-specific polymerase chain reaction and cycle sequencing. Am J Trop Med Hyg. 1998;58:578–586. doi: 10.4269/ajtmh.1998.58.578. [DOI] [PubMed] [Google Scholar]
- Chung YK. Pang FY. Dengue virus infection rate in field populations of female Aedes aegypti and Aedes albopictus in Singapore. Trop Med Int Health. 2002;7:322–330. doi: 10.1046/j.1365-3156.2002.00873.x. [DOI] [PubMed] [Google Scholar]
- Ciota AT. Lovelace AO. Jones SA. Payne A, et al. Adaptation of two flaviviruses results in differences in genetic heterogeneity and virus adaptability. J Gen Virol. 2007;88:2398–2406. doi: 10.1099/vir.0.83061-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delwart EL. Panganiban AT. Role of reticuloendotheliosis virus envelope glycoprotein in superinfection interference. J Virol. 1989;63:273–280. doi: 10.1128/jvi.63.1.273-280.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Roode JC. Pansini R. Cheesman SJ. Helinski ME, et al. Virulence and competitive ability in genetically diverse malaria infections. Proc Natl Acad Sci USA. 2005;102:7624–7628. doi: 10.1073/pnas.0500078102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Simone TS. Nogueira RM. Araujo ES. Guimaraes FR, et al. Dengue virus surveillance: the co-circulation of DENV-1, DENV-2 and DENV-3 in the State of Rio de Janeiro, Brazil. Trans R Soc Trop Med Hyg. 2004;98:553–562. doi: 10.1016/j.trstmh.2003.09.003. [DOI] [PubMed] [Google Scholar]
- Durbin AP. Karron RA. Sun W. Vaughn DW, et al. Attenuation and immunogenicity in humans of a live dengue virus type-4 vaccine candidate with a 30 nucleotide deletion in its 3′-untranslated region. Am J Trop Med Hyg. 2001;65:405–413. doi: 10.4269/ajtmh.2001.65.405. [DOI] [PubMed] [Google Scholar]
- Ebel GD. Carricaburu J. Young D. Bernard KA, et al. Genetic and phenotypic variation of West Nile virus in New York, 2000–2003. Am J Trop Med Hyg. 2004;71:493–500. [PubMed] [Google Scholar]
- el Hussein A. Ramig RF. Holbrook FR. Beaty BJ. Asynchronous mixed infection of Culicoides variipennis with bluetongue virus serotypes 10 and 17. J Gen Virol. 1989;70:3355–3362. doi: 10.1099/0022-1317-70-12-3355. [DOI] [PubMed] [Google Scholar]
- Endy TP. Nisalak A. Chunsuttiwat S. Libraty DH, et al. Spatial and temporal circulation of dengue virus serotypes: a prospective study of primary school children in Kamphaeng Phet, Thailand. Am J Epidemiol. 2002;156:52–59. doi: 10.1093/aje/kwf006. [DOI] [PubMed] [Google Scholar]
- Fouque F. Garinci R. Gaborit P. Epidemiological and entomological surveillance of the co-circulation of DEN-1, DEN-2 and DEN-4 viruses in French Guiana. Trop Med Int Health. 2004;9:41–46. doi: 10.1046/j.1365-3156.2003.01166.x. [DOI] [PubMed] [Google Scholar]
- Geib T. Sauder C. Venturelli S. Hassler C, et al. Selective virus resistance conferred by expression of Borna disease virus nucleocapsid components. J Virol. 2003;77:4283–4290. doi: 10.1128/JVI.77.7.4283-4290.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler DJ. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev. 1998;11:480–496. doi: 10.1128/cmr.11.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler DJ. Dengue/dengue haemorrhagic fever: history and current status. Novartis Found Symp. 2006;277:3–16. doi: 10.1002/0470058005.ch2. [DOI] [PubMed] [Google Scholar]
- Hanley KA. Manlucu LR. Gilmore LE. Blaney JE, Jr, et al. A trade-off in replication in mosquito versus mammalian systems conferred by a point mutation in the NS4B protein of dengue virus type 4. Virology. 2003;312:222–232. doi: 10.1016/s0042-6822(03)00197-1. [DOI] [PubMed] [Google Scholar]
- Henchal EA. Gentry MK. McCown JM. Brandt WE. Dengue virus-specific and flavivirus group determinants identified with monoclonal antibodies by indirect immunofluorescence. Am J Trop Med Hyg. 1982;31:830–836. doi: 10.4269/ajtmh.1982.31.830. [DOI] [PubMed] [Google Scholar]
- Igarashi A. Isolation of a Singh's Aedes albopictus cell clone sensitive to dengue and chikungunya viruses. J Gen Virol. 1978;40:531–544. doi: 10.1099/0022-1317-40-3-531. [DOI] [PubMed] [Google Scholar]
- Karpf AR. Lenches E. Strauss EG. Strauss JH, et al. Superinfection exclusion of alphaviruses in three mosquito cell lines persistently infected with Sindbis virus. J Virol. 1997;71:7119–7123. doi: 10.1128/jvi.71.9.7119-7123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YM. Tscherne DM. Yun SI. Frolov I, et al. Dual mechanisms of pestiviral superinfection exclusion at entry and RNA replication. J Virol. 2005;79:3231–3242. doi: 10.1128/JVI.79.6.3231-3242.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linthicum KJ. Platt K. Myint KS. Lerdthusnee K, et al. Dengue 3 virus distribution in the mosquito Aedes aegypti: an immunocytochemical study. Med Vet Entomol. 1996;10:87–92. doi: 10.1111/j.1365-2915.1996.tb00086.x. [DOI] [PubMed] [Google Scholar]
- Lohmann V. Hoffmann S. Herian U. Penin F, et al. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J Virol. 2003;77:3007–3019. doi: 10.1128/JVI.77.5.3007-3019.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorono-Pino MA. Cropp CB. Farfan JA. Vorndam AV, et al. Common occurrence of concurrent infections by multiple dengue virus serotypes. Am J Trop Med Hyg. 1999;61:725–730. doi: 10.4269/ajtmh.1999.61.725. [DOI] [PubMed] [Google Scholar]
- Mackenzie JS. Gubler DJ. Petersen LR. Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat Med. 2004;10:S98–S109. doi: 10.1038/nm1144. [DOI] [PubMed] [Google Scholar]
- Pepin KM. Lambeth KL. Hanley KA. Asymmetric competitive suppression between strains of dengue viruses. BMC Microbiol. 2008;8:28. doi: 10.1186/1471-2180-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales C. Mateo R. Mateu MG. Domingo E. Insights into RNA virus mutant spectrum and lethal mutagenesis events: replicative interference and complementation by multiple point mutants. J Mol Biol. 2007;369:985–1000. doi: 10.1016/j.jmb.2007.03.074. [DOI] [PubMed] [Google Scholar]
- Rico-Hesse R. Microevolution and virulence of dengue viruses. Adv Virus Res. 2003;59:315–341. doi: 10.1016/s0065-3527(03)59009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie SA. Long S. Smith G. Pyke A, et al. Entomological investigations in a focus of dengue transmission in Cairns, Queensland, Australia, by using the sticky ovitraps. J Med Entomol. 2004;41:1–4. doi: 10.1603/0022-2585-41.1.1. [DOI] [PubMed] [Google Scholar]
- Romero-Vivas CM. Leake CJ. Falconar AK. Determination of dengue virus serotypes in individual Aedes aegypti mosquitoes in Colombia. Med Vet Entomol. 1998;12:284–288. doi: 10.1046/j.1365-2915.1998.00117.x. [DOI] [PubMed] [Google Scholar]
- Rudnick A. Studies of the ecology of dengue in Malaysia: a preliminary report. J Med Entomol. 1965;2:203–208. doi: 10.1093/jmedent/2.2.203. [DOI] [PubMed] [Google Scholar]
- Rudnick A. Ecology of dengue virus. Asian J Infect Dis. 1978;2:156–160. [Google Scholar]
- Rudnick A. Dengue virus ecology in Malaysia. Inst Med Res Malyas Bull. 1986;23:51–152. [Google Scholar]
- Rudnick A. Tan EE. Lucas JK. Omar MB. Mosquito-borne haemorrhagic fever in Malaya. Br Med J. 1965;1:1269–1272. doi: 10.1136/bmj.1.5445.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar MI. Richardson JH. Sanchez-Vargas I. Olson KE, et al. Dengue virus type 2: replication and tropisms in orally infected Aedes aegypti mosquitoes. BMC Microbiol. 2007;7:9. doi: 10.1186/1471-2180-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saluzzo JF. Cornet M. Adam C. Eyraud M, et al. [Dengue 2 in eastern Senegal: serologic survey in simian and human populations. 1974–85] Bull Soc Pathol Exot Filiales. 1986;79:313–322. [PubMed] [Google Scholar]
- Scott TW. Amerasinghe PH. Morrison AC. Lorenz LH, et al. Longitudinal studies of Aedes aegypti (Diptera: Culicidae) in Thailand and Puerto Rico: blood feeding frequency. J Med Entomol. 2000;37:89–101. doi: 10.1603/0022-2585-37.1.89. [DOI] [PubMed] [Google Scholar]
- Shinjoh M. Omoe K. Saito N. Matsuo N, et al. In vitro growth profiles of respiratory syncytial virus in the presence of influenza virus. Acta Virol. 2000;44:91–97. [PubMed] [Google Scholar]
- Shurtleff AC. Beasley DW. Chen JJ. Ni H, et al. Genetic variation in the 3′ non-coding region of dengue viruses. Virology. 2001;281:75–87. doi: 10.1006/viro.2000.0748. [DOI] [PubMed] [Google Scholar]
- Simon KO. Cardamone JJ., Jr Whitaker-Dowling PA. Youngner JS, et al. Cellular mechanisms in the superinfection exclusion of vesicular stomatitis virus. Virology. 1990;177:375–379. doi: 10.1016/0042-6822(90)90494-c. [DOI] [PubMed] [Google Scholar]
- Singh IR. Suomalainen M. Varadarajan S. Garoff H, et al. Multiple mechanisms for the inhibition of entry and uncoating of superinfecting Semliki Forest virus. Virology. 1997;231:59–71. doi: 10.1006/viro.1997.8492. [DOI] [PubMed] [Google Scholar]
- Singh KRP. Cell cultures derived from larvae of Aedes albopictus (Skuse) and Aedes aegypti (L.) Curr Sci. 1967;36:506–508. [Google Scholar]
- Smith DR. Adams AP. Kenney JL. Wang E, et al. Venezuelan equine encephalitis virus in the mosquito vector Aedes taeniorhynchus: infection initiated by a small number of susceptible epithelial cells and a population bottleneck. Virology. 2007;372:176–186. doi: 10.1016/j.virol.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srisuphanunt M. Sithiprasasna R. Patpoparn S. Attatippaholkun W, et al. ELISA as an alternative tool for epidemiological surveillance for dengue in mosquitoes: a report from Thailand. J Vector Borne Dis. 2007;44:272–276. [PubMed] [Google Scholar]
- Steck FT. Rubin H. The mechanism of interference between an avian leukosis virus and Rous sarcoma virus. I. Establishment of interference. Virology. 1966;29:628–641. doi: 10.1016/0042-6822(66)90287-x. [DOI] [PubMed] [Google Scholar]
- Sundin DR. Beaty BJ. Interference to oral superinfection of Aedes triseriatus infected with La Crosse virus. Am J Trop Med Hyg. 1988;38:428–432. doi: 10.4269/ajtmh.1988.38.428. [DOI] [PubMed] [Google Scholar]
- Thavara U. Siriyasatien P. Tawatsin A. Asavadachanukorn P, et al. Double infection of heteroserotypes of dengue viruses in field populations of Aedes aegypti and Aedes albopictus (Diptera: Culicidae) and serological features of dengue viruses found in patients in southern Thailand. Southeast Asian J Trop Med Public Health. 2006;37:468–476. [PubMed] [Google Scholar]
- Troyer JM. Hanley KA. Whitehead SS. Strickman D, et al. A live attenuated recombinant dengue-4 virus vaccine candidate with restricted capacity for dissemination in mosquitoes and lack of transmission from vaccinees to mosquitoes. Am J Trop Med Hyg. 2001;65:414–419. doi: 10.4269/ajtmh.2001.65.414. [DOI] [PubMed] [Google Scholar]
- Tscherne DM. Evans MJ. von Hahn T. Jones CT, et al. Super-infection exclusion in cells infected with hepatitis C virus. J Virol. 2007;81:3693–3703. doi: 10.1128/JVI.01748-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsetsarkin KA. Vanlandingham DL. McGee CE. Higgs S. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog. 2007;3:e201. doi: 10.1371/journal.ppat.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urdaneta L. Herrera F. Pernalete M. Zoghbi N, et al. Detection of dengue viruses in field-caught Aedes aegypti (Diptera: Culicidae) in Maracay, Aragua State, Venezuela by type-specific polymerase chain reaction. Infect Genet Evol. 2005;5:177–184. doi: 10.1016/j.meegid.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Vasilakis N. Holmes EC. Fokam EB. Faye O, et al. Evolutionary processes among sylvatic dengue type 2 viruses. J Virol. 2007a;81:9591–9595. doi: 10.1128/JVI.02776-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilakis N. Shell EJ. Fokam EB. Mason PW, et al. Potential of ancestral sylvatic dengue-2 viruses to re-emerge. Virology. 2007b;358:402–412. doi: 10.1016/j.virol.2006.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E. Ni H. Xu R. Barrett AD, et al. Evolutionary relationships of endemic/epidemic and sylvatic dengue viruses. J Virol. 2000;74:3227–3234. doi: 10.1128/jvi.74.7.3227-3234.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WK. Chao DY. Lin SR. King CC, et al. Concurrent infections by two dengue virus serotypes among dengue patients in Taiwan. J Microbiol Immunol Infect. 2003;36:89–95. [PubMed] [Google Scholar]
- Wargo AR. Huijben S. de Roode JC. Shepherd J, et al. Competitive release and facilitation of drug-resistant parasites after therapeutic chemotherapy in a rodent malaria model. Proc Natl Acad Sci USA. 2007;104:19914–19919. doi: 10.1073/pnas.0707766104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westaway EG. Flavivirus replication strategy. Adv Virus Res. 1987;33:45–90. doi: 10.1016/s0065-3527(08)60316-4. [DOI] [PubMed] [Google Scholar]
- Yuwono J. Suharyono W. Koiman I. Tsuchiya Y, et al. Seroepidemiological survey on dengue and Japanese encephalitis virus infections in Asian monkeys. Southeast Asian J Trop Med Public Health. 1984;15:194–200. [PubMed] [Google Scholar]