Abstract
LIN-35 is the single C. elegans ortholog of the mammalian pocket protein family members, pRb, p107, and p130. To gain insight into the roles of pocket proteins during development, a microarray analysis was performed with lin-35 mutants. Stage-specific regulation patterns were revealed, indicating that LIN-35 plays diverse roles at distinct developmental stages. LIN-35 was found to repress the expression of many genes involved in cell proliferation in larvae, an activity that is carried out in conjunction with E2F. In addition, LIN-35 was found to regulate neuronal genes during embryogenesis and targets of the intestinal-specific GATA transcription factor, ELT-2, at multiple developmental stages. Additional findings suggest that LIN-35 functions in cell cycle regulation in embryos in a manner that is independent of E2F. A comparison of LIN-35-regulated genes with known fly and mammalian pocket-protein targets revealed a high degree of overlap, indicating strong conservation of pocket protein functions in diverse phyla. Based on microarray results and our refinement of the C. elegans E2F consensus sequence, we were able to generate a comprehensive list of putative E2F-regulated genes in C. elegans. These results implicate a large number of genes previously unconnected to cell cycle control as having potential roles in this process.
Keywords: lin-35, retinoblastoma, pRb, p107, p130, pocket proteins, C. elegans, development, microarray
INTRODUCTION
The mammalian retinoblastoma protein (pRb) was amongst the first tumor suppressor proteins to be identified (Dryja et al., 1986; Friend et al., 1986; Lee et al., 1987), and loss or inactivation of pRb is thought to play a causative role in the majority of human cancers (Bindra and Glazer, 2006; Knudsen and Knudsen, 2006; Sherr, 2004; Sherr and McCormick, 2002; Yamasaki, 2003). Many studies on pRb, p107, and p130 have demonstrated their importance in the regulation of cell cycle progression, largely by binding E2F transcription factor family members using a cleft or “pocket” domain. This leads to the repression of E2F target genes and the consequent suppression of S-phase entry by inhibiting E2Fs promoting target gene expression (E2F1–3) or by augmenting E2Fs acting as transcriptional repressors (E2F4–6) (Dannenberg et al., 2000; Du and Dyson, 1999; Reed, 1997; Sage et al., 2000).
Although the interaction of pocket proteins with E2F family members is the most thoroughly characterized function of pRb-family members, they are far from the only transcription factors known to bind pocket proteins (Lipinski and Jacks, 1999; Morris and Dyson, 2001). For example, pRb has been reported to interact with C/EBPε, to promote the terminal differentiation of granulocytes and adipocytes (Chen et al., 1996; Gery et al., 2004), with GATA-1 to induce erythrocyte differentiation (Rekhtman et al., 2003), and with MyoD, to promote myocyte differentiation (Gu et al., 1993). pRb also antagonizes the cell-differentation inhibitor Id2, allowing for the terminal differentiation of several cell types (Iavarone et al., 1994; Lasorella et al., 2000). In addition, both pRb and p107 have been shown to act through the basic helix-loop-helix transcription factor, NeuroD1, which is critical for early development of the neural system in Xenopus (Batsche et al., 2005).
Notably, the majority of reports to date describe activities that have been observed in tissue culture systems and significantly less is known about pocket protein functions in the context of intact developing organisms. This lack of supporting in vivo data is in part due to the lethality of many pRb family knockouts in mice and flies (Clarke et al., 1992; Jacks et al., 1992; Lee et al., 1992; Dimova et al., 2003; Du and Dyson, 1999; Wikenheiser-Brokamp, 2006). C. elegans has only a single pocket protein family member, lin-35, which is not essential for viability. In addition, C. elegans has only one DP homolog and three E2F homologs, further simplifying the analysis of the role of pocket proteins in this organism. Studies have demonstrated that lin-35 also functions in G1–S-phase regulation during development (Boxem and van den Heuvel, 2001; Fay, 2005; Fay et al., 2002), and plays a number of diverse roles outside of cell cycle control. For example, lin-35, has been shown to repress vulval developmental fates (Ceol and Horvitz, 2001; Lu and Horvitz, 1998), and to play roles in pharynx and vulva morphogenesis (Bender et al., 2006; Fay et al., 2003; Fay et al., 2004), asymmetric cell division (Cui et al., 2004), somatic gonad cell fate (Bender et al., 2004), larval growth (Cardoso et al., 2005; Chesney et al., 2006; Cui et al., 2004), germline gene repression (Wang et al., 2005), and even RNA interference (Lehner et al., 2006; Wang et al., 2005).
In this study, a microarray analysis was performed using lin-35 null mutants to determine the role of this protein at different developmental stages in the organism and to identify which proteins and pathways it may be acting through. A number of differentially-regulated genes identified in this study were homologous to mammalian genes also regulated by pocket proteins, suggesting a strong conservation of function of pRb-family members between nematodes and mammals. We have also refined the nematode E2F consensus binding site and have generated a compelling list of putative E2F targets in this organism. This, along with the discovery of several novel motifs that could represent binding sites for additional LIN-35 binding partners, may substantially expand the network of pRb-family regulated genes and help to elucidate the routes by which they function.
MATERIALS AND METHODS
Strains and maintenance
All C. elegans strains were maintained according to established protocols (Stiernagle). All experiments were carried out at 20°C. Strains used in these studies included the following: N2 (wild type) and of lin- 35(n745) (Lu and Horvitz, 1998), which had been additionally backcrossed by our laboratory 5X.
Staging of organisms
Wild-type and mutant worms were staged by bleaching gravid adults to collect eggs, which were hatched onto NGM plates in the absence of food. After hatching, worms were transferred to NGM plates with bacteria (OP50). RNA harvested for embryonic stage worms was obtained from a total preparation of embryos. As a result of extracting embryos from young adult stage worms, predominantly early stage embryos were obtained. The differences in embryos between strains were similar to the differences between replicates in the same strain. For RNA from L1 stage larvae, collection occurred ~1 hour prior to the apoptosis of T.ppp, which occurred ~10 or 11 hours after hatching in N2 and lin-35 mutants, respectively (Sulston and Horvitz, 1977). For L4 stage larvae, a distinct Christmas tree vulval morphology was used as a biomarker.
RNA extraction and microarrays
Embryos or larvae were washed off of NGM plates using M9. RNA was extracted using phenol followed by phenol:chloroform:isoamyl alcohol. RNA was precipitated overnight at 4° C with 4M lithium acetate and ethanol. RNA was DNase treated, purified on RNEasy minicolumns (QIAGEN), and verified to be free of genomic DNA contamination by quantitative reverse transcription-mediated real time PCR (qRT-PCR). cDNA synthesis, cRNA synthesis, labeling, fragmentation, GeneChip hybridization, and scanning were performed according to specifications from Affymetrix. Triplicate chips were run for each strain at each stage. RNA and cDNA qualities were verified at each step using capillary electrophoresis (Bioanalyzer; Agilent Technologies). Gene expression values were determined using Robust Multiarray Analysis software (Irizarry et al., 2003). Relevant expression differences were identified using a modified Wilcoxon-Rank test (for details see Results).
qRT-PCR analysis
cDNA synthesis reactions were performed with SuperScriptII (Invitrogen), and reactions contained 5 mg RNA were purified as above. Approximately 20 ng of cDNA was used for each qRT-PCR reaction. qRT-PCR reactions were performed in an iCycler iQ (Bio-Rad), using a SYBR Green fluorophore. Control reactions were performed using actin-specific primers, and threshold cycles for all reactions were normalized using a D/DCt method.
Regulatory motif analysis
To identify candidate regulatory sequences in the 5′ noncoding regions of putative C. elegans homologs of p107/p130-target genes, 1 kilobase of sequence data upstream of the start codon of each gene was retrieved from C. elegans genomic information using Regulatory Sequence Analysis Tools software (http://embnet.ccg.unam.mx/rsa-tools/). Regions of DNA with low sequence complexity were masked using RepeatMasker (http://repeatmasker.org), and then searched for overrepresented motifs using the program MEME (Multiple Em for Motif Elicitation) (http://meme.sdsc.edu/meme/intro.html). This analysis was performed locally, using background models of the fourth order. Motif sequence logos were created using an online program (http://weblogo.berkeley.edu/logo.cgi).
To verify the relevance of motifs returned by the software, a PWM was generated for each motif by MEME. For the E2F consensus site, this matrix was applied to all 1949 responsive genes, and RSAT software was used to examine the relative representation of the motif in genes within the list of responsive genes as compared with the entire genome. This analysis was performed with two different types of cutoff ranges, using scores obtained by applying the PWM to the relevant genes. First, successive ranges of cutoff scores were used. Score ranges were obtained using cutoff scores between the highest score obtained and sequentially lower scores (yielding sequentially larger ranges with lower cutoffs). Second, interval score cutoffs were examined. These were obtained using PWM score intervals (with a range of two between selected points).
For the other reported motifs, sequences were derived and PWM were generated as above. Motifs were reported if they exhibited significant (p < 0.01) overrepresentation in genes that were in the cluster (see below) within which the motif was identified compared to the remaining non-E2F genes, overrepresentation within the non-E2F regulated genes as compared with E2F regulated genes, and overrepresentation within the non-E2F responsive genes as compared with the entire genome. The cutoff scores for these calculations were set by the lowest score obtained by applying the PWM to the genes that the motif was identified within.
Clustering
Gene expression levels were normalized to gene-specific relative maxima across both strains and all stages (six points per gene). Genes were then divided into ten clusters using profiles that were specific for strains and stages. Clustering was performed using MultiExperiment Viewer software (http://www.tm4.org/mev.html), with a number of different clustering algorithms tested, including Hierarchical Clustering, K-Means/K-Medians Support, Self-Organizing Maps, K-Means/K-Medians Clustering, and Quantitative Threshold Clustering. K-Means/K-Medians clustering yielded groupings of genes that were the closest to the average expression line for each cluster, so this method was used to further divide the differentially expressed genes in each cluster into subclusters according to their fold-change profile. This division was based on relative expression level of the genes in the cluster.
RESULTS
Time points and primary data analysis
To identify genes that are responsive to LIN-35 during development, RNA was harvested from synchronized lin-35(n745) mutant and wild type (N2) worms at embryonic, L1, and L4 stages. Our specific time points were chosen in order to provide information about LIN-35 activities at several distinct developmental stages and because previous studies have implicated biological roles for LIN-35 during these stages (also see Introduction). To ensure the precise developmental synchronization of animal populations for these studies, we used several stage-specific developmental markers as opposed to chronological timing, which can be variable between strains and individual replicates (see Materials and methods). RNA was purified from each stage and used to generate cRNA, which was fragmented and hybridized to C. elegans Affymetrix GeneChips®. Three independent biological replicates were performed for each stage and expression values for each gene were generated using Robust Multiarray Analysis software (Irizarry et al., 2003).
To determine relevant changes in gene expression, a modified Wilcoxon-Rank test was performed using a minimal quotient of >1.15, which has been shown to efficiently eliminate false positives and to yield results with high biological relevance (Braatsch et al., 2004; Harpster et al., 2006; Moskvin et al., 2005; Pappas et al., 2004; Zeller et al., 2005). In addition, a minimal mean- and median-fold change limit for individual genes was set at 1.5, which was stringent enough to return an analytically manageable number of genes while still retaining sufficient sensitivity. Using these criteria, a false discovery rate of <1.5% was calculated, suggesting that these applied conditions were highly effective in identifying statistically significant, differentially-regulated genes (data not shown).
Overview of stage-specific transcriptional responses
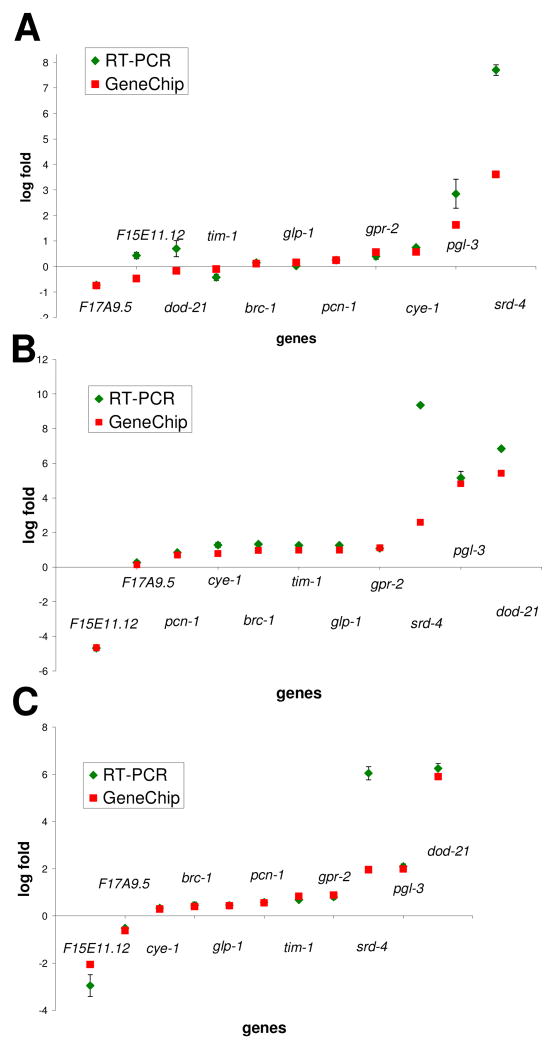
In this array study, a total of 1,949 genes showed altered expression in the lin-35 mutant background as compared with wild type. There were 642 differentially regulated genes at embryonic stages, 945 at L1, and 1108 at the L4 stage; a full list of these genes can be located in Fig. S1. Importantly, a number of genes previously demonstrated to be regulated by pRb–E2F complexes in multiple species, including human, mouse, and Drosophila, were amongst the targets identified (e.g. cyclin A, cyclin E, thymidylate synthase, BRCA, mcm-2, mcm-3, mcm-4, and mcm-5). Moreover, consistent with the notion that LIN-35/pRB functions primarily as a transcriptional co-repressor, the large majority of identified responsive genes were upregulated in the lin-35 null background; 81% in embryos, 70% at L1, and 64% at L4. To further validate the array findings, eleven genes with different developmental expression patterns and different biological functions were selected as targets for RT-PCR (Fig. 1). Although the microarray and RT-PCR data are not identical in all cases, they correlate well, strengthening the legitimacy of the array results.
Fig. 1. Comparison of RT-PCR and GeneChip Data.
RNA from (A) embryonic, (B) L1, and (C) L4-stage worms was used for RT-PCR. At least 6 reactions were performed for each gene. Error bars represent standard error (SE) for all replicates. Genes with well-characterized differential regulation showed appropriate expression levels in both GeneChip® and RT-PCR data (e.g. cye-1, pcn-1, pgl-3). Discrepancies in expression levels (e.g. srd-4) are likely due to a single primer set used for RT-PCR and averaging of multiple primer sets for microarray data.
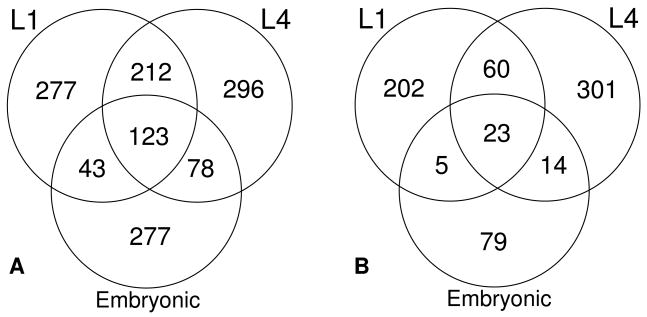
Of the total 1,306 upregulated, responsive genes, only 123 (9%) were found to be differentially regulated at all three developmental stages (Fig. 2A). This relative lack of overlap is not simply due to a paucity of differentially-regulated genes at any given stage, but more likely reflects distinct biological roles for LIN-35 at specific stages of development. In binary comparisons, L1 and L4 stages showed the greatest overlap with 33% (335/1029), despite the greater temporal separation of these stages as compared with L1 larvae and embryos. Furthermore, the overlapped genes showed a strong correlation in their patterns of differential expression (Fig. S2). In contrast, L1 and L4 stages showed only a 16% (166/1010) and 20% (201/1029) overlap with embryos respectively (Fig. 2A). Furthermore, the behavior of genes in the embryonic population did not strongly correlate with the expression patterns of these genes in larvae; a large number of genes that were upregulated in embryos were downregulated in larval stages (Fig S2). These data suggest substantive differences in the nature of LIN-35 function at different developmental stages, particularly in larvae and embryos.
Fig. 2. lin-35 Responsive Genes at Different Stages.
Venn diagrams representing overlap between upregulated (A) and downregulated (B) genes responding to the lin-35 mutation at L1, L4 and embryonic stages.
Considerably fewer genes were observed to be downregulated in lin-35 mutants: 19% in embryos, 30% in L1, and 36% in L4. The increase in the number of genes specifically downregulated across developmental stages, from 79 in embryos to 301 in L4 stage worms, argues that some of this downregulation may be a result of non-specific causes. Furthermore, the degree of overlap observed in downregulated genes compared to upregulated genes (Fig. 2B) also suggests that much of the observed downregulation may be due to secondary effects.
With these analyses in hand, the responsive genes were divided into functional categories based on gene ontologies (GOs) and literature searches. Consistent with the general lack of overlap between certain developmental stages (Fig. 2), stage-specific profiles emerged. Both L1 and L4 stages showed a statistically significant enrichment for responsive genes characterized as having “extended cell cycle” functions, including genes involved in cell cycle control, DNA replication and metabolism, mitosis, and meiosis (Fig. S3). Nearly all (93%) of the differentially-regulated genes belonging to this extended cell cycle category were upregulated. The strong bias of enrichment and regulation of cell cycle-related genes indicates that LIN-35 plays an important role in controlling these processes during larval development.
In embryos, a significantly larger number of genes associated with neurogenesis and neurotransmission were present as compared with larval stages (Fig. S3). This finding suggests a previously unknown role for LIN-35 in the nervous system during embryonic development. In stark contrast with larval stages, genes involved in cell cycle, DNA metabolism, and cell maintenance were not statistically overrepresented at this stage (Fig. S3). These latter results indicate that LIN-35 may not play a major role in cell cycle regulation during embryogenesis.
Categorical analysis was further corroborated by evaluating stage-specific responses within the context of gene mountains (Kim et al., 2001). Mountain analysis first groups co-regulated genes, using a meta-array approach, and then attempts to ascribe functions to the mounts based on representative genes with established functions or known expression patterns. L1- and L4-stage mountain distributions showed expression patterns consistent with enrichment of meiosis and mitosis, and retinoblastoma complex members (Fig. S4B, C). Mountain analysis of embryos showed a dramatic overrepresentation of responsive genes in a mount associated with neuronal genes (Fig. S4A). These results corroborate those from those of the GO analysis and strengthen the hypothesis that LIN-35 may play an unrecognized role in the nervous system of C. elegans embryos.
Two mountains (7 and 11) described to possess germline genes were also overrepresented. Genes in both mountains were strongly enriched in E2F sites (Fig. S4), consistent with the involvement of LIN-35 in the direct regulation of these genes (also see below) and with previous reports demonstrating LIN-35 repression of germline-associated genes in somatic tissues (Wang et al., 2005).
Comparison of pocket protein targets from different species
Genes previously demonstrated to be responsive to mammalian pRb (Finocchiaro et al., 2005) or to p107/p130 (Balciunaite et al., 2005) were used to identify best genome sequence matches from C. elegans. Genes were considered to be putative orthologs if a reciprocal search returned the original pocket-protein target in H. sapiens. Based on this approach, >50% of the mammalian pocket-protein responsive genes were found to contain likely C. elegans orthologs. Cross-referencing these genes with the list of LIN-35-responsive genes revealed that ~30% of the putative mammalian orthologs were also targets for regulation by LIN-35, a percentage that is highly statistically significant (Fig. S5B). Furthermore, similar levels of overlap were observed for both pRb- and p107/p130-target genes with the identified LIN-35-responsive genes. This indicates that LIN-35 may carry out ancestral-type functions that are common to all three mammalian family members. This finding is also consistent with the placement of LIN-35 equally distant from all three mammalian pocket proteins and both Drosophila pocket proteins and as being located in an ancestral position to all five (Fig. S5A).
The majority of overlap between LIN-35 and the mammalian pocket proteins came from targets that were differentially regulated at either the L1 stage or at both the L1 and L4 stages. Polymerase subunits, replication factors, cyclins, and histones were among the overlapping genes that are targets of the human and C. elegans pocket proteins (see Fig. S6 for a full list). As has previously been noted, many of these targets are known to act in cell cycle control and DNA replication, further demonstrating a conservation of function between LIN-35 and the mammalian pocket proteins.
In contrast, only nine of the 94 genes overlapping between LIN-35 and the mammalian pocket proteins were differentially regulated in embryos. This is consistent with previous data, such as the lack of conserved targets between embryonic and larval stages, further indicating distinct functions for LIN-35 in embryos that are unconnected to the cell cycle. Similar to embryos, LIN-35-responsive genes in the germline (Chi and Reinke, 2006), also failed to show high levels of overlap with the mammalian targets, indicating that LIN-35 plays a unique role in this tissue as well (Fig. S5). In both that study and in this one, responsive genes in embryos and the germline appeared to play a more prominent role in differentiation than in the extended cell cycle functions. We also note that the overlap of the mammalian pocket-protein targets with genes responsive to the Drosophila pocket proteins, though statistically significant, was much weaker than that observed for LIN-35. This may reflect more divergent functions for the Drosophila proteins or may be a reflection of the different experimental approaches used in these studies.
Search for evolutionarily-conserved Rb family binding partners
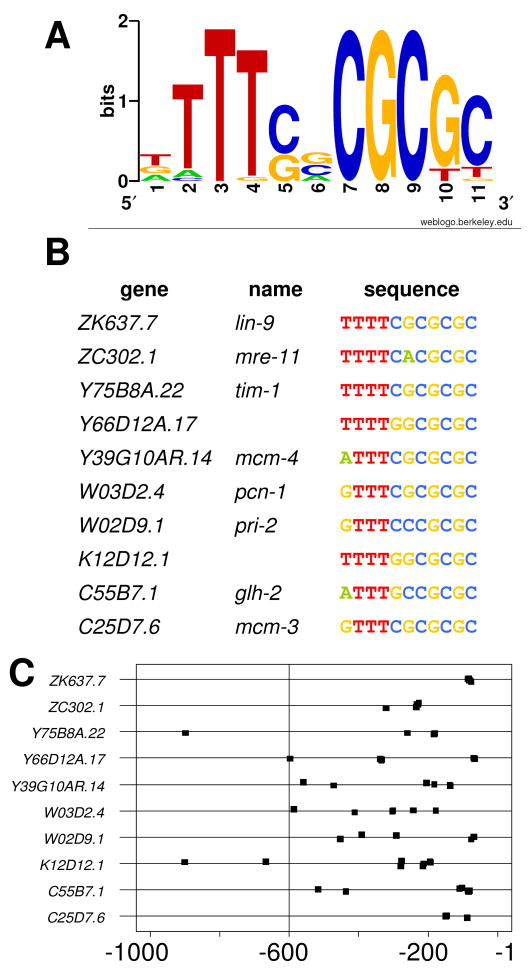
In order to find conserved Rb-family binding partners, the LIN-35-responsive orthologs of the mammalian p107/p130-responsive genes were searched for overrepresented motifs. Genes overlapping with p107/p130 targets were chosen instead of pRb-responsive genes because this data was derived from ChIP experiments rather than from a meta-analysis of multiple microarray experiments, thereby increasing the probability that the derived targets would be directly regulated by LIN-35. One kilobase of upstream sequence for each gene was retrieved from a C. elegans genomic database, regions of low sequence complexity were masked, and an unbiased search for overrepresented motifs was performed using MEME (see Materials and methods). This analysis returned a single motif (TTTSSCGCGC) that was enriched in both the genes that overlapped with mammals and in the entire list of lin-35-responsive genes (Fig. 3A). This sequence bore a strong similarity to the consensus E2F binding site (TTTSGCGC) from mammals and also to a recently described consensus binding site for E2F in C. elegans (TTCSCGCS) (Chi and Reinke, 2006). Given that this was the only motif enriched among this group of genes, E2F may be the only pocket protein binding partner in these organisms that is strongly evolutionarily conserved. While other conserved Rb-interacting proteins may exist and may play roles in limited temporal and/or spatial circumstances, their absence from this analysis suggests that they are not involved in organism-wide gene regulation.
Fig. 3. Refinement of the C. elegans E2F-Binding Site Motif.
The C. elegans E2F-binding site consensus motif (A) was refined using an unbiased set of conserved genes (see text for details). Example genes (B) show clustering of this motif toward their transcriptional start sites (C).
E2F-regulated genes in C. elegans
A position-weight matrix (PWM) was derived for the putative C. elegans E2F consensus binding site (described above; see Materials and Methods). A PWM determines a weighted probability of particular nucleotides occurring at individual motif positions based on the degree of conservation. This PWM was used to search the promoter regions of the 1949 LIN-35-responsive genes. Scores were generated for each gene containing a putative E2F motif, with higher scores indicating a better match. Several genes known to function in DNA replication and mitosis (including mcm-3 and -4, pcn-1, and pri-2) obtained high scores using this matrix.
The motif was found to be significantly (p<0.01) overrepresented in our list of LIN-35-responsive genes at all three developmental stages (see Materials and Methods for criteria), suggesting that this motif has biological significance. Furthermore, a strong correlation was observed between the PWM score and the probability that a given gene would be upregulated in the lin-35 null mutant background. For example, amongst the 50 best-scoring LIN-35-responsive genes from the L1 stage, all genes were observed to be upregulated in lin-35 mutants (p≪0.01) compared to 70% of genes among all L1-stage responsive genes. In contrast, the presence of a high-scoring E2F motif was not found to be a reliable predictor of gene behavior in embryos, further indicating that the binding factor(s) acting through this motif are functioning differently at this stage.
To generate a list of putative E2F-target genes from our dataset, several criteria were applied. First, candidate genes had to have a minimum score of 4 (based on several selective criteria, data not shown) from PWM application. Second, the nucleotides at positions 3, 5, 7, 8, and 9 had to conform to the refined motif (Fig. 3A). Third, the gene had to have at least one consensus E2F site within 600 base pairs of the transcriptional start site, as has been previously shown to return biologically relevant data (Chi and Reinke, 2006). Of the 1949 responsive genes on the list, 452 met all three criteria. The presence of many known E2F targets (including cyclin A, cyclin E, thymidylate synthase, and mcm genes), demonstrates the validity of this algorithm (see Fig. S7 for a complete list).
The 452 genes identified shared several important characteristics. First, they demonstrated enrichment in cell cycle and DNA maintenance functions. Genes with this consensus site also often contained multiple copies (Fig. 3B) and these motifs were generally biased toward transcriptional start sites (Fig. 3C). Finally, an examination of homologs in C. briggsae (a species closely related to C. elegans), revealed that 75% of these genes contained at least one E2F binding site in their proximal promoter regions, suggesting that this regulatory pathway is well conserved.
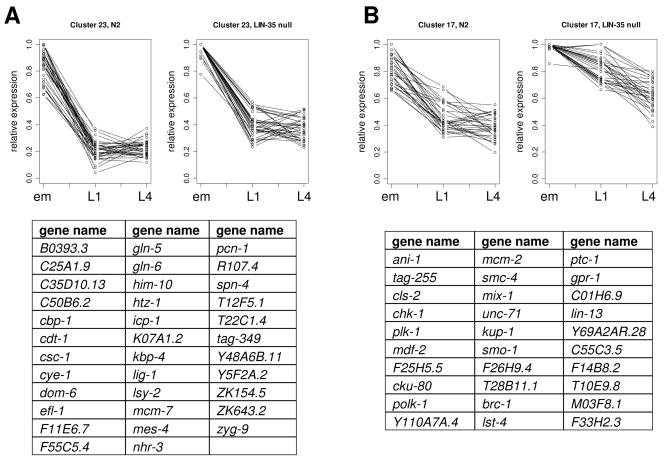
As a means for further analysis, the putative E2F target genes were divided into 25 clusters based on their expression profiles using a K-means/K-medians clustering (KMC) algorithm (see Materials and Methods) (Fig. S8). Of these, 197 genes fell into nine clusters strongly enriched for cell cycle genes (p ≪ 0.01). The gross expression patterns exhibited by these genes in wild-type animals showed a striking similarity between the clusters (Fig. 4), which was attenuated in null mutants. This pattern suggests that LIN-35 is used to regulate these genes upon entry into larval stages of development. Decreased expression of these genes in lin-35 mutants suggests that other transcriptional regulators could be acting in parallel to lin-35 for this process.
Fig. 4. Expression Profiles for Genes with E2F Binding Sites.
Two representative clusters (A and B) of responsive genes with E2F binding sites that are enriched in genes with cell cycle functions are shown. Graphs on the left of each panel represent relative expression levels for each gene in N2, graphs on the right represent relative expression levels for the same genes in lin-35 mutants. Also shown are lists of the genes contained in the clusters. Panel (A) represents the cluster with the best average score when a PWM is applied, panel (B) represents a cluster with a lower average score. Lines are intended to facilitate visualization of cluster patterns and are not intended to represent proximal chronological steps.
It is noteworthy that many genes not previously annotated to play a role in cell cycle and replication functions were also located in these nine clusters. For example, gpr-2, a protein regulating chromosomal and spindle movements, possesses a number of high-scoring E2F sites and was upregulated at all three observed stages. TIM-1 (timeless), another protein upregulated at both larval stages, participates in the regulation of developmental timing in other systems (Gotter, 2006; Hardin, 2005; Myers et al., 1995). A number of transcription factors, most of which have not been previously connected to cell cycle control (e.g., cbp-1, mdl-1, lin-59, C18G1.2), were also found. The existence of nearly two hundred proteins in these clusters potentially expands the network of Rb/E2F cell cycle functions quite considerably.
Identification of four potential non-E2F LIN-35 regulatory sites
The remaining 1,497 responsive genes that did not contain consensus E2F binding sites were divided into clusters and subclusters using a KMC algorithm (see Materials and Methods). Upstream sequence data from each gene in these subclusters was retrieved, masked, and searched for overrepresented motifs (see Materials and Methods). In order to identify biologically relevant motifs amongst those returned by the software, a number of significance thresholds (p < 0.01) were applied (also see Materials and Methods). Among these, the motif had to be significantly overrepresented in the cluster compared to the list of non-E2F-responsive genes and also had to be overrepresented in the non-E2F responsive genes compared to the 452 E2F-responsive genes and to the entire C. elegans genome.
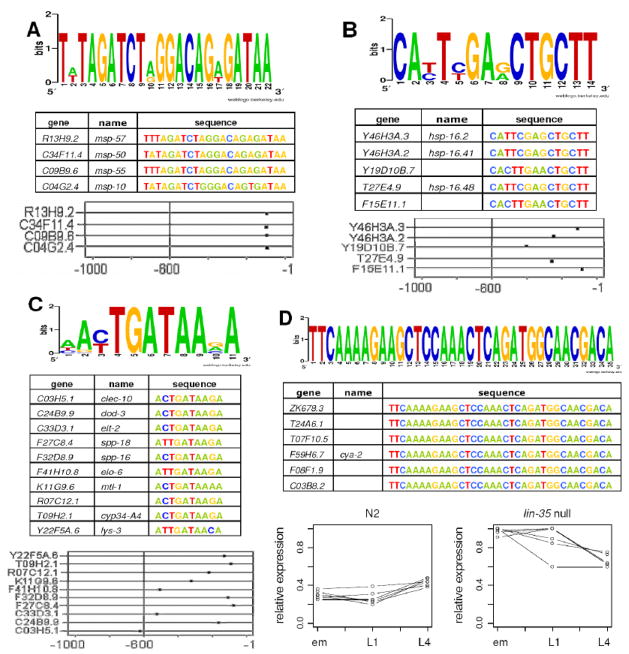
This algorithm was used to identify two very precise motifs present exclusively in responsive genes that were downregulated at the L1 stage. The first motif (TWTAGATCTRGGACAGWGATAA, Fig. 5A) was found in four genes encoding major sperm proteins (MSP). This motif was long, very highly conserved, and was found in less than 10 genes in the entire genome. This motif was also located at nearly the same position with respect to the transcription initiation site in all of these genes, a relatively uncommon feature amongst promoter binding sites. The second motif was preferentially located in heat shock proteins (Fig. 5B). This motif (CAYTYGARCTGCTT) was located in twelve genes in the genome, and was found in five genes in the subcluster from which it was identified. These five genes were the only responsive genes containing this motif. This motif was also highly conserved and seemed to show a bias toward the -1 promoter position. The identification of both a significant proportion of the genes in the genome as well as the clustering together of all responsive genes containing these motifs demonstrates the validity of using this sequential clustering technique.
Fig. 5. Identification of Non-E2F Binding Elements in lin-35-Responsive Genes.
(A) A motif enriched in four major sperm proteins that were downregulated at the L1 stage. (B) A motif enriched in heat-shock proteins that were downregulated at larval stages. (C) A motif with a GATA core that was identified in 528 genes, examples of which are listed. This motif is frequently located in intestinal genes and is clustered toward transcriptional start sites. (D) A motif that was identified in six cyclinA-like that were strongly upregulated in embryos, but did not show a localization bias. Lines in panel D are intended to facilitate visualization of cluster patterns and are not intended to represent proximal chronological steps.
A more general motif (AYTGATAAVA) with a GATA core was identified in a large number of the responsive genes (Fig. 5C). This motif, independently discovered by an unbiased search of clusters (as described above), nearly perfectly matches a recently refined consensus ELT-2 GATA motif (McGhee et al., 2006). ELT-2 is known to be the major regulator of intestinal development in C. elegans (Fukushige et al., 1999). As transcription factors that bind GATA sites are known to be capable of enhancing transcription from distances much farther than one kilobase, upstream regions up to three kilobases were examined for this site in the complete set of 1,949 responsive genes. A total of 528 genes were identified containing this consensus motif. Searching a serial analysis of gene expression (SAGE) library prepared from intestinal tissue of dissected glp-4 adult C. elegans worms (SWAG1) returned approximately half of these genes in common (McGhee et al., 2006), and 57% of the putative C. briggase orthologs of these genes were found to contain the GATA consensus site. Furthermore, the observed overlap between the two lists is particularly significant since different developmental stages were examined in the different experiments (embryos and L1 and L4 larvae in this study and adults in the SAGE library). These results are also consistent with the observed enrichment of genes located in an intestinal mount at all examined stages (see Fig. S4 ). These findings suggest that LIN-35 may act through ELT-2 to control intestinal-associated gene expression. Finally, this is also consistent with a demonstrated role for LIN-35/Rb in pharyngeal development (Fay et al., 2004), as a statistically significant overlap (p < 0.05) between our list of differentially expressed genes and genes involved in foregut development was observed (Gaudet et al., 2004).
In addition to the motifs already mentioned, a fourth, very specific, motif was located by clustering genes that were responsive at the embryonic stage (Fig. 5D). Although the thirty-five nucleotide motif obtained (TTCAAAAGAAGCTCCAAACTCAGATGGCAACGACA) was completely conserved, it did not show a location bias toward the transcriptional start site (data not shown). Six of the seven genes in the genome containing this site showed a response in lin-35 mutants at the embryonic stage. Five of these genes were annotated as cyclin A-like genes and the sixth, annotated as an uncharacterized protein, returned cyclin A as the best result from a BLAST search. Expression levels of these genes were very dissimilar in N2 and lin-35 null worms, with expression of these genes being significantly higher in the mutants. Additionally, only two of these genes contain consensus E2F binding sites. The absence of consensus binding sites in the remaining genes, combined with their expression patterns in wild-type and mutant worms, strongly suggests that these genes are being regulated by LIN-35, but that this regulation is largely taking place through a non-E2F transcription factor. As these genes are annotated as cyclins, this suggests that LIN-35 may play a role in cell cycle regulation in the embryo independently of E2F.
DISCUSSION
In this study, a total of 1949 genes were found to be responsive to a null mutation in lin-35, the sole pocket protein family member in C. elegans. While it is somewhat surprising that these organisms can function with misregulation of upwards of five hundred genes at each developmental stage, they appear to suffer little more than a developmental lag. Their ability to survive this misregulation is possibly due to compensatory changes in the regulated expression of other genes, alterations in protein activities, and differences in polypeptide clearance. This is in marked contrast with other developmental models, such as mouse and fruit fly, where null alleles of pRb and its fly homolog, dRbF1, are lethal (Clarke et al., 1992; Du and Dyson, 1999; Jacks et al., 1992; Lee et al., 1992).
Despite the lack of an overt phenotype, our analysis demonstrates a remarkable conservation of function between LIN-35/pRb and pocket proteins from other organisms. LIN-35/pRb regulates a large number of genes involved in cell cycle progression, replication, and mitosis, most of which are upregulated in the lin-35 null mutant background (Fig. 2, Fig. S1, Fig. S3), similar to findings in humans and flies (Balciunaite et al., 2005; Black et al., 2003; Dimova et al., 2003; Finocchiaro et al., 2005; Markey et al., 2002; Muller et al., 2001). In fact, the greater overlap observed between the mammalian pocket protein targets and LIN-35 versus Drosophila RbF, attests to the strength of this system for analyzing pRb family functions (Fig. S5).
Our results also strongly indicate that LIN-35/Rb carries out cell cycle functions largely by acting through E2F family transcription factors, consistent with findings in other organisms (Du and Pogoriler, 2006; Korenjak and Brehm, 2005; Stevaux et al., 2005). Interestingly, the observed misregulation of genes with cell cycle functions in lin-35 mutants is much more prevalent during larval stages than in embryos, where expression levels of genes containing E2F binding motifs remain relatively unchanged, suggesting that LIN-35/Rb-E2F complexes are largely superfluous for this role in embryos. In accordance with these observations, CyclinD, which is known to act with cyclin-dependent kinases to inhibit pocket protein activity, is dispensable in worm and mouse embryos (Lukas et al., 1995; Park and Krause, 1999). Most interestingly, our results also provide compelling evidence that LIN-35/Rb may control the expression of certain cell cycle genes, specifically those encoding cyclin A family members, during embryogenesis through a mechanism that is independent of E2F binding (Fig 5D). We also note that similar to embryos, recent microarray studies have show LIN-35/Rb to play only a minor role in cell cycle regulation in the germline (Chi and Reinke, 2006; Wikenheiser-Brokamp, 2006). Collectively, these findings indicate that cell cycle regulation occurs via distinct mechanisms in different tissues of C. elegans and at different times in development.
Since the E2F family of transcription factors is one of the most thoroughly studied groups of pRb binding factors, we undertook an in-depth analysis of the LIN-35/Rb responsive genes in order to further delineate the C. elegans E2F motif and to generate a high-confidence list of over 450 putative targets of the LIN-35/Rb-E2F complex in C. elegans. Of these genes, nearly 200 were clustered groups that were strongly enriched in genes with cell cycle functions. Many of these genes are directly involved in cell cycle control and replication and division processes, such as CDKs, cyclins, and helicases, and regulators of the mitosis. Interestingly, a substantial number of the differentially-regulated genes containing this site have no previous connection to these processes, and their identification may serve as a valuable resource for finding genes with previously unappreciated roles in cell cycle control and progression. It is also important to note that some known pRb/E2F-regulated genes did not make our list E2F targets. For example, ribonucleotide reductase (rnr-1), failed to make the list of responsive genes because its fold-change was only 1.4, which was below our cutoff. Furthermore, bub-1, another known pRb/E2F target that was identified as being responsive to lin-35 failed to make the list because its E2F site did not match sufficiently close to the derived consensus. We contend that the existence of false negatives among our data set was a necessary tradeoff in order to minimize the occurrence of false positives.
In addition to a role in cell cycle regulation, our findings indicate that LIN-35/Rb may play a role in neurological development and neurotransmission. This hypothesized function seems to be important predominantly in embryos, since the proportion of genes annotated with these functions drops precipitously by the L1 stage. While this function was somewhat unexpected, pocket proteins have been previously implicated in the differentiation of neural cells in mammals and amphibians (Batsche et al., 2005; Ferguson and Slack, 2001; Slack and Miller, 1996), suggesting that a role in the nervous system may be conserved.
Our analysis also yielded four additional motifs that may constitute target sites for LIN-35 regulation. One of these motifs was located in the upstream regions of major sperm proteins. Another motif is found in the promoter regions of heat shock proteins, which is consistent with reports of pRb playing a role in stress erythropoiesis in mammals (Spike et al., 2004). As discussed above, a third motif was located in genes differentially regulated in embryos that were annotated to be cyclin-A-related. A fourth motif, containing a GATA core, has recently been identified as the binding site for ELT-2 (McGhee et al., 2006), the major regulator of intestinal development in worms. This finding is unlikely to be the result of LIN-35/pRb–E2F transcriptional regulation of the elt-2, however, as the upstream regulatory region of this gene is devoid of consensus E2F binding sites. In agreement with this finding, an E2F-independent role of pRb in intestinal differentiation in mice has been observed (Haigis et al., 2006). Further, a region of pRb overlapping the E2F binding site (Dick and Dyson, 2003) has been shown to be tethered to another GATA transcription factor, GATA-1, resulting in repression of its activity (Rekhtman et al., 2003). Thus, a role in the regulation GATA targets by pocket proteins may also be conserved.
While we do not currently know whether the identified role of LIN-35/pRb in regulating genes involved in intestinal and neuronal differentiation is direct, future experiments will address this question, as well as verifying the roles of some of the other genes predicted to function in cell cycle regulation. Regardless of whether this regulation is direct, similar findings in mammals and amphibians suggest that this role of LIN-35/pRb is biologically relevant and is highly conserved across widely disparate species. This fact, combined with the recapitulation of the regulation of homologous genes by pocket proteins in both worms and in mammals, demonstrates the utility of C. elegans as a model for determining pocket protein functions with whole organisms in vivo.
Supplementary Material
Colors codes are as follows: yellow – embryonic stage only, light green – L1 stage only, blue – L4 stage only, dark green – L1 and L4 stages, magenta – embryonic and L1 stages, purple – embryonic and L4 stages, red – all stages, pink – genes containing an E2F consensus site. Column headed “mountain” refers to mountains described in Kim et al., 2001.
Expression patterns of genes responsive at (A) embryonic, (B) L1, and (C) L4 stages and their behavior at the other two stages. Each line represents the fold-change for a single gene (normalized to N2, logarithmic scale). Lines are colored red if the gene is upregulated and blue if it is downregulated at the representative stage for each panel (embryonic in panel A, L1 in panel B, and L4 in panel C). Each gene retained this color in the other two stages (e.g., L1 and L4 for panel A) for each panel, independently of the observed regulation at these stages.
Responsive genes from (A, E) embryonic stage, (B, F) L1 stage, (C, G) L4 stage, and (D, H) genes containing E2F sites were divided into categories according to known or putative functions. Panels A–D represent upregulated responsive genes and panels E–H represent downregulated responsive genes. Individual subcategories of genes involved in cell cycle regulation that were statistically overrepresented (p ≪ 0.01) at L1 and L4 stages include chromosome organization and biogenesis (GO: 0051276), DNA metabolism (GO:0006259), DNA repair (GO: 0006281), and DNA replication (GO: 0006260). The cell cycle category included genes involved in cell cycle regulation, DNA replication, repair, and metabolism, mitosis, meiosis, and cell division.
Responsive genes from (A) embryos, (B) L1 stage, (C) L4 stage, and (D) genes with E2F sites were divided into mountains (Kim et al., 2001). Percentages of responsive genes were compared to corresponding percentages of total genes from the genome within each mountain.
(A) A phylogenetic tree including known human, Drosophila melanogaster, and C. elegans pocket proteins was built using distance methods (topological clustering) from multiple sequence alignment. The numbers represent relative distance between the proteins. Overlap of (B) pRb-responsive genes, (C) p107/p130-responsive genes, and (D) all human pocket proteins with LIN-35 responsive genes from this study, LIN-35- and E2F/DP-responsive genes in the C. elegans germline (Chi and Reinke, 2006), and dRbF1/2-responsive genes (Dimova et al., 2003).
Colors codes are as described in Fig. S1.
Colors codes are as described in Fig. S1. Genes are sorted by their PWM score. Genes in bold are those most likely to have cell cycle function according to cluster analysis (See Results and Fig. S8).
Clusters obtained by KMC algorithm, ranked (top to bottom) by PWM score from E2F consensus site. Clusters 23, 17, 20, 25, 7, 4, and 12 were strongly enriched in cell cycle genes. Lines are intended to facilitate visualization of cluster patterns and are not intended to represent proximal chronological steps.
Acknowledgments
We thank Oleg Moskovin for help with many aspects of our analysis, Mark Gomelsky for reagents, and James DeGregori for advice. We also thank Jim McGhee for sharing unpublished data, and Bifeng Gao and Tom Evans for assistance with microarray experiments. This work was supported by GM066868-01A1 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Balciunaite E, Spektor A, Lents NH, Cam H, Te Riele H, Scime A, Rudnicki MA, Young R, Dynlacht BD. Pocket protein complexes are recruited to distinct targets in quiescent and proliferating cells. Mol Cell Biol. 2005;25:8166–78. doi: 10.1128/MCB.25.18.8166-8178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batsche E, Moschopoulos P, Desroches J, Bilodeau S, Drouin J. Retinoblastoma and the related pocket protein p107 act as coactivators of NeuroD1 to enhance gene transcription. J Biol Chem. 2005;280:16088–95. doi: 10.1074/jbc.M413427200. [DOI] [PubMed] [Google Scholar]
- Bender AM, Kirienko NV, Olson SK, Esko JD, Fay DS. lin-35/Rb and the CoREST ortholog spr-1 coordinately regulate vulval morphogenesis and gonad development in C. elegans. Dev Biol. 2006 doi: 10.1016/j.ydbio.2006.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender AM, Wells O, Fay DS. lin-35/Rb and xnp-1/ATR-X function redundantly to control somatic gonad development in C. elegans. Dev Biol. 2004;273:335–49. doi: 10.1016/j.ydbio.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Bindra RS, Glazer PM. Basal Repression of BRCA1 by Multiple E2Fs and Pocket Proteins at Adjacent E2F Sites. Cancer Biol Ther. 2006;5 doi: 10.4161/cbt.5.10.3454. [DOI] [PubMed] [Google Scholar]
- Black EP, Huang E, Dressman H, Rempel R, Laakso N, Asa SL, Ishida S, West M, Nevins JR. Distinct gene expression phenotypes of cells lacking Rb and Rb family members. Cancer Res. 2003;63:3716–23. [PubMed] [Google Scholar]
- Boxem M, van den Heuvel S. lin-35 Rb and cki-1 Cip/Kip cooperate in developmental regulation of G1 progression in C. elegans. Development. 2001;128:4349–59. doi: 10.1242/dev.128.21.4349. [DOI] [PubMed] [Google Scholar]
- Braatsch S, Moskvin OV, Klug G, Gomelsky M. Responses of the Rhodobacter sphaeroides transcriptome to blue light under semiaerobic conditions. J Bacteriol. 2004;186:7726–35. doi: 10.1128/JB.186.22.7726-7735.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm A, Miska EA, McCance DJ, Reid JL, Bannister AJ, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- Bremner R, Chen D, Pacal M, Livne-Bar I, Agochiya M. The RB protein family in retinal development and retinoblastoma: new insights from new mouse models. Dev Neurosci. 2004;26:417–34. doi: 10.1159/000082284. [DOI] [PubMed] [Google Scholar]
- Cardoso C, Couillault C, Mignon-Ravix C, Millet A, Ewbank JJ, Fontes M, Pujol N. XNP-1/ATR-X acts with RB, HP1 and the NuRD complex during larval development in C. elegans. Dev Biol. 2005;278:49–59. doi: 10.1016/j.ydbio.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Ceol CJ, Horvitz HR. dpl-1 DP and efl-1 E2F act with lin-35 Rb to antagonize Ras signaling in C. elegans vulval development. Mol Cell. 2001;7:461–73. doi: 10.1016/s1097-2765(01)00194-0. [DOI] [PubMed] [Google Scholar]
- Chen PL, Riley DJ, Chen Y, Lee WH. Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/EBPs. Genes Dev. 1996;10:2794–804. doi: 10.1101/gad.10.21.2794. [DOI] [PubMed] [Google Scholar]
- Chesney MA, Kidd AR, 3rd, Kimble J. gon-14 functions with class B and class C synthetic multivulva genes to control larval growth in Caenorhabditis elegans. Genetics. 2006;172:915–28. doi: 10.1534/genetics.105.048751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi W, Reinke V. Promotion of oogenesis and embryogenesis in the C. elegans gonad by EFL-1/DPL-1 (E2F) does not require LIN-35 (pRB) Development. 2006;133:3147–57. doi: 10.1242/dev.02490. [DOI] [PubMed] [Google Scholar]
- Clarke AR, Maandag ER, van Roon M, van der Lugt NM, van der Valk M, Hooper ML, Berns A, te Riele H. Requirement for a functional Rb-1 gene in murine development. Nature. 1992;359:328–30. doi: 10.1038/359328a0. [DOI] [PubMed] [Google Scholar]
- Cui M, Fay DS, Han M. lin-35/Rb cooperates with the SWI/SNF complex to control Caenorhabditis elegans larval development. Genetics. 2004;167:1177–85. doi: 10.1534/genetics.103.024554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannenberg JH, van Rossum A, Schuijff L, te Riele H. Ablation of the retinoblastoma gene family deregulates G(1) control causing immortalization and increased cell turnover under growth-restricting conditions. Genes Dev. 2000;14:3051–64. doi: 10.1101/gad.847700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta P, Padmanabhan J, Chellappan S. Rb function in the apoptosis and senescence of non-neuronal and neuronal cells: role in oncogenesis. Curr Mol Med. 2006;6:719–29. doi: 10.2174/1566524010606070719. [DOI] [PubMed] [Google Scholar]
- De Falco G, Comes F, Simone C. pRb: master of differentiation. Coupling irreversible cell cycle withdrawal with induction of muscle-specific transcription. Oncogene. 2006;25:5244–9. doi: 10.1038/sj.onc.1209623. [DOI] [PubMed] [Google Scholar]
- Dick FA, Dyson N. pRB contains an E2F1-specific binding domain that allows E2F1-induced apoptosis to be regulated separately from other E2F activities. Mol Cell. 2003;12:639–49. doi: 10.1016/s1097-2765(03)00344-7. [DOI] [PubMed] [Google Scholar]
- Dimova DK, Stevaux O, Frolov MV, Dyson NJ. Cell cycle-dependent and cell cycle-independent control of transcription by the Drosophila E2F/RB pathway. Genes Dev. 2003;17:2308–20. doi: 10.1101/gad.1116703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryja TP, Rapaport JM, Joyce JM, Petersen RA. Molecular detection of deletions involving band q14 of chromosome 13 in retinoblastomas. Proc Natl Acad Sci U S A. 1986;83:7391–4. doi: 10.1073/pnas.83.19.7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W, Dyson N. The role of RBF in the introduction of G1 regulation during Drosophila embryogenesis. Embo J. 1999;18:916–25. doi: 10.1093/emboj/18.4.916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W, Pogoriler J. Retinoblastoma family genes. Oncogene. 2006;25:5190–200. doi: 10.1038/sj.onc.1209651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay DS. The cell cycle and development: lessons from C. elegans. Semin Cell Dev Biol. 2005;16:397–406. doi: 10.1016/j.semcdb.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Fay DS, Keenan S, Han M. fzr-1 and lin-35/Rb function redundantly to control cell proliferation in C. elegans as revealed by a nonbiased synthetic screen. Genes Dev. 2002;16:503–17. doi: 10.1101/gad.952302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay DS, Large E, Han M, Darland M. lin-35/Rb and ubc-18, an E2 ubiquitin-conjugating enzyme, function redundantly to control pharyngeal morphogenesis in C. elegans. Development. 2003;130:3319–30. doi: 10.1242/dev.00561. [DOI] [PubMed] [Google Scholar]
- Fay DS, Qiu X, Large E, Smith CP, Mango S, Johanson BL. The coordinate regulation of pharyngeal development in C. elegans by lin-35/Rb, pha-1, and ubc-18. Dev Biol. 2004;271:11–25. doi: 10.1016/j.ydbio.2004.03.022. [DOI] [PubMed] [Google Scholar]
- Ferguson KL, Slack RS. The Rb pathway in neurogenesis. Neuroreport. 2001;12:A55–62. doi: 10.1097/00001756-200107030-00001. [DOI] [PubMed] [Google Scholar]
- Finocchiaro G, Mancuso F, Muller H. Mining published lists of cancer related microarray experiments: identification of a gene expression signature having a critical role in cell-cycle control. BMC Bioinformatics. 2005;6 Suppl 4:S14. doi: 10.1186/1471-2105-6-S4-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friend SH, Bernards R, Rogelj S, Weinberg RA, Rapaport JM, Albert DM, Dryja TP. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature. 1986;323:643–6. doi: 10.1038/323643a0. [DOI] [PubMed] [Google Scholar]
- Fukushige T, Hendzel MJ, Bazett-Jones DP, McGhee JD. Direct visualization of the elt-2 gut-specific GATA factor binding to a target promoter inside the living Caenorhabditis elegans embryo. Proc Natl Acad Sci U S A. 1999;96:11883–8. doi: 10.1073/pnas.96.21.11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet J, Muttumu S, Horner M, Mango SE. Whole-genome analysis of temporal gene expression during foregut development. PLoS Biol. 2004;2:e352. doi: 10.1371/journal.pbio.0020352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gery S, Gombart AF, Fung YK, Koeffler HP. C/EBPepsilon interacts with retinoblastoma and E2F1 during granulopoiesis. Blood. 2004;103:828–35. doi: 10.1182/blood-2003-01-0159. [DOI] [PubMed] [Google Scholar]
- Gotter AL. A Timeless debate: resolving TIM’s noncircadian roles with possible clock function. Neuroreport. 2006;17:1229–33. doi: 10.1097/01.wnr.0000233092.90160.92. [DOI] [PubMed] [Google Scholar]
- Gu W, Schneider JW, Condorelli G, Kaushal S, Mahdavi V, Nadal-Ginard B. Interaction of myogenic factors and the retinoblastoma protein mediates muscle cell commitment and differentiation. Cell. 1993;72:309–24. doi: 10.1016/0092-8674(93)90110-c. [DOI] [PubMed] [Google Scholar]
- Haigis K, Sage J, Glickman J, Shafer S, Jacks T. The related retinoblastoma (pRb) and p130 proteins cooperate to regulate homeostasis in the intestinal epithelium. J Biol Chem. 2006;281:638–47. doi: 10.1074/jbc.M509053200. [DOI] [PubMed] [Google Scholar]
- Hardin PE. The circadian timekeeping system of Drosophila. Curr Biol. 2005;15:R714–22. doi: 10.1016/j.cub.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Harpster MH, Bandyopadhyay S, Thomas DP, Ivanov PS, Keele JA, Pineguina N, Gao B, Amarendran V, Gomelsky M, McCormick RJ, et al. Earliest changes in the left ventricular transcriptome postmyocardial infarction. Mamm Genome. 2006;17:701–15. doi: 10.1007/s00335-005-0120-1. [DOI] [PubMed] [Google Scholar]
- Iavarone A, Garg P, Lasorella A, Hsu J, Israel MA. The helix-loop-helix protein Id-2 enhances cell proliferation and binds to the retinoblastoma protein. Genes Dev. 1994;8:1270–84. doi: 10.1101/gad.8.11.1270. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Jacks T, Fazeli A, Schmitt EM, Bronson RT, Goodell MA, Weinberg RA. Effects of an Rb mutation in the mouse. Nature. 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- Kim SK, Lund J, Kiraly M, Duke K, Jiang M, Stuart JM, Eizinger A, Wylie BN, Davidson GS. A gene expression map for Caenorhabditis elegans. Science. 2001;293:2087–92. doi: 10.1126/science.1061603. [DOI] [PubMed] [Google Scholar]
- Knudsen ES, Knudsen KE. Retinoblastoma tumor suppressor: where cancer meets the cell cycle. Exp Biol Med (Maywood) 2006;231:1271–81. doi: 10.1177/153537020623100713. [DOI] [PubMed] [Google Scholar]
- Korenjak M, Brehm A. E2F-Rb complexes regulating transcription of genes important for differentiation and development. Curr Opin Genet Dev. 2005;15:520–7. doi: 10.1016/j.gde.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Lasorella A, Noseda M, Beyna M, Yokota Y, Iavarone A. Id2 is a retinoblastoma protein target and mediates signalling by Myc oncoproteins. Nature. 2000;407:592–8. doi: 10.1038/35036504. [DOI] [PubMed] [Google Scholar]
- Lee EY, Chang CY, Hu N, Wang YC, Lai CC, Herrup K, Lee WH, Bradley A. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature. 1992;359:288–94. doi: 10.1038/359288a0. [DOI] [PubMed] [Google Scholar]
- Lee MH, Williams BO, Mulligan G, Mukai S, Bronson RT, Dyson N, Harlow E, Jacks T. Targeted disruption of p107: functional overlap between p107 and Rb. Genes Dev. 1996;10:1621–32. doi: 10.1101/gad.10.13.1621. [DOI] [PubMed] [Google Scholar]
- Lee WH, Bookstein R, Hong F, Young LJ, Shew JY, Lee EY. Human retinoblastoma susceptibility gene: cloning, identification, and sequence. Science. 1987;235:1394–9. doi: 10.1126/science.3823889. [DOI] [PubMed] [Google Scholar]
- Lehner B, Calixto A, Crombie C, Tischler J, Fortunato A, Chalfie M, Fraser AG. Loss of LIN-35, the Caenorhabditis elegans ortholog of the tumor suppressor p105Rb, results in enhanced RNA interference. Genome Biol. 2006;7:R4. doi: 10.1186/gb-2006-7-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski MM, Jacks T. The retinoblastoma gene family in differentiation and development. Oncogene. 1999;18:7873–82. doi: 10.1038/sj.onc.1203244. [DOI] [PubMed] [Google Scholar]
- Lu X, Horvitz HR. lin-35 and lin-53, two genes that antagonize a C. elegans Ras pathway, encode proteins similar to Rb and its binding protein RbAp48. Cell. 1998;95:981–91. doi: 10.1016/s0092-8674(00)81722-5. [DOI] [PubMed] [Google Scholar]
- Lukas J, Bartkova J, Rohde M, Strauss M, Bartek J. Cyclin D1 is dispensable for G1 control in retinoblastoma gene-deficient cells independently of cdk4 activity. Mol Cell Biol. 1995;15:2600–11. doi: 10.1128/mcb.15.5.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo RX, Postigo AA, Dean DC. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–73. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain JP, Troalen F, Trouche D, Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–5. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- Markey MP, Angus SP, Strobeck MW, Williams SL, Gunawardena RW, Aronow BJ, Knudsen ES. Unbiased analysis of RB-mediated transcriptional repression identifies novel targets and distinctions from E2F action. Cancer Res. 2002;62:6587–97. [PubMed] [Google Scholar]
- McGhee JD, Sleumer MC, Bilenky M, Wong K, McKay SJ, Goszczynski B, Tian H, Krich ND, Khattra J, Holt RA, et al. The ELT-2 GATA-factor and the global regulation of transcription in the C. elegans intestine. Dev Biol. 2006 doi: 10.1016/j.ydbio.2006.10.024. [DOI] [PubMed] [Google Scholar]
- McLear JA, Garcia-Fresco G, Bhat MA, Van Dyke TA. In vivo inactivation of pRb, p107 and p130 in murine neuroprogenitor cells leads to major CNS developmental defects and high seizure rates. Mol Cell Neurosci. 2006;33:260–73. doi: 10.1016/j.mcn.2006.07.012. [DOI] [PubMed] [Google Scholar]
- Morris EJ, Dyson NJ. Retinoblastoma protein partners. Adv Cancer Res. 2001;82:1–54. doi: 10.1016/s0065-230x(01)82001-7. [DOI] [PubMed] [Google Scholar]
- Moskvin OV, Gomelsky L, Gomelsky M. Transcriptome analysis of the Rhodobacter sphaeroides PpsR regulon: PpsR as a master regulator of photosystem development. J Bacteriol. 2005;187:2148–56. doi: 10.1128/JB.187.6.2148-2156.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H, Bracken AP, Vernell R, Moroni MC, Christians F, Grassilli E, Prosperini E, Vigo E, Oliner JD, Helin K. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 2001;15:267–85. doi: 10.1101/gad.864201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan G, Jacks T. The retinoblastoma gene family: cousins with overlapping interests. Trends Genet. 1998;14:223–9. doi: 10.1016/s0168-9525(98)01470-x. [DOI] [PubMed] [Google Scholar]
- Mulligan GJ, Wong J, Jacks T. p130 is dispensable in peripheral T lymphocytes: evidence for functional compensation by p107 and pRB. Mol Cell Biol. 1998;18:206–20. doi: 10.1128/mcb.18.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers MP, Wager-Smith K, Wesley CS, Young MW, Sehgal A. Positional cloning and sequence analysis of the Drosophila clock gene, timeless. Science. 1995;270:805–8. doi: 10.1126/science.270.5237.805. [DOI] [PubMed] [Google Scholar]
- Pappas CT, Sram J, Moskvin OV, Ivanov PS, Mackenzie RC, Choudhary M, Land ML, Larimer FW, Kaplan S, Gomelsky M. Construction and validation of the Rhodobacter sphaeroides 2.4.1 DNA microarray: transcriptome flexibility at diverse growth modes. J Bacteriol. 2004;186:4748–58. doi: 10.1128/JB.186.14.4748-4758.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M, Krause MW. Regulation of postembryonic G(1) cell cycle progression in Caenorhabditis elegans by a cyclin D/CDK-like complex. Development. 1999;126:4849–60. doi: 10.1242/dev.126.21.4849. [DOI] [PubMed] [Google Scholar]
- Reed SI. Control of the G1/S transition. Cancer Surv. 1997;29:7–23. [PubMed] [Google Scholar]
- Rekhtman N, Choe KS, Matushansky I, Murray S, Stopka T, Skoultchi AI. PU.1 and pRB interact and cooperate to repress GATA-1 and block erythroid differentiation. Mol Cell Biol. 2003;23:7460–74. doi: 10.1128/MCB.23.21.7460-7474.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet. 2000;25:338–42. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- Sage J, Mulligan GJ, Attardi LD, Miller A, Chen S, Williams B, Theodorou E, Jacks T. Targeted disruption of the three Rb-related genes leads to loss of G(1) control and immortalization. Genes Dev. 2000;14:3037–50. doi: 10.1101/gad.843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider JW, Gu W, Zhu L, Mahdavi V, Nadal-Ginard B. Reversal of terminal differentiation mediated by p107 in Rb-/- muscle cells. Science. 1994;264:1467–71. doi: 10.1126/science.8197461. [DOI] [PubMed] [Google Scholar]
- Sherr CJ. Principles of tumor suppression. Cell. 2004;116:235–46. doi: 10.1016/s0092-8674(03)01075-4. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, McCormick F. The RB and p53 pathways in cancer. Cancer Cell. 2002;2:103–12. doi: 10.1016/s1535-6108(02)00102-2. [DOI] [PubMed] [Google Scholar]
- Slack RS, Miller FD. Retinoblastoma gene in mouse neural development. Dev Genet. 1996;18:81–91. doi: 10.1002/(SICI)1520-6408(1996)18:1<81::AID-DVG9>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Spike BT, Dirlam A, Dibling BC, Marvin J, Williams BO, Jacks T, Macleod KF. The Rb tumor suppressor is required for stress erythropoiesis. Embo J. 2004;23:4319–29. doi: 10.1038/sj.emboj.7600432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevaux O, Dimova DK, Ji JY, Moon NS, Frolov MV, Dyson NJ. Retinoblastoma family 2 is required in vivo for the tissue-specific repression of dE2F2 target genes. Cell Cycle. 2005;4:1272–80. doi: 10.4161/cc.4.9.1982. [DOI] [PubMed] [Google Scholar]
- Stiernagle T WormBook. Maintenance of C. elegans. The C. elegans Research Community, WormBook; Feb 11, 2006. http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110–56. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Wang D, Kennedy S, Conte D, Jr, Kim JK, Gabel HW, Kamath RS, Mello CC, Ruvkun G. Somatic misexpression of germline P granules and enhanced RNA interference in retinoblastoma pathway mutants. Nature. 2005;436:593–7. doi: 10.1038/nature04010. [DOI] [PubMed] [Google Scholar]
- Wikenheiser-Brokamp KA. Retinoblastoma family proteins: insights gained through genetic manipulation of mice. Cell Mol Life Sci. 2006;63:767–80. doi: 10.1007/s00018-005-5487-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, de Bruin A, Saavedra HI, Starovic M, Trimboli A, Yang Y, Opavska J, Wilson P, Thompson JC, Ostrowski MC, et al. Extra-embryonic function of Rb is essential for embryonic development and viability. Nature. 2003;421:942–7. doi: 10.1038/nature01417. [DOI] [PubMed] [Google Scholar]
- Yamasaki L. Role of the RB tumor suppressor in cancer. Cancer Treat Res. 2003;115:209–39. doi: 10.1007/0-306-48158-8_9. [DOI] [PubMed] [Google Scholar]
- Zeller T, Moskvin OV, Li K, Klug G, Gomelsky M. Transcriptome and physiological responses to hydrogen peroxide of the facultatively phototrophic bacterium Rhodobacter sphaeroides. J Bacteriol. 2005;187:7232–42. doi: 10.1128/JB.187.21.7232-7242.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HS, Dean DC. Rb-mediated chromatin structure regulation and transcriptional repression. Oncogene. 2001;20:3134–8. doi: 10.1038/sj.onc.1204338. [DOI] [PubMed] [Google Scholar]
- Zhang HS, Gavin M, Dahiya A, Postigo AA, Ma D, Luo RX, Harbour JW, Dean DC. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell. 2000;101:79–89. doi: 10.1016/S0092-8674(00)80625-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Colors codes are as follows: yellow – embryonic stage only, light green – L1 stage only, blue – L4 stage only, dark green – L1 and L4 stages, magenta – embryonic and L1 stages, purple – embryonic and L4 stages, red – all stages, pink – genes containing an E2F consensus site. Column headed “mountain” refers to mountains described in Kim et al., 2001.
Expression patterns of genes responsive at (A) embryonic, (B) L1, and (C) L4 stages and their behavior at the other two stages. Each line represents the fold-change for a single gene (normalized to N2, logarithmic scale). Lines are colored red if the gene is upregulated and blue if it is downregulated at the representative stage for each panel (embryonic in panel A, L1 in panel B, and L4 in panel C). Each gene retained this color in the other two stages (e.g., L1 and L4 for panel A) for each panel, independently of the observed regulation at these stages.
Responsive genes from (A, E) embryonic stage, (B, F) L1 stage, (C, G) L4 stage, and (D, H) genes containing E2F sites were divided into categories according to known or putative functions. Panels A–D represent upregulated responsive genes and panels E–H represent downregulated responsive genes. Individual subcategories of genes involved in cell cycle regulation that were statistically overrepresented (p ≪ 0.01) at L1 and L4 stages include chromosome organization and biogenesis (GO: 0051276), DNA metabolism (GO:0006259), DNA repair (GO: 0006281), and DNA replication (GO: 0006260). The cell cycle category included genes involved in cell cycle regulation, DNA replication, repair, and metabolism, mitosis, meiosis, and cell division.
Responsive genes from (A) embryos, (B) L1 stage, (C) L4 stage, and (D) genes with E2F sites were divided into mountains (Kim et al., 2001). Percentages of responsive genes were compared to corresponding percentages of total genes from the genome within each mountain.
(A) A phylogenetic tree including known human, Drosophila melanogaster, and C. elegans pocket proteins was built using distance methods (topological clustering) from multiple sequence alignment. The numbers represent relative distance between the proteins. Overlap of (B) pRb-responsive genes, (C) p107/p130-responsive genes, and (D) all human pocket proteins with LIN-35 responsive genes from this study, LIN-35- and E2F/DP-responsive genes in the C. elegans germline (Chi and Reinke, 2006), and dRbF1/2-responsive genes (Dimova et al., 2003).
Colors codes are as described in Fig. S1.
Colors codes are as described in Fig. S1. Genes are sorted by their PWM score. Genes in bold are those most likely to have cell cycle function according to cluster analysis (See Results and Fig. S8).
Clusters obtained by KMC algorithm, ranked (top to bottom) by PWM score from E2F consensus site. Clusters 23, 17, 20, 25, 7, 4, and 12 were strongly enriched in cell cycle genes. Lines are intended to facilitate visualization of cluster patterns and are not intended to represent proximal chronological steps.