Abstract
Quality of botanical food is increasingly assessed by the content of multiple bioactive compounds. In this study we report, for the first time, an HPLC fingerprinting method for the quality evaluation of Rubus suavissimus leaves possessing multiple bioactivities. Five constituents, gallic acid, rutin, ellagic acid, rubusoside, and steviol monoside were quantified and used in developing qualitative chromatographic fingerprints. The limits of detection and quantification ranged from 0.29 μg/mL to 37.86 μg/mL. The relative standard deviations (RSDs) of intra- and inter-day precisions were no more than 3.14% and 3.01%, respectively. The average recoveries were between 93.1% and 97.5%. The developed method was validated in analyzing fourteen leaf samples with satisfactory results. The contents of the five marker compounds accounted for an average of about 6% w/w with a variability of 16% among the fourteen samples collected from a single site and year. Gallic acid was the least whereas steviol monoside the most variable compounds among the fourteen leaf samples. The characteristic compound rubusoside that is responsible for the sweet taste accounted for 5% of leaf weight. The validated method can now be used to quantitatively and qualitatively assess the quality of Rubus suavissimus leaves as traditional beverage or potential medicines.
Keywords: HPLC-PDA, quality assessment, Rubus suavissimus, similarity test, temporal variations
INTRODUCTION
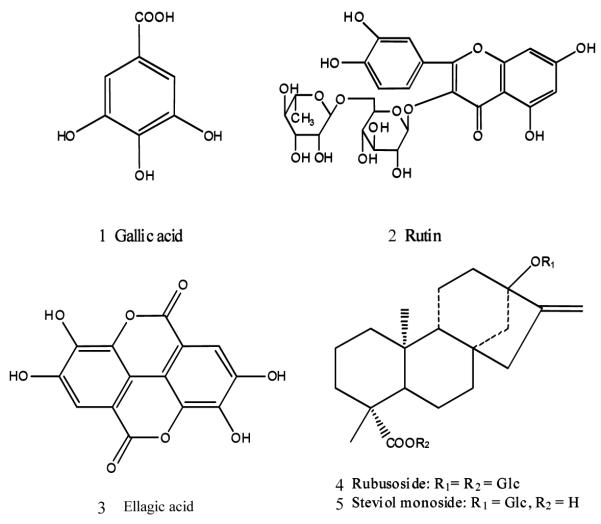
Rubus suavissimus S. Lee (Rosaceae) is a shrub widely grown in southwestern China, leaves of which are usually used as an herbal tea, a different leaf tea from the well-known green tea (Camellia sinensis). It is particularly referred as Tian-cha or Sweet Tea in China, because of its natural sweetness. In addition to being consumed as an herbal tea, it has also been made into healthy drink because of the recent pharmacological studies that revealed significant bioactivities such as anti-angiogenic (1) and anti-allergic activities (2). Moreover, investigations over the chemical constituents of Rubus suavissimus provided new knowledge that gallotannins, ellagitannins, flavonoids and diterpenes are the major classes of constituents (3-8). These classes of compounds represented by gallic acid, ellagic acid, rutin, rubusoside, and steviol monoside (Figure 1) were found to be dominant and responsible for the biological activities (9).
Figure 1.
Chemical structures of the three phenols and two diterpene glycosides in the leaves of Rubus suavissimus.
Traditionally, the quality of the sweet tea is measured by its sweetness. This makes sense for an herbal tea where it got its name. The sweetness of the leaves is provided by the diterpene glucosides such as rubusoside and steviol monoside. Because of the recent scientific revelation of multiple bioactivities, the quality of the leaves is now measured by other bioactive compounds in addition to the characteristic diterpene glycosides. For examples, gallic acid is one of the active compounds that have potent anti-angiogenic activity (1). Ellagic acid is another bioactive compound reported as antioxidant (10), anticancer (11, 12) and anti-inflammatory (13, 14). Rutin is the third bioactive compound as an antioxidant (15, 16) and anti-inflammatory (17). In our preliminary analyses over the leaves, these bioactive compounds were all present. Since they are representative compounds for the four classes mentioned above and have various bioactivities, they were chosen as chemical markers. Although most of these compounds are common in some plants, it is the presence of them altogether in one botanical part (leaf) in a ratio naturally made that may have contributed to its multiple and perhaps concerted bioactivities. Therefore, measurement of whole leaf constituents is of great interest and importance through chromatographic fingerprint analysis and determination of multiple characteristic compounds.
To develop reliable and highly representative chromatographic fingerprints and quantitative analyses for any raw botanical food, it takes a good biological and chemical convergence. First, the development must choose plant materials that represent an authentic chemical composition for a given species. Although it is highly challenging or may not be possible to know exactly and completely what a plant produces chemically, it is sufficient for investigators to know the major constituents. Second, with the authentic plant material, a reliable and repeatable method must be validated chemically to allow future uses and references. Based on these judgments, a study site located in the major production area where the sweet tea is being grown was selected. This site provided all the sweet tea leaf samples previously to the authors, which have been consistently proven to be effective angiogenic inhibitors (1). Therefore, it is justified to use this source for our current investigation.
Increasingly, it is discovered that a bioactive plant contains multiple bioactive compounds and taking them apart may diminish the overall bioactivity (18). This advantage of concerted actions of a single plant extract has been taken and embraced by many users of herbal remedies throughout the world. The U.S Food and Drug Administration’s recent decision to enact the botanical drug pathway further illustrates broader acceptance of this concept and willingness to take this potential advantage. Although a Rubus suavissimus leaf extract has been repeatedly found bioactive (anti-angiogenic) in our investigations, the challenge to controlling batch-to-batch variations prompts this investigation. This study aims at establishing a reliable and sensitive quality assessment method for this botanical food plant that begins at its origin of raw leaf material. The present study reports a validated HPLC method for simultaneous determination of five representative bioactive components and chromatographic fingerprints.
MATERIALS AND METHODS
Plant materials
Fourteen samples of Rubus suavissimus (Chinese sweet tea plant) leaves were acquired during a growing season from a cultivation farm in Guizhou Province, China and were authenticated by the corresponding author. A voucher specimen was deposited in the Herbarium of Louisiana State University (Accession number: 131298).
Standards and chemicals
Gallic acid (GA), rutin (RUT), and ellagic acid (EGA) were purchased from Sigma Chemical Company (St. Louis, MO, USA). The reference standards of rubusoside (RUB) and steviol monoside (SM) were isolated in our own lab and were identified by spectral data (UV, MS, 1H NMR, 13C NMR and 2D-NMR). Both RUB and SM have purities greater than 98% by HPLC—PDA analyses based on a peak area normalization method. HPLC grade acetonitrile and water were obtained from Mallinckrodt (Phillipsburg, NJ, USA). ACS grade phosphoric acid was provided by Fisher Scientific (Fair Lawn, NJ, USA).
HPLC—PDA analysis
An HPLC system consisting of a Waters (Milford, MA, USA) 600 pump, a 717 auto-sampler, and a 2996 UV-vis Photodiode Array (PDA) Detector was used for all analyses. HPLC separation was performed on an Alltech Prevail C18 column (250 mm × 4.6 mm i.d.; 5 μm) together with a YMC C18 guard column (7.5 mm × 4.6 mm i.d.; 5 μm), which was placed in a column heater set at 25°C. The mobile phase consisted of solvent A (0.17% phosphoric acid in acetonitrile) and solvent B (0.17% phosphoric acid in water). The gradient elution program was as follows: from 0 to 65 min, A followed a linear change from 5% to 30%; from 65 to 85 min, A linearly changed from 30% to 60%; from 85 to 90 min, A linearly changed from 60% to 70%; and from 90 to 100 min, A was isocratic at 70%. The flow rate was set at 1.0 mL/min. The injection volume was 10 μL. The wavelength of PDA detector ranged from 200 to 400 nm. Because gallic acid, ellagic acid, and rutin had strong UV absorption at 254 nm whereas rubusoside and steviol monoside had strong UV absorption at 205 nm, each chromatogram was derived by combining chromatograms at 254 nm and 205 nm.
Preparation of standard solution
Accurately weighed appropriate amounts of the reference compounds were mixed and dissolved in methanol in a 50-mL volumetric flask. Concentration of the five compounds in the stock solution was 0.05 mg/mL (1; GA), 0.024 mg/mL (2; RUT), 0.206 mg/mL (3; EGA), 1.404 mg/mL (4; RUB), and 0.120 mg/mL (5; SM), respectively. Stock solution containing these compounds was stored at 4°C in the dark before HPLC analysis.
Sample preparation
All leaf samples were dried at 60°C for 12 h, and then were pulverized to fine powder, which passed through a 100 mesh screen. One gram of each powdered sample was accurately weighed into a 250-mL flask then 100 mL water was added. After the weight of the whole flask was recorded, sample was heated at reflux for two hours. The original solvent weight was restored after sample was cooled to room temperature. After centrifugation at 10000 × g for 10 min, 25 mL of the supernatant was transferred to a 50-mL flask and was concentrated to dryness using rotary evaporator under reduced pressure. The dried residue was dissolved in methanol and transferred to a 10-mL volumetric flask. This methanol solution was filtered through a 0.2-μm syringe filter (Nalgene, NY, USA) prior to HPLC analysis. For each sample, the complete assay procedure was conducted in triplicate and the standard deviation was calculated.
Calibration curves, limits of detection and quantification
External standard calibrations were established at five data points covering the concentration range of each compound according to the level estimated in the plant samples. Working solutions were prepared by stepwise dilution of the stock solution with methanol. Triplicate analyses were performed for each concentration. Calibration curves were constructed from peak areas versus compound concentrations. The limits of detection (LOD) and quantification (LOQ) for each marker compound under the present chromatographic conditions were determined at a signal-to-noise ratio (S/N) of 3 and 10, respectively.
Precision, reproducibility, and accuracy
To assess the intra-day precision of the method, the standard solution of assayed compounds was injected six times within a day. The inter-day precision was determined with the same standard solution over 3 days by three injections per day. Reproducibility of the method was verified by analyzing five extracts of identical sample. A recovery test was used to evaluate the accuracy of this method. Exact amounts of the five reference compounds were added to a 0.5 g leaf sample and then was extracted and analyzed as described above. Average recoveries were calibrated by the formula: recovery (%) = {(amount found—original amount)/amount spiked} ×100%, and relative standard deviation or RSD (%) = (SD/mean) ×100%.
Similarity analysis
Similarity tests among the fourteen samples were performed based on the relative retention time (RRT) and relative peak area (RPA) using the professional software named Similarity Evaluation System for Chromatographic Fingerprint of Traditional Chinese Medicine (2004A). The matching amongst the fingerprints of samples was performed by a multipoint calibration mode based on the retention time and UV spectra. In this test, all fourteen samples were examined to generate a reference chromatogram as the representative standard fingerprint and the similarity of each chromatogram against this standard chromatogram was then calculated. Furthermore, RRT and RPA of each characteristic peak related to a reference peak were also constructed for quantitative measurement of the chemical composition of a R. suavissimus leaf sample.
RESULTS AND DISCUSSION
Selection of sample preparation method
The efficiency of the extraction procedure was tested by using different extraction methods and by sequentially varying the composition of the solvents (methanol, 70% and 50% aqueous methanol, and water). The result showed that water yielded more compounds of interest than other solvents used. The amounts of the selected marker compounds prepared by refluxing extraction, ultrasonic extraction, and Soxhlet extraction were compared. For all the five analytes, the highest extraction efficiency was achieved by refluxing extraction. The refluxing time was then tested and it was found that 2 h was all it maximally took to exhaustively extract the five marker compounds with 100 mL water.
Optimization of chromatographic conditions
In the development of HPLC method for the fingerprint analysis and determination of representative compounds of the sweet tea plant, several solvent systems (methanol-water, methanol-water-formic acid, methanol-water-acetic acid, methanol-water-phosphoric acid, acetonitrile-water, acetonitrile-water-formic acid, acetonitrile-water-acetic acid, acetonitrile-water-phosphoric acid) and separation columns (Symmetry C18 5 μm 4.6 ×150 mm, Symmetry C18 5 μm 4.6×250 mm, SunFire C18 3.5 μm 4.6 ×150 mm, Atlantis 3.5 μm 4.6 ×150 mm, Luna C18 5μm 4.6×250 mm, and Alltech Prevail C18 5 μm 4.6×250 mm) were evaluated and compared. The result showed that 0.17% v/v phosphoric acid in the solvent system could reduce the ionization of the phenol group and gave more symmetrical peaks and better separation than non-acid system. The Prevail C18 ODS column provided better separation of components in the plant than other brands of C18 columns. Detection wavelength was also optimized in this work. Due to the lack of chromophore in chemical structures, diterpene glycosides show mainly terminal absorptions such as 205 nm in their UV spectra (19, 20), whereas phenols generally show strong UV absorptions at 254 nm. Considering the significant difference of UV absorption characteristics between diterpene glycosides and phenols, a combined wavelength (205 and 254 nm) was used in post-run data processing to generate a single chromatogram for each sample instead of two separate chromatograms, each with a single wavelength, in the quantification and chromatographic fingerprints analyses. All five markers had baseline separations to allow accurate integrations for quantifications. It was found that the selection of the combined UV wavelength not only satisfied the quantitative analysis of the five marker compounds but also offered a more informative HPLC chromatogram in a single chromatogram and a single run than the use of multiple chromatograms in displaying the same major constituents. It is a much simpler way to assess the quality of this plant from both angles of marker compounds and non-marker components.
Method validation
All the correlation coefficients (r2) of the calibration curves for the five marker compounds were greater than 0.999. As shown in Table 1, the LOD values were 1.24, 1.15, 0.29, 11.36, and 3.85 μg/mL for compounds 1-5, respectively, while the LOQ values were 3.45, 2.78, 2.41, 37.86, and 11.09 μg/mL for compounds 1-5, respectively. The RSDs of the intra-day and inter-day precisions were found to be 3.14% and 3.01%, respectively (Table 2). The reproducibility of the experiment was evaluated by a series of five experiments on the same plant material with identical extraction and analytical methods. The result showed the reproducibility of the method was satisfactory with the RSDs below 3.67% for any of the five compounds (Table 2). The recovery of the compounds (1-5) was determined by the addition of a standard mixture solution at a concentration close to what would be expected in the actual plant leaf samples and the mean recovery rate was found to be in the range of 93.08% and 97.50% with satisfactory RSDs in the range of 2.35% and 4.90% (Table 3).
Table 1.
Calibration curve parameters, limit of detection (LOD) and limit of quantification (LOQ) for the five marker compounds identified in the Chinese sweet tea plant.
| Compound | Calibration curve | Correlation coefficient (r2) |
Linear range (μg/mL) |
LOD (μg/mL) (n=6) |
LOQ (μg/mL) (n=6) |
|---|---|---|---|---|---|
| Gallic acid | y = 89905x - 27934 | 0.9994 | 5-50 | 1.24 | 3.45 |
| Rutin | y = 60463x - 27542 | 0.9999 | 4-20 | 1.15 | 2.78 |
| Ellagic acid | y = 130689x + 43120 | 0.9994 | 20-200 | 0.29 | 2.41 |
| Rubusoside | y = 6278x + 45650 | 0.9998 | 140-1400 | 11.36 | 37.86 |
| Steviol monoside | y = 8124x - 6071 | 0.9998 | 12-120 | 3.85 | 11.09 |
Table 2.
Precision and reproducibility of the HPLC method developed for the Chinese sweet tea plant.
| Compound | Precision | Reproducibility | ||||
|---|---|---|---|---|---|---|
| Intraday (n = 6) | Interday (n = 9) | Mean (mg/g) | RSD (%) | |||
| Mean (μg/mL) |
RSD (%) | Mean (μg/mL) |
RSD (%) | |||
| Gallic acid | 28.92 | 2.96 | 29.05 | 0.39 | 1.30 | 2.70 |
| Rutin | 12.82 | 3.14 | 12.70 | 3.01 | 0.88 | 3.43 |
| Ellagic acid | 115.15 | 2.00 | 111.06 | 0.73 | 7.44 | 3.67 |
| Rubusoside | 880.40 | 0.90 | 872.72 | 0.43 | 48.80 | 1.63 |
| Steviol monoside | 71.68 | 2.65 | 72.94 | 1.63 | 0.78 | 2.83 |
Table 3.
Recovery rate of the five marker compounds using the developed HPLC method for the Chinese sweet tea plant.
| Compound | Original mean (mg) |
Spiked mean (mg) |
Detected mean (mg) |
Recovery (%) |
RSD (%) (n=5) |
|---|---|---|---|---|---|
| Gallic acid | 0.66 | 0.45 | 1.09 | 95.56 | 3.99 |
| Rutin | 0.50 | 0.40 | 0.89 | 97.50 | 4.57 |
| Ellagic acid | 4.58 | 3.00 | 7.40 | 94.00 | 4.90 |
| Rubusoside | 25.07 | 25.00 | 48.34 | 93.08 | 2.35 |
| Steviol monoside | 0.36 | 0.40 | 0.75 | 97.50 | 3.23 |
Analysis for the marker compounds in the plant material
The developed quantitative method above was applied to simultaneously analyze the fourteen samples of R. suavissimus collected at different harvesting time on a growing season in Guizhou, China. The five reference compounds were eluted at 11.2 min, 53.9 min, 57.3 min, 80.6 min, and 86.3 min, respectively as seen in the chromatogram developed at combined dual wavelengths (Figure 2 Upper). In a representative sample, these five compounds were eluted in the same retention time and had good baseline separations from other components of the sample (Figure 2 Lower). The five marker compounds combined had an average content of 5.9% (w/w) composed of 2.2% gallic acid, 1.9% rutin, 12.0% ellagic acid, 82.5% rubusoside, and 1.4% steviol monoside (Table 4). Clearly, rubusoside is a dominant compound. The other 94% was obviously other extractable components mostly seen in the fingerprints and non extractable components (e.g., structural components such as fibers) in the plant materials. The contents of the five compounds in the fourteen plant samples were variable but the variability was dependent on each individual compound (Table 4). Among these marker compounds, the content of steviol monoside varied the greatest with 29% among the fourteen samples. Gallic acid content was the least variable among the fourteen samples with only slightly over 12%. Overall, the sum of the five markers had approximately 16% variation among the fourteen leaf samples.
Figure 2.
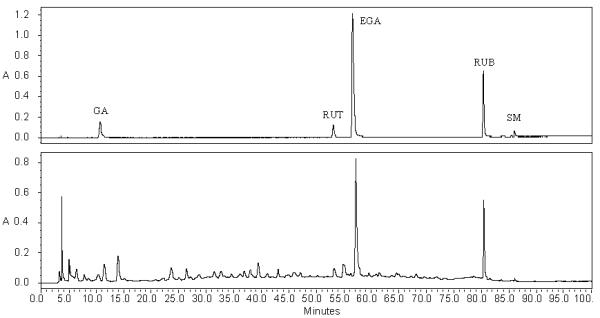
HPLC chromatograms of a standard solution containing the five marker compounds (upper) and a sample solution (lower). GA, gallic acid; RUT, rutin; EGA, ellagic acid; RUB, rubusoside; and SM, steviol monoside.
Table 4.
Content (%) of each and total of the five marker compounds in the fourteen Rubus suavissimus samples collected from a single site and different months of a growing season.
| Sample No. |
Harvest time |
Sample name |
Gallic acid |
Rutin | Ellagic acid |
Rubusoside | Steviol monoside |
Total |
|---|---|---|---|---|---|---|---|---|
| 1 | 3/2006 | SLT030106 | 0.150 | 0.133 | 0.683 | 4.846 | 0.111 | 5.92 |
| 2 | 3/2006 | SLT030406 | 0.162 | 0.101 | 0.459 | 3.519 | 0.063 | 4.30 |
| 3 | 4/2006 | SLT040506 | 0.141 | 0.102 | 0.742 | 5.167 | 0.058 | 6.21 |
| 4 | 5/2006 | SLT050606 | 0.127 | 0.100 | 0.798 | 4.993 | 0.072 | 6.09 |
| 5 | 5/2006 | SLT111206 | 0.121 | 0.104 | 0.740 | 4.948 | 0.076 | 5.99 |
| 6 | 6/2006 | SLT060206 | 0.120 | 0.107 | 0.788 | 5.496 | 0.105 | 6.62 |
| 7 | 6/2006 | SLT060706 | 0.120 | 0.141 | 0.896 | 5.322 | 0.082 | 6.56 |
| 8 | 7/2006 | SLT070806 | 0.119 | 0.148 | 0.920 | 5.618 | 0.074 | 6.88 |
| 9 | 7/2006 | SLT121306 | 0.130 | 0.124 | 0.741 | 5.497 | 0.096 | 6.59 |
| 10 | 8/2006 | SLT080906 | 0.124 | 0.054 | 0.516 | 3.776 | 0.076 | 4.55 |
| 11 | 8/2006 | SLT021406 | 0.119 | 0.145 | 0.809 | 6.350 | 0.057 | 7.48 |
| 12 | 9/2006 | SLT090306 | 0.110 | 0.118 | 0.713 | 4.473 | 0.044 | 5.46 |
| 13 | 9/2006 | SLT091006 | 0.104 | 0.082 | 0.526 | 3.868 | 0.097 | 4.68 |
| 14 | 10/2006 | SLT101106 | 0.119 | 0.096 | 0.657 | 4.296 | 0.128 | 5.30 |
| Mean | 0.13 | 0.11 | 0.71 | 4.87 | 0.08 | 5.90 | ||
| Std dev | 0.02 | 0.03 | 0.14 | 0.80 | 0.02 | 0.94 | ||
| %variation | 12.25 | 23.44 | 19.71 | 16.47 | 28.81 | 15.93 | ||
Since the fourteen leaf samples came from the same site but collected in different months, the variations observed in the content reflected most likely the temporal variations. Temporal variations in natural compounds are fairly common. For examples, camptothecin content declined steadily at 11% within a growth season (21), the contents of saponins, dencichine, flavonoid and polysaccharide in root of Panax notoginseng showed a seasonal variation (22), and the influence of harvest time on vitamin C and flavonoids in lemon tree was found (23). Obviously temporal variations are associated with the plant materials and can contribute to variations of the finished botanical products early on. Depending on what is the interest, e.g., gallic acid vs. steviol monoside, the total variations of the whole extract can vary to different degrees ranging from slight 12% to severe 29%, based on the findings of this study. In this particular circumstance, if gallic acid is the active marker, combining all harvests from the entire growth season would provide a beginning raw leaf material of 0.13% w/w gallic acid content with a 12% variation. If gallic acid must be maximized, using the March leaf material would provide a 0.16% material, some 23% higher than the combined bulk. On the other hand, if steviol monoside is the desired active compound, a variation up to 29% would be expected with a mean content of 0.71% w/w. The characteristic rubusoside content averaged almost 5% with about 16% variation. Rubusoside content in the leaves appeared to be fairly stable across temporal dimensions, although June and July might offer slightly higher content. Additional investigations over the temporal and spatial variations of the Chinese sweet tea plant are warranted.
Analysis of chromatographic fingerprints of the plant extract
The developed HPLC method was applied to assess the quality of the sweet tea plant from the angle of a chromatographic fingerprint using a chemometric software widely applied in chromatographic fingerprint analysis of TCM and related products (24-28). Chromatograms of the fourteen sweet tea leaf samples collected at different harvesting times in the same cultivation farm in Guizhou were analyzed with this software to generate a reference chromatogram. Thirty-three common characteristic peaks in the fourteen chromatograms were selected. Among them, five characteristic peaks were identified by matching their retention time (RT) and UV spectra to the respective reference compounds. In this study, peak no. 24 (RT= 57.3 min, ellagic acid) was chosen as the internal reference peak because it was present in the middle of the chromatogram and a maximum peak (Figure 3). Based on this internal reference, RRT and RPA were constructed for each peak in the chromatogram of each sweet tea leaf sample (Tables 5 and 6). The RRT and RPA for each peak served to normalize each chromatogram. Normalized chromatograms were used to generate a reference chromatogram for comparison of similarities of various samples. Although the contents of the five markers in the fourteen leaf samples varied with harvest time, the chemical profiles among them were very similar. That is, the chromatographic fingerprints showed similar compositions and full sets of detectable components, in which not only most of the common characteristic peaks were present but also the peak-to-peak distribution patterns were stable and consistent. As a result, similarities among the fourteen samples were greater than 0.98 (Figure 4). Thus it could be concluded that the samples from the same location have sufficient similarity in chemical composition to ensure the quality stability of the sweet tea leaf materials. Furthermore, the established fingerprint method for sweet tea plant is convenient and feasible as a tool for species authentication and quality assessment of the raw material.
Figure 3.
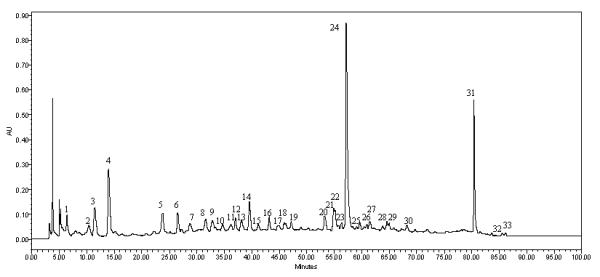
A representative fingerprint of the leaves of Rubus suavissimus showing 33 common peaks. Peak 3 is gallic acid, Peak 20 is rutin, Peak 24 is ellagic acid, Peak 31 is rubusoside, and Peak 33 is steviol monoside.
Table 5.
The relative retention time (RRT) of chromatographic peaks in the chromatograms of fourteen Rubus suavissimus leaf samples over a reference compound of ellagic acid (peak 24).
| Peak No. |
Sample number |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | |
| 1 | 0.137 | 0.135 | 0.142 | 0.139 | 0.136 | 0.138 | 0.141 | 0.141 | 0.140 | 0.137 | 0.137 | 0.137 | 0.140 | 0.140 |
| 2 | 0.183 | 0.180 | 0.183 | 0.182 | 0.180 | 0.182 | 0.184 | 0.183 | 0.182 | 0.182 | 0.182 | 0.183 | 0.183 | 0.181 |
| 3 | 0.201 | 0.197 | 0.201 | 0.200 | 0.199 | 0.199 | 0.202 | 0.201 | 0.200 | 0.201 | 0.201 | 0.201 | 0.200 | 0.199 |
| 4 | 0.246 | 0.240 | 0.243 | 0.244 | 0.243 | 0.243 | 0.245 | 0.246 | 0.244 | 0.245 | 0.245 | 0.246 | 0.244 | 0.243 |
| 5 | 0.415 | 0.413 | 0.415 | 0.414 | 0.412 | 0.414 | 0.416 | 0.416 | 0.415 | 0.415 | 0.415 | 0.415 | 0.415 | 0.414 |
| 6 | 0.464 | 0.462 | 0.464 | 0.463 | 0.462 | 0.463 | 0.465 | 0.465 | 0.464 | 0.464 | 0.464 | 0.464 | 0.464 | 0.463 |
| 7 | 0.503 | 0.501 | 0.503 | 0.502 | 0.501 | 0.503 | 0.504 | 0.504 | 0.503 | 0.503 | 0.504 | 0.503 | 0.503 | 0.503 |
| 8 | 0.552 | 0.550 | 0.552 | 0.551 | 0.550 | 0.552 | 0.552 | 0.552 | 0.552 | 0.551 | 0.552 | 0.552 | 0.552 | 0.551 |
| 9 | 0.574 | 0.573 | 0.573 | 0.573 | 0.571 | 0.573 | 0.574 | 0.574 | 0.574 | 0.573 | 0.573 | 0.573 | 0.573 | 0.572 |
| 10 | 0.606 | 0.605 | 0.605 | 0.606 | 0.605 | 0.605 | 0.606 | 0.607 | 0.606 | 0.606 | 0.606 | 0.607 | 0.606 | 0.606 |
| 11 | 0.633 | 0.631 | 0.635 | 0.632 | 0.631 | 0.633 | 0.635 | 0.637 | 0.636 | 0.632 | 0.633 | 0.633 | 0.632 | 0.633 |
| 12 | 0.647 | 0.645 | 0.646 | 0.646 | 0.645 | 0.646 | 0.647 | 0.648 | 0.647 | 0.646 | 0.647 | 0.647 | 0.646 | 0.646 |
| 13 | 0.665 | 0.667 | 0.665 | 0.665 | 0.664 | 0.665 | 0.666 | 0.666 | 0.665 | 0.665 | 0.665 | 0.668 | 0.665 | 0.665 |
| 14 | 0.691 | 0.691 | 0.691 | 0.691 | 0.690 | 0.691 | 0.692 | 0.692 | 0.692 | 0.691 | 0.691 | 0.691 | 0.691 | 0.691 |
| 15 | 0.720 | 0.720 | 0.720 | 0.720 | 0.719 | 0.720 | 0.720 | 0.721 | 0.720 | 0.720 | 0.720 | 0.720 | 0.720 | 0.719 |
| 16 | 0.754 | 0.742 | 0.754 | 0.754 | 0.753 | 0.754 | 0.754 | 0.755 | 0.754 | 0.753 | 0.754 | 0.754 | 0.754 | 0.754 |
| 17 | 0.787 | 0.771 | 0.785 | 0.791 | 0.791 | 0.791 | 0.791 | 0.788 | 0.797 | 0.786 | 0.790 | 0.781 | 0.785 | 0.786 |
| 18 | 0.807 | 0.806 | 0.801 | 0.804 | 0.804 | 0.803 | 0.802 | 0.803 | 0.811 | 0.806 | 0.807 | 0.807 | 0.807 | 0.803 |
| 19 | 0.823 | 0.823 | 0.824 | 0.824 | 0.823 | 0.824 | 0.824 | 0.825 | 0.835 | 0.823 | 0.824 | 0.824 | 0.824 | 0.824 |
| 20 | 0.931 | 0.929 | 0.931 | 0.932 | 0.931 | 0.931 | 0.932 | 0.933 | 0.931 | 0.931 | 0.932 | 0.932 | 0.930 | 0.931 |
| 21 | 0.960 | 0.958 | 0.959 | 0.960 | 0.960 | 0.959 | 0.959 | 0.960 | 0.960 | 0.959 | 0.959 | 0.959 | 0.958 | 0.959 |
| 22 | 0.964 | 0.963 | 0.964 | 0.964 | 0.966 | 0.964 | 0.965 | 0.964 | 0.964 | 0.964 | 0.965 | 0.964 | 0.964 | 0.964 |
| 23 | 0.984 | 0.983 | 0.984 | 0.985 | 0.984 | 0.984 | 0.985 | 0.984 | 0.985 | 0.983 | 0.984 | 0.984 | 0.984 | 0.984 |
| 24 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
| 25 | 1.042 | 1.042 | 1.043 | 1.043 | 1.042 | 1.042 | 1.043 | 1.042 | 1.043 | 1.042 | 1.043 | 1.042 | 1.043 | 1.042 |
| 26 | 1.065 | 1.064 | 1.065 | 1.066 | 1.065 | 1.065 | 1.066 | 1.065 | 1.066 | 1.064 | 1.066 | 1.065 | 1.065 | 1.066 |
| 27 | 1.074 | 1.074 | 1.074 | 1.076 | 1.075 | 1.074 | 1.075 | 1.074 | 1.075 | 1.074 | 1.075 | 1.074 | 1.075 | 1.075 |
| 28 | 1.129 | 1.128 | 1.129 | 1.130 | 1.130 | 1.129 | 1.130 | 1.129 | 1.130 | 1.128 | 1.130 | 1.129 | 1.129 | 1.130 |
| 29 | 1.136 | 1.136 | 1.136 | 1.137 | 1.136 | 1.136 | 1.137 | 1.136 | 1.137 | 1.135 | 1.136 | 1.136 | 1.136 | 1.137 |
| 30 | 1.192 | 1.192 | 1.193 | 1.193 | 1.187 | 1.193 | 1.193 | 1.191 | 1.193 | 1.191 | 1.193 | 1.192 | 1.193 | 1.193 |
| 31 | 1.404 | 1.406 | 1.407 | 1.407 | 1.407 | 1.407 | 1.407 | 1.405 | 1.408 | 1.404 | 1.405 | 1.404 | 1.407 | 1.408 |
| 32 | 1.459 | 1.461 | 1.463 | 1.463 | 1.463 | 1.462 | 1.462 | 1.460 | 1.463 | 1.459 | 1.460 | 1.459 | 1.462 | 1.464 |
| 33 | 1.503 | 1.505 | 1.506 | 1.503 | 1.507 | 1.506 | 1.506 | 1.504 | 1.507 | 1.503 | 1.504 | 1.503 | 1.506 | 1.508 |
Table 6.
The relative peak area (RPA) of chromatographic peaks in the chromatograms of fourteen Rubus suavissimus leaf samples over a reference compound of ellagic acid (peak 24).
| Peak No. |
Sample number |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | |
| 1 | 0.067 | 0.074 | 0.029 | 0.006 | 0.034 | 0.010 | 0.006 | 0.006 | 0.005 | 0.034 | 0.018 | 0.012 | 0.013 | 0.006 |
| 2 | 0.038 | 0.031 | 0.065 | 0.064 | 0.059 | 0.058 | 0.109 | 0.117 | 0.079 | 0.081 | 0.067 | 0.081 | 0.082 | 0.093 |
| 3 | 0.196 | 0.308 | 0.167 | 0.142 | 0.146 | 0.135 | 0.108 | 0.115 | 0.120 | 0.213 | 0.101 | 0.137 | 0.174 | 0.171 |
| 4 | 0.214 | 0.549 | 0.341 | 0.358 | 0.210 | 0.341 | 0.305 | 0.226 | 0.268 | 0.383 | 0.265 | 0.213 | 0.424 | 0.304 |
| 5 | 0.104 | 0.064 | 0.116 | 0.105 | 0.102 | 0.106 | 0.178 | 0.167 | 0.119 | 0.150 | 0.104 | 0.142 | 0.143 | 0.184 |
| 6 | 0.106 | 0.197 | 0.110 | 0.097 | 0.084 | 0.102 | 0.052 | 0.049 | 0.073 | 0.120 | 0.059 | 0.048 | 0.117 | 0.093 |
| 7 | 0.048 | 0.085 | 0.050 | 0.040 | 0.031 | 0.042 | 0.018 | 0.017 | 0.025 | 0.020 | 0.023 | 0.015 | 0.022 | 0.026 |
| 8 | 0.043 | 0.049 | 0.062 | 0.053 | 0.062 | 0.067 | 0.068 | 0.063 | 0.067 | 0.043 | 0.062 | 0.042 | 0.059 | 0.070 |
| 9 | 0.053 | 0.043 | 0.071 | 0.045 | 0.056 | 0.057 | 0.104 | 0.092 | 0.067 | 0.044 | 0.062 | 0.046 | 0.058 | 0.072 |
| 10 | 0.044 | 0.053 | 0.040 | 0.030 | 0.030 | 0.041 | 0.027 | 0.017 | 0.032 | 0.031 | 0.027 | 0.013 | 0.037 | 0.035 |
| 11 | 0.052 | 0.069 | 0.036 | 0.032 | 0.031 | 0.030 | 0.029 | 0.034 | 0.024 | 0.030 | 0.058 | 0.026 | 0.017 | 0.031 |
| 12 | 0.155 | 0.172 | 0.065 | 0.054 | 0.047 | 0.055 | 0.026 | 0.028 | 0.032 | 0.029 | 0.055 | 0.036 | 0.033 | 0.034 |
| 13 | 0.059 | 0.050 | 0.067 | 0.064 | 0.065 | 0.086 | 0.060 | 0.069 | 0.082 | 0.044 | 0.082 | 0.030 | 0.038 | 0.060 |
| 14 | 0.110 | 0.159 | 0.133 | 0.123 | 0.110 | 0.125 | 0.112 | 0.108 | 0.145 | 0.152 | 0.092 | 0.073 | 0.157 | 0.181 |
| 15 | 0.045 | 0.025 | 0.032 | 0.027 | 0.031 | 0.034 | 0.039 | 0.038 | 0.028 | 0.028 | 0.036 | 0.035 | 0.030 | 0.041 |
| 16 | 0.049 | 0.051 | 0.062 | 0.056 | 0.051 | 0.064 | 0.034 | 0.036 | 0.073 | 0.039 | 0.043 | 0.015 | 0.054 | 0.065 |
| 17 | 0.012 | 0.009 | 0.037 | 0.015 | 0.014 | 0.025 | 0.024 | 0.019 | 0.032 | 0.015 | 0.006 | 0.053 | 0.034 | 0.015 |
| 18 | 0.072 | 0.010 | 0.034 | 0.026 | 0.027 | 0.022 | 0.088 | 0.069 | 0.038 | 0.031 | 0.014 | 0.046 | 0.021 | 0.046 |
| 19 | 0.047 | 0.033 | 0.039 | 0.033 | 0.026 | 0.029 | 0.051 | 0.047 | 0.026 | 0.044 | 0.024 | 0.060 | 0.059 | 0.059 |
| 20 | 0.115 | 0.129 | 0.081 | 0.075 | 0.057 | 0.080 | 0.093 | 0.093 | 0.076 | 0.052 | 0.082 | 0.098 | 0.091 | 0.101 |
| 21 | 0.076 | 0.071 | 0.082 | 0.078 | 0.103 | 0.078 | 0.088 | 0.077 | 0.047 | 0.053 | 0.020 | 0.076 | 0.070 | 0.064 |
| 22 | 0.100 | 0.108 | 0.092 | 0.074 | 0.037 | 0.089 | 0.053 | 0.054 | 0.103 | 0.023 | 0.091 | 0.121 | 0.118 | 0.117 |
| 23 | 0.028 | 0.049 | 0.025 | 0.026 | 0.028 | 0.035 | 0.023 | 0.032 | 0.024 | 0.015 | 0.018 | 0.031 | 0.032 | 0.036 |
| 24 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
| 25 | 0.037 | 0.033 | 0.022 | 0.022 | 0.024 | 0.029 | 0.021 | 0.023 | 0.024 | 0.022 | 0.041 | 0.031 | 0.031 | 0.030 |
| 26 | 0.034 | 0.022 | 0.014 | 0.013 | 0.012 | 0.016 | 0.017 | 0.015 | 0.017 | 0.008 | 0.026 | 0.023 | 0.024 | 0.015 |
| 27 | 0.047 | 0.029 | 0.034 | 0.033 | 0.027 | 0.032 | 0.037 | 0.034 | 0.030 | 0.020 | 0.038 | 0.037 | 0.042 | 0.032 |
| 28 | 0.040 | 0.024 | 0.043 | 0.036 | 0.027 | 0.042 | 0.048 | 0.038 | 0.027 | 0.030 | 0.038 | 0.039 | 0.046 | 0.039 |
| 29 | 0.013 | 0.008 | 0.023 | 0.024 | 0.020 | 0.031 | 0.026 | 0.031 | 0.019 | 0.013 | 0.016 | 0.049 | 0.035 | 0.035 |
| 30 | 0.037 | 0.026 | 0.024 | 0.024 | 0.015 | 0.026 | 0.023 | 0.025 | 0.025 | 0.012 | 0.038 | 0.028 | 0.019 | 0.021 |
| 31 | 0.428 | 0.464 | 0.420 | 0.378 | 0.405 | 0.421 | 0.352 | 0.369 | 0.345 | 0.438 | 0.364 | 0.378 | 0.434 | 0.450 |
| 32 | 0.009 | 0.004 | 0.006 | 0.005 | 0.005 | 0.006 | 0.004 | 0.004 | 0.003 | 0.003 | 0.006 | 0.003 | 0.003 | 0.003 |
| 33 | 0.011 | 0.010 | 0.010 | 0.004 | 0.012 | 0.007 | 0.006 | 0.006 | 0.006 | 0.007 | 0.010 | 0.006 | 0.006 | 0.007 |
Figure 4.
The overlay HPLC fingerprints of the fourteen Rubus suavissimus samples (S1-S14) by a similarity evaluation system that assesses sample similarity against a generated reference chromatogram (R).
The main purpose of this study was to establish an HPLC fingerprint for Rubus suavissimus using the samples collected from a site in the plant’s origin. This fingerprint was to show and represent as many components as possible in the raw materials in order to set a sound basis for future quality assessment and control. To achieve that goal a longer analysis time (100 minutes) was adopted. Although 100 min retention time seemed to be longer than most for quantitative analysis purposes, it is often acceptable for fingerprint analysis. For example, in the development of chromatographic fingerprint for Rheum tanguticum, the fingerprinting analysis time was 142 min (29). Had the purposes of this investigation been limited to quantitatively measure the five compounds, the analysis time would have been much shorter. However, our purposes also included fingerprint analysis, which required not only baseline separation of the five known compounds but also other unknown components of a whole leaf sample. Needless to say, there is room to shorten the analytical time to achieve the same fingerprint quality by using other means such as the faster speed UPLC. However, this fingerprinting method and the quality it presents set the basis for adaptations.
The entire fingerprint of sweet tea plant was divided into three retention time zones to further illustrate the distribution of various classes of compounds in the chromatogram (Figure 5) as follows: Zone A (0-20 min) contained four peaks (peaks 1-4) featuring peak 3 (gallic acid) and 4 (a possible gallic acid analogue) and representing the gallotannins according to a previous investigation (1). Since gallic acid is an anti-angiogenic agent, Zone A would possess anti-angiogenic property as well. Zone B (20 to 70 min) consisted of 26 of the 33 total peaks and is most abundant and diverse in chemical composition. Two classes of compounds were identified in Zone B and most of them are flavonoids and ellagitannins by comparing their UV spectra with those of rutin (peak 20) and ellagic acid (peak 24). Zone C (70 to 90 min) showing only three observed peaks (peak 31-33) contained the characteristic compounds of this plant — the diterpene glycosides, represented by peak 31 (rubusoside) and peak 33 (steviol monoside) while peak 32 can be also tentatively identified as another diterpene glycoside based on its UV spectrum. Rubusoside (peak 31) was the most dominant and abundant diterpene glycoside. In fact, rubusoside in the plant leaves was reported as high as 5% w/w (6, 30) and our results were in full agreement. Rubusoside and other diterpene glycosides in this zone are signature compounds for the sweet tea plant and can be used effectively to chemically authenticate the raw materials from this plant.
Figure 5.
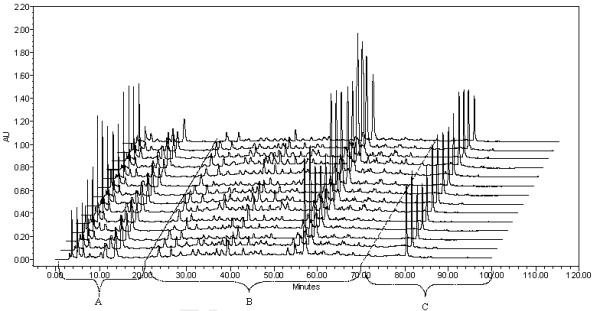
HPLC Fingerprints of the fourteen samples viewed as three zones. Zone A containing the antiangiogenic agent gallic acid is defined as an active zone; Zone B is viewed as the anchoring fingerprint zone showing abundant and diverse ellagitannins and flavonoids; Zone C is characteristic of this plant featuring the diterpene glycosides that give this plant the name of sweet tea plant.
ACKNOWLEDGEMENTS
The project described was supported primarily by Grant Number R21AT002882 from the National Center for Complementary & Alternative Medicine. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Complementary & Alternative Medicine or the National Institutes of Health. The LSU Agricultural Center also provided partial monetary support from the Technology Transfer Fund.
LITERATURE CITED
- (1).Liu Z, Schwimer J, Liu D, Lewis J, Greenway FL, York DA, Woltering EA. Gallic acid is partially responsible for the antiangiogenic activities of Rubus leaf extract. Phytother. Res. 2006;20:806–813. doi: 10.1002/ptr.1966. [DOI] [PubMed] [Google Scholar]
- (2).Feng J, Xin N. Advance in studies on chemical constituents and pharmacological action of Rubus suavissimus S. Lee and Lithocarpus polystachyus Rehd. Lishizhen Medicine and Materia Medica Res. 2007;18:1089–1090. [Google Scholar]
- (3).Ohtani K, Aikawa Y, Kasai R, Chou WH, Yamasaki K, Tanaka O. Minor diterpene glycosides from sweet leaves of Rubus suavissimus. Phytochemistry. 1992;31:1553–1559. [Google Scholar]
- (4).Li H, Tanaka T, Zhang YJ, Yang CR, Kouno I. Rubusuaviins A-F, Monomeric and oligomeric ellagitannins from Chinese sweet tea and their α-amylase inhibitory activity. Chem. Pharm. Bull. 2007;55:1325–1331. doi: 10.1248/cpb.55.1325. [DOI] [PubMed] [Google Scholar]
- (5).Sugimoto N, Kikuchi H, Yamazaki T, Maitani T. Polyphenolic constituents from the leaves of Rubus suavissimus. Nat. Med. 2001;55:219–223. [Google Scholar]
- (6).Tanaka T, Kawamura K, Kitahara T, Kohda H, Tanaka O. Ent-labdane-type diterpene glucosides from leaves of Rubus chingii. Phytochemistry. 1984;23:615–621. [Google Scholar]
- (7).Wang JX, Lv HC. Studies on the chemical constituents of Rubus suavissimus S. Lee. Zhongyaocai. 2007;30:800–802. [PubMed] [Google Scholar]
- (8).Hirono S, Chou WH, Kasai R. Sweet and bitter diterpene-glucosides from leaves of Rubus suavissimus. Chem. Pharm. Bull. 1990;38:1743–1744. [Google Scholar]
- (9).Ohtani K, Aikawa Y, Ishikawa H, Kasai R, Kitahata S, Mizutani K, Doi S, Nakaura M, Tanaka O. Futher study on the 1,4-alpha-transglucosylation of rubusoside, a sweet steviol bisglucoside from Rubus suavissimus. Agric. Biol. Chem. 1991;55:449–453. [PubMed] [Google Scholar]
- (10).Devipriya N, Sudheer AR, Menon VP. Dose-response effect of ellagic acid on circulatory antioxidants and lipids during alcohol-induced toxicity in experimental rats. Fundam Clin. Pharmacol. 2007;21:621–630. doi: 10.1111/j.1472-8206.2007.00551.x. [DOI] [PubMed] [Google Scholar]
- (11).Bell C, Hawthorne S. Ellagic acid, pomegranate and prostate cancer -- a mini review. J. Pharm. Pharmacol. 2008;60:139–144. doi: 10.1211/jpp.60.2.0001. [DOI] [PubMed] [Google Scholar]
- (12).Zhang Y, Seeram NP, Lee R, Feng L, Heber D. Isolation and identification of strawberry phenolics with antioxidant and human cancer cell antiproliferative properties. J. Agric. Food Chem. 2008;56:670–675. doi: 10.1021/jf071989c. [DOI] [PubMed] [Google Scholar]
- (13).Papoutsi Z, Kassi E, Chinou I, Halabalaki M, Skaltsounis LA, Moutsatsou P. Walnut extract (Juglans regia L.) and its component ellagic acid exhibit anti-inflammatory activity in human aorta endothelial cells and osteoblastic activity in the cell line KS483. Br. J. Nutr. 2008;99:715–722. doi: 10.1017/S0007114507837421. [DOI] [PubMed] [Google Scholar]
- (14).Rogerio AP, Fontanari C, Borducchi E, Keller AC, Russo M, Soares EG, Albuquerque DA, Faccioli LH. Anti-inflammatory effects of Lafoensia pacari and ellagic acid in a murine model of asthma. Eur. J. Pharmacol. 2008;580:262–270. doi: 10.1016/j.ejphar.2007.10.034. [DOI] [PubMed] [Google Scholar]
- (15).Vukics V, Kery A, Bonn GK, Guttman A. Major flavonoid components of heartsease (Viola tricolor L.) and their antioxidant activities. Anal. Bioanal. Chem. 2008;390:1917–1925. doi: 10.1007/s00216-008-1885-3. [DOI] [PubMed] [Google Scholar]
- (16).Kartika H, Li QX, Wall MM, Nakamoto ST, Iwaoka WT. Major phenolic acids and total antioxidant activity in Mamaki leaves, Pipturus albidus. J. Food Sci. 2007;72:696–701. doi: 10.1111/j.1750-3841.2007.00530.x. [DOI] [PubMed] [Google Scholar]
- (17).Fang SH, Rao YK, Tzeng YM. Anti-oxidant and inflammatory mediator’s growth inhibitory effects of compounds isolated from Phyllanthus urinaria. J. Ethnopharmacol. 2008;116:333–340. doi: 10.1016/j.jep.2007.11.040. [DOI] [PubMed] [Google Scholar]
- (18).Liu Z, Schwimer J, Liu D, Greenway FL, Anthony CT, Woltering EA. Black raspberry extract and fractions contain angiogenesis inhibitors. J. Agric. Food Chem. 2005;53:3909–3915. doi: 10.1021/jf048585u. [DOI] [PubMed] [Google Scholar]
- (19).Lin CZ, Zhu CC, Yang JY. Determination of two ent-kaurane diterpenoids in Isodon lophanthoides var. gerardianus. Lishizhen Medicine Materia Medica Res. 2007;18:1549–1551. [Google Scholar]
- (20).Gui MY, Li XW, Wang BZ, Xu JQ, Jin YR. Content of diterpenoids in Rabdosia excisa in different growth periods via RP-HPLC. J. Jilin University. 2007;45:125–127. [Google Scholar]
- (21).Liu Z, Carpenter SB, Bourgeois WJ, Yu Y, Constantin RJ, Falcon MJ, Adams JC. Variation in the secondary metabolite camptothecin in relation to tissue age and season in Camptotheca acuminata (Nyssaceae) Tree Physiol. 1998;18:265–270. doi: 10.1093/treephys/18.4.265. [DOI] [PubMed] [Google Scholar]
- (22).Dong TT, Cui XM, Song ZH, Zhao KJ, Ji ZN, Lo CK, Tsim KW. Chemical assessment of roots of Panax notoginseng in China: regional and seasonal variations in its active constituents. J. Agric. Food Chem. 2003;51:4617–4623. doi: 10.1021/jf034229k. [DOI] [PubMed] [Google Scholar]
- (23).González-Molina E, Moreno DA, García-Viguera C. Genotype and harvest time influence the phytochemical quality of Fino lemon juice (Citrus limon (L.) Burm. F.) for industrial use. J. Agric. Food Chem. 2008;56:1669–1675. doi: 10.1021/jf073282w. [DOI] [PubMed] [Google Scholar]
- (24).Chen C, Zhang H, Xiao W, Yong ZP, Bai N. High-performance liquid chromatographic fingerprint analysis for different origins of sea buckthorn berries. J. Chromatogr. A. 2007;1154:250–259. doi: 10.1016/j.chroma.2007.03.097. [DOI] [PubMed] [Google Scholar]
- (25).Xie PS, Chen SB, Liang YZ, Wang XH, Tian RT, Upton R. Chromatographic fingerprint analysis-a rational approach for quality assessment of traditional Chinese herbal medicine. J. Chromatogr. A. 2006;1112:171–180. doi: 10.1016/j.chroma.2005.12.091. [DOI] [PubMed] [Google Scholar]
- (26).Yang LW, Wu DH, Tang X, Peng W, Wang XR, Ma Y, Su WW. Fingerprint quality control of Tianjihuang by high-performance liquid chromatography—photodiode array detection. J. Chromatogr. A. 2005;1070:35–42. doi: 10.1016/j.chroma.2005.02.081. [DOI] [PubMed] [Google Scholar]
- (27).Chen Y, Yan Y, Xie MY, Nie SP, Liu W, Gong XF, Wang YX. Development of a chromatographic fingerprint for the chloroform extracts of Ganoderma lucidum by HPLC and LC—MS. J. Pharma. Biomed. Anal. 2008;47:469–477. doi: 10.1016/j.jpba.2008.01.039. [DOI] [PubMed] [Google Scholar]
- (28).Han C, Shen Y, Chen JH, Lee FSC, Wang XR. HPLC fingerprinting and LC—TOF-MS analysis of the extract of Pseudostellaria heterophylla (Miq.) Pax root. J. Chromatogr. B. 2008;862:125–131. doi: 10.1016/j.jchromb.2007.11.041. [DOI] [PubMed] [Google Scholar]
- (29).Wei. Jin, Ri-Li Ge, Quan-Jia Wei. Development of high-performance liquid chromatographic fingerprint for the quality control of Rheum tanguticum Maxim. ex Balf. J. Chromatogr. A. 2006;1132:320–324. doi: 10.1016/j.chroma.2006.08.022. [DOI] [PubMed] [Google Scholar]
- (30).Seto T, Tanaka T, Tanaka O, Naruhashi N. β Glucosyl esters of 19α hydroxyursolic acid derivatives in leaves of Rubus species. Phytochemistry. 1984;23:2829–2834. [Google Scholar]