Abstract
Nuclear pore complexes (NPCs) facilitate nucleocytoplasmic transport. These massive assemblies comprise an eight-fold symmetric scaffold of architectural proteins and central-channel phenylalanine-glycine-repeat proteins forming the transport barrier. We determined the Nup85•Seh1 structure, a module in the heptameric Nup84 complex. Structural, biochemical, and genetic analyses position the Nup84 complex in two peripheral NPC rings. We establish a conserved tripartite element, the ancestral coatomer element ACE1, that reoccurs in several nucleoporins and vesicle coat proteins, providing structural evidence of coevolution from a common ancestor. We identify interactions that define the organization of the Nup84 complex based on comparison with vesicle coats and confirmed the sites by mutagenesis. We propose the NPC scaffold, like vesicle coats, is composed of polygons with vertices and edges forming a membrane-proximal lattice providing docking sites for additional nucleoporins.
Exchange of macromolecules across the nuclear envelope is exclusively mediated by NPCs (1–3). Whereas much progress has been made understanding the soluble factors mediating nucleocytoplasmic transport, the structure of the ~40–60 MDa NPC itself is still largely enigmatic. Cryo-electrontomography (cryo-ET) and cryo-electronmicroscopy (cryo-EM) have established the NPC structure at low resolution (4–6). Crystal structures of scaffold NPC components are emerging (7–10), but the resolution gap still precludes fitting into the cryo-ET structure. Overall, the NPC has eight-fold rotational symmetry with an outer diameter of ~100 nm and a core scaffold ring ~30 nm wide. The central FG-repeat containing transport channel measures ~40 nm in diameter, defining the maximum size of substrates (11).
The modularity of the NPC assembly suggests a path toward a high-resolution structure (12). Of the ~30 bona fide nucleoporins (Nups) that comprise the NPC, only a core subset is stably attached (13). In S. cerevisiae, this core includes two essential complexes; the heptameric Nup84 complex and the heteromeric Nic96-containing complex (hereafter called the Nic96 complex; unless noted all proteins are from S. cerevisiae). The Nup84 complex is composed of one copy each of Nup84, Nup85, Nup120, Nup133, Nup145C, Sec13 and Seh1. It self-assembles from recombinant proteins in vitro and forms a branched Y-shaped structure (14). Deletion or depletion of individual components of the Nup84 complex leads to severe assembly defects in many organisms (15–17). The Nic96 complex is less well characterized, but appears to contain the architectural nucleoporins Nup157/170, Nup188, Nup192, Nup53, and Nup59 (18–20). β-propellers and stacked α-helical domains form the building blocks of the constituents of the Nup84 and Nic96 complexes (12, 21). Because vesicle coats (including COPI, COPII, and clathrin) share similar elements, a common ancestry has been hypothesized despite very low sequence homology and the absence of experimental structural evidence (22).
A recent computer-generated model of the NPC, based on a plethora of primary data from different sources, places the Nup84 complex at the NPC periphery, sandwiching the Nic96 complex in the center (23). In contrast, a model solely based on the structure of the Nup145C•Sec13 heterodimer and crystal packing interactions has been proposed and is diametrically opposed to the computer model (9).
We solved the structure of a complex of Nup85 residues 1–564 (of 744) and intact Seh1 (referred to as Nup85•Seh1) at 3.5 Å (Table S1). Seh1 and Nup85 form distinct units in a tightly associated complex (Fig. 1 & S1). Seh1 folds into an open six-bladed β-propeller structure. The blades fan out consecutively around a central axis, typical for canonical β-propeller structures (24). Between blades 1 and 6, the N-terminus of Nup85 is inserted and forms a three-stranded blade that completes the Seh1 propeller in trans. Following its N-terminal insertion blade, Nup85 forms a compact cuboid structure composed of 20 helices, with two distinct modules, referred to as ‘crown’ and ‘trunk’. Helices α1–α3 (residues 100–200) meander along one side of the trunk; the other side is formed by helices α12–α19 (residues 362–509) running in the opposite direction in an anti-parallel zig-zag to the C-terminus. The trunk elements are separated by an intervening crown composed of helices α4–α11 (residues 201–361) that form a distinct bundle that caps one end of the trunk. Helices α5–α10 in the crown module are almost perpendicular to the helices in the trunk.
Fig. 1. Structure of the Nup85•Seh1 complex.
The structure of the heterodimeric Nup85•Seh1 complex is shown. Nup85 has a trunk (orange, helices α1–α3 and α12–α20) and a crown (blue, helices α4–α11) module. The β-strands at the extreme N-terminus of Nup85 form an insertion blade, which complete the Seh1 (green) β-propeller.
In the asymmetric unit of the crystal, two heterodimers are aligned along a non-crystallographic dyad generating patches of contacts (Fig. S2). This interaction is unlikely to be functionally meaningful, as the contact residues are poorly conserved in orthologs. Moreover, analysis of Nup85•Seh1 by analytical ultracentrifugation (AUC) showed a single species of ~104 kDa with a hydrodynamic radius of 4.4 nm (Fig. S3). This hydrodynamic radius is close to the theoretical value calculated from the atomic coordinates using HYDROPRO (25) and reflects the elongated shape of the 103 kDa Nup85•Seh1 complex (a spherical protein of 220 kDa would have the same radius). Gel filtration also showed that Nup85•Seh1 is a single 103 kDa heterodimer at concentrations up to 20 mg/ml (Fig. S4). Hence, we restrict our analysis to this heterodimer.
The connectivity and topology of secondary structure elements and the three-dimensional folds of Nup85•Seh1 and Nup145C•Sec13 (9) are remarkably similar (Fig. 2A), despite very low sequence identity between Nup85 and Nup145C (10%) and moderate identity between Seh1 and Sec13 (32%). Like Nup85, Nup145C has an N-terminal three-stranded β-sheet that provides a seventh blade to close open β-propeller of Sec13. The trunk and crown modules of Nup145C are also similar to those in Nup85, although their relative orientation is modestly different in the two proteins.
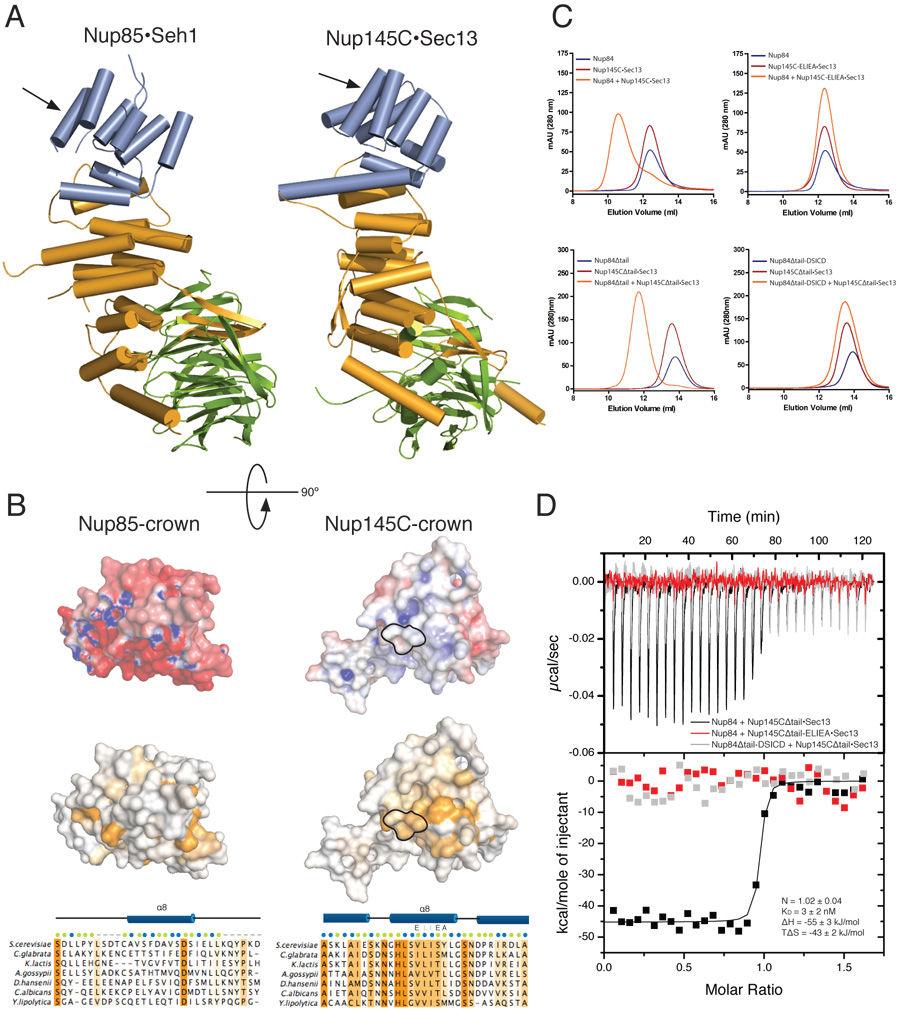
Fig. 2. Comparison of Nup85•Seh1 and Nup145C•Sec13 and identification of the Nup84•Nup145C crown-crown binding interface.
(A) The topologies of the Nup85•Seh1 (left) and Nup145C•Sec13 (right, PDB code 3BG1 (9)) complexes are shown, illustrating an overall similarity with three shared structural elements — trunk, crown, and β-propeller. Colors are assigned as in Figure 1. (B) Surface representations of the crowns of Nup85 and Nup145C are shown colored according to electrostatic surface potentials (top) and sequence conservation (bottom) in a view rotated 90° from (A). Sequence conservation is based on the phylogenetic tree of budding yeasts (39) and is colored from white (not conserved) to orange (conserved). A partial sequence alignment of helix α8 (indicated by arrows in (A)) is also shown with surface exposed residues indicated by green dots, residues buried in the hydrophobic core by blue dots, and residues not modeled in the structure by dashes. Mutations made in this helix in Nup145C are shown above the sequence alignment and the corresponding residues are outlined in the surface representations of Nup145C. (C) In the upper panels gel filtration data of Nup84 alone, Nup145C•Sec13 (wild type or -ELIEA mutant) alone, and Nup84 plus Nup145C•Sec13 (wild type or -ELIEA mutant) are shown. The shift in the Nup84 plus wild type Nup145C•Sec13 chromatogram indicates complex formation and is absent in the case of the -ELIEA mutant. In the lower panels gel filtration data of Nup145C•Sec13 alone, Nup84 alone (wild type or -DSICD mutant) alone, and Nup145C•Sec13 plus Nup84 (wild type or -DSICD mutant) are shown. The shift in the Nup145C•Sec13 plus wild type Nup84 chromatogram indicates complex formation and is absent in the case of the -DSICD mutant. (D) Isothermal titration calorimetry data illustrating high-affinity binding for wild-type Nup145C•Sec13 and Nup84 (black). Experimental values for N, KD, ΔH, and TΔS are shown. In contrast, binding is lost for both crown-surface mutants Nup84-DSICD (grey) and Nup145C-ELIEA (red).
The most conserved regions of Nup85 are involved in the interaction with Seh1. The corresponding interface between Nup145C and Sec13 is also well conserved, but Nup145C has an additional highly conserved surface on the crown module, around helix α8, that is not observed in Nup85 (Fig. 2B). This region is reasonably polar and poorly conserved in Nup85 but highly conserved and distinctly hydrophobic in Nup145C, suggesting a protein-protein interaction site. Nup84 and Nup120 bind to roughly opposite sides of Nup145C•Sec13 in the Y-shaped complex (14), and the C-terminal helical region of Nup145C is necessary for binding Nup120 (Fig. S5). Thus, we hypothesized that the α8 crown surface of Nup145C is the binding site for Nup84.
To test this hypothesis, we mutated the Nup145C sequence “VLISY” in α8 to “ELIEA,” introducing two negative charges and eliminating a conserved aromatic side chain on the crown surface (Fig. 2B). The overall structure of Nup145C did not appear to be perturbed by this modification: (i) Nup145C-ELIEA•Sec13 bound to Nup120 to form a 1:1 complex indistinguishable from Nup145C•Sec13 in gel-filtration experiments (Fig. S6); (ii) Nup145C•Sec13 and Nup145C-ELIEA•Sec13 complexes had comparable thermostability (Fig. S7) and (iii) showed identical behavior in gel filtration (Fig. 2C). The ELIEA mutation completely eliminated Nup84 binding. In isothermal-titration-calorimetry (ITC) experiments, Nup84 bound wild-type Nup145C•Sec13 tightly (KD = 3 ± 2 nM; 1:1 stoichiometry) but not Nup145C-ELIEA•Sec13 (Fig. 2D). Similarly, Nup84 formed a stable complex with Nup145C•Sec13 but not with Nup145C-ELIEA•Sec13 in gel-filtration (Fig. 2C). We conclude that the Nup84 binding site on Nup145C includes the exposed surface of helix α8.
To determine the consequences of abolishing the Nup84 binding site on Nup145C in vivo, we introduced the Nup145C-ELIEA mutation into the NUP145 gene in yeast. Strains carrying NUP145-ELIEA in a ΔNUP145/NUP84-GFP or ΔNUP145/NUP133-GFP background displayed a marked defect in incorporating Nup84-GFP and Nup133-GFP into the NPC ( Fig. 3 & S8). Compared to wild type, a significantly larger fraction of GFP-tagged proteins were found in the cytoplasm, indicating that the Nup84 binding interface on Nup145C is crucial in recruiting both Nup133 and Nup84 to the NPC (Fig. S10). In addition, nuclear pores were clustered into discrete foci on the nuclear envelope of the strains expressing Nup145C-ELIEA, indicative of severe NPC assembly defects and similar to Nup84 and Nup133 null strains (26, 27). Cells expressing wild-type Nup145C demonstrated the expected punctate nuclear rim staining in both Nup84-GFP and Nup133-GFP strains. Thus, disruption of the Nup84 binding site on Nup145C affects NPC assembly and function and causes loss of Nup84 and Nup133 from pores. The loss of Nup133 can be rationalized because it is attached to the Y-shaped Nup84 complex through a binary interaction with Nup84 (8, 14). Some Nup84 and Nup133 proteins remain associated with nuclear pores in the Nup145C-ELIEA expressing strains, arguing for the existence of additional weaker attachment sites for both proteins in the NPC. It has been shown that an ALPS membrane-binding motif is present in Nup133 (28). Because Nup133 and Nup84 are tightly associated (8), the ALPS motif might be weakly functional in recruiting Nup133•Nup84 to the NPC even when the Nup84•Nup145C interaction is compromised.
Fig. 3. Elimination of the Nup84 binding site on Nup145C results in nuclear pore assembly defects in vivo.
NUP145/NUP84-GFP and NUP145-ELIEA/NUP84-GFP were grown at 24 °C and visualized by fluorescence microscopy. DIC, GFP-fluorescence, DNA (visualized with Hoechst dye), and false-colored overlay (GFP fluorescence – green, DNA – blue) images of the same field are shown in columns from left to right.
Based on lattice packing observed in crystals of Nup145C•Sec13, Hsia et al. (9) proposed that Nup145C•Sec13 and Nup85•Seh1 each form heterooctameric poles that span the entire NPC in a “concentric cylinder” model of NPC structure. However, the Nup145C•Sec13 lattice contacts involved in the putative heterooctamer overlap with the crown surface of Nup145C shown here to be the Nup84-binding site. Additionally, Nup145C•Sec13 and Nup85•Seh1 behave nearly identically during gel filtration, indicative of heterodimers when their large hydrodynamic radii are taken into account (Fig. 2 & Fig S4). AUC experiments confirmed that Nup85•Seh1 is a heterodimer in solution (Fig. S3). Thus, the heterooctameric pole model (9) is inconsistent with our results.
The structural similarity between Nup85 and Nup145C extends to at least three other proteins (Fig. 4 & Fig. S11). First, the architectural nucleoporin Nic96 (10) shares a common structural core (Fig. S11) but has a distinct N-terminus (Fig. 4). The shared cores mutually superimpose with an rmsd of 3.0–3.5 Å. Nic96 has a trunk module (α1–α3 and α12–α19), a crown module (α4–α11), and an N-terminal coiled-coil extension (instead of the insertion blade of Nup145C and Nup85) that tethers it to the FG-containing Nsp1 complex (29). Apart from the N-terminal differences the three proteins differ mainly in the relative orientation of the crown and trunk modules. Although a previous comparison of Nup145C to the COPII coat component Sec31 did not reveal a strong similarity (9), comparison with Nup85, Nup145C and Nic96 shows that Sec31 has corresponding trunk (α1–α3 and α12–α18) and crown (α4–α11) modules. Sec31 homodimerizes to create an “edge element” in the COPII coat by an internal domain-swap between two crown modules (30). This domain swap results in a mixed crown module that is identical in topology to the unmixed crowns in Nup85, Nup145C, and Nic96 (30). Structural prediction using Phyre (31) also places Nup84 in the Nup85/Nup145C/Nic96/Sec31 group. Similarity extends beyond the trunk and crown modules to a ‘tail’ module that has been structurally characterized in the C-terminal domain of human Nup107 (homolog of yNup84) and in Nic96 (8, 10) (Fig. S11C). The last three helices in the tail module of Nup107 form the interaction site with Nup133 (8). In Nic96 this region is predicted to be protein binding site as well (10). Because we find this characteristic tripartite structural element of crown, trunk and tail in architectural proteins of the NPC and the COPII coat, we term it Ancestral Coatomer Element 1 (ACE1).
Fig. 4. Architecture of ACE1.
(A) ACE1 containing proteins are shown as cylinders and sheets. Crowns are shown in blue, trunks in orange, tails in green, and other domains in gray. Modules with predicted structures are shown half-transparent. (PDB codes: 2QX5, Nic96; 3BG1, Nup145C; 3CQC, Nup107 (Nup84 homolog); 2PM6, Sec31) (B) Cartoon illustrating the similarity and modular nature of the ACE1 element. The N-terminal elaborations are for Nic96 a coiled-coil domain that interacts with the Nsp1 complex, for Nup85 the Seh1-interacting insertion blade, for Nup145C the Sec13-interacting insertion blade preceded by an autocatalytic cleavage domain and Nup145N, and for Sec31 the Sec13-interacting insertion blade is preceded by its own N-terminal 7-bladed β-propeller. Sec31 has a unique proline rich insertion C-terminal to its trunk module followed by a conserved region predicted to be α-helical.
Can we predict ACE1 functional sites from established interactions? Analogy to Sec31 monomers in the COPII edge element suggests that Nup145C and Nup84 might interact crown to crown. Based on the Phyre-model and structural alignment we constructed a surface point mutant replacing two conserved hydrophobic residues on helix α8 of the Nup84 crown with aspartate (Nup84-ISICM to Nup84-DSICD) (Fig. S7, S12). Nup84-DSICD disrupts Nup145C binding in a manner analogous to Nup145C-ELIEA severing Nup84 binding, as shown by gel filtration and ITC (Fig. 2C,D) Thus, the Nup84•Nup145C interface is a crown-crown interaction involving α8 helices as in Sec31 homodimerization. Additionally, we found that the tail modules of Nup145C and Nup85 are necessary for interaction with Nup120 in a manner analogous to the human Nup107 interaction site for human Nup133 (8) (Fig. S5, S6).
Here we have shown that ACE1 is abundant in the two main scaffolding subcomplexes of the NPC. To date, Nic96 is the only ACE1 protein in which all three modules (crown, trunk and tail) are structurally defined. In Nic96, the three modules form a continuous, rigid hydrophobic core (10, 32). In the other four experimental structures, only a subset of the modules are present. We speculate that the three modules within ACE1 can allow hinge movements, utilized to different extents in specific family members.
ACE1 is different from regular α-helical repeat structures, including HEAT-repeats and TPR-repeats (as discussed in (10)). The α-helical modules that compose ACE1 are distinctly irregular, most notably with elements that fold back onto themselves forming a U-turn within the crown module. The trunk is composed of two zig-zaging helical units running in opposite directions. We propose that this architecture confers rigidity to the trunk and thus distinguishes it from regular helical repeat structures that are often inherently flexible (33). As a consequence of the specific arrangement of the helices in ACE1, several helices in trunk and crown are encased by neighboring helices and thus have a characteristic hydrophobic character (typically helices α6 and α10). This pattern of hydrophobic helices may help to find additional ACE1 proteins. Several sequence elements, notably in the crown and at the predicted hinge regions, distinguish ACE1 from other α-helical domains (10). Nevertheless, these characteristics are subtle enough to remain undetected in typical primary sequence (i.e. BLAST) searches and candidate proteins need to be examined using all available tools, including phylogeny and secondary and tertiary structure analysis.
Based on distance constraints and stoichiometric considerations, the heptameric Y-shaped Nup84 complex has been placed in two concentric eight-membered rings on the nucleoplasmic and cytoplasmic faces of the NPC (23). But how is it oriented and how is it connected to the inner ring of the scaffold? Nup133 is anchored to the structural scaffold by its interaction with Nup84, positioning it at the periphery of the pore (8). Nup84 is the link between Nup133 and Nup145C. Thus, we position the extended arm of the Y composed of Nup145C•Sec13, Nup84, and Nup133 facing outward (Fig. 5). Excluding the Nup133•Nup84 pair, the remaining pentamer forms a roughly symmetrical triskelion that conceptually resembles the vertex elements that form polygonal cages in vesicle coats. Cryo-EM samples showed that the triskelion measures approximately 20 nm between the tips (14). An eight-membered ring of the Y-complex around the central transport channel has a ~50 nm diameter if the edges were to touch at the tips. Alternatively, the Y complexes might connect through a yet unidentified adaptor protein.
Fig. 5. Lattice model for the Nup84 complex and the structural scaffold of the nuclear pore complex.
The ACE1 proteins Nup85, Nup145C, Nup84, Sec31, and Nic96 are colored according to Figure 4. (A) Schematic diagram of COPII outer coat organization. The Sec31•Sec13 cuboctahedron composed of 24 edge elements (Sec31•Sec13 heterotetramers) is shown unwrapped and laid flat in 2 dimensions. The Sec31•Sec31 crown-crown interactions make edge elements while propeller-propeller interactions are vertex elements (30). (B) Schematic diagram of the predicted lattice-like organization of the structural scaffold of the NPC. The entire scaffold (8 spokes) is illustrated unwrapped and laid flat in two dimensions. The Nup84 complex comprises the nuclear and cytoplasmic rings, while the Nic96-containing complex makes up the inner ring. The relative position and interactions between the seven proteins in the Nup84 complex are shown with Sec13, Seh1, Nup133, and Nup120 colored in gray. The remainder of the complex (Nup157/170, Nup188, and Nup192) is illustrated in gray. The illustration is not meant to predict relative positions of proteins or structure of the inner ring per se, but shows the lattice-like organization of the structural scaffold similar to vesicle coating complexes.
Two of the three interface types observed in the outer COPII coat are also found in the NPC coat (Fig. 5B,C). Nup145C and Nup84 heterodimerize via their crown modules similar to Sec31 homodimerization and the insertion of a seventh blade into an incomplete propeller domain is a recurring theme in Sec13•Sec31, Sec13•Nup145C, and Seh1•Nup85. Because Nic96 shares an ACE1 element, we predict that the inner scaffold ring is branched and lattice-like as are the peripheral rings. We postulate that the Nup84 and Nic96 complexes are both vertex elements in the assembly of the NPC structural scaffold. This would generate a lattice-like NPC coat similar to clathrin and COP coats (30, 34) (Fig. 5B). This model explains how the relatively small mass of nucleoporin subcomplexes fills the large volume observed for the scaffold structure of the NPC (23) and is generally consistent with low-resolution images of NPCs (4, 6). Notably, COP and clathrin cages do not directly contact membranes, but use adaptor protein (AP) complexes to span the ~10 nm gap between the surfaces (35). Consistent with a related architecture, a similar sized gap has been observed between the scaffold ring and membrane surface in intact NPCs (4).
The modular nature of COP and clathrin coats enables the construction of assemblies varying in composition and size (36, 37), because the polygonal elements can be arranged in different ways. If the same principle applies to the NPC, it could explain the existence of a subset of NPCs that do not obey eight-fold rotational symmetry (38) or further allow for the assembly of NPCs of distinct composition, possibly tailored to more specific tasks. These possibilities are now testable and will bring us closer to fully understanding the many functions of the NPC.
Supplementary Material
Materials and Methods
Figs S1 to S13
Tables S1, S2
References
References and Notes
- 1.Tran EJ, Wente SR. Cell. 2006;125:1041. doi: 10.1016/j.cell.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 2.Weis K. Cell. 2003;112:441. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- 3.D'Angelo MA, Hetzer MW. Trends Cell Biol. 2008;18:456. doi: 10.1016/j.tcb.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beck M, Lucic V, Forster F, Baumeister W, Medalia O. Nature. 2007;449:611. doi: 10.1038/nature06170. [DOI] [PubMed] [Google Scholar]
- 5.Drummond SP, Rutherford SA, Sanderson HS, Allen TD. Can. J. Physiol. Pharmacol. 2006;84:423. doi: 10.1139/y05-148. [DOI] [PubMed] [Google Scholar]
- 6.Stoffler D, Feja B, Fahrenkrog B, Walz J, Typke D, Aebi U. J. Mol. Biol. 2003;328:119. doi: 10.1016/s0022-2836(03)00266-3. [DOI] [PubMed] [Google Scholar]
- 7.Berke IC, Boehmer T, Blobel G, Schwartz TU. J. Cell Biol. 2004;167:591. doi: 10.1083/jcb.200408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boehmer T, Jeudy S, Berke IC, Schwartz TU. Mol. Cell. 2008;30:721. doi: 10.1016/j.molcel.2008.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsia KC, Stavropoulos P, Blobel G, Hoelz A. Cell. 2007;131:1313. doi: 10.1016/j.cell.2007.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeudy S, Schwartz TU. J. Biol. Chem. 2007;282:34904. doi: 10.1074/jbc.M705479200. [DOI] [PubMed] [Google Scholar]
- 11.Pante N, Kann M. Mol. Biol. Cell. 2002;13:425. doi: 10.1091/mbc.01-06-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwartz TU. Curr. Opin. Struct. Biol. 2005;15:221. doi: 10.1016/j.sbi.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 13.Rabut G, Doye V, Ellenberg J. Nat. Cell Biol. 2004;6:1114. doi: 10.1038/ncb1184. [DOI] [PubMed] [Google Scholar]
- 14.Lutzmann M, Kunze R, Buerer A, Aebi U, Hurt E. EMBO J. 2002;21:387. doi: 10.1093/emboj/21.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fabre E, Hurt E. Annu. Rev. Genet. 1997;31:277. doi: 10.1146/annurev.genet.31.1.277. [DOI] [PubMed] [Google Scholar]
- 16.Galy V, Mattaj IW, Askjaer P. Mol. Biol. Cell. 2003;14:5104. doi: 10.1091/mbc.E03-04-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harel A, et al. Mol. Cell. 2003;11:853. doi: 10.1016/s1097-2765(03)00116-3. [DOI] [PubMed] [Google Scholar]
- 18.Lusk CP, Makhnevych T, Marelli M, Aitchison JD, Wozniak RW. J. Cell Biol. 2002;159:267. doi: 10.1083/jcb.200203079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marelli M, Aitchison JD, Wozniak RW. J. Cell Biol. 1998;143:1813. doi: 10.1083/jcb.143.7.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zabel U, Doye V, Tekotte H, Wepf R, Grandi P, Hurt EC. J. Cell Biol. 1996;133:1141. doi: 10.1083/jcb.133.6.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Devos D, et al. Proc. Natl. Acad. Sci. U. S. A. 2006;103:2172. doi: 10.1073/pnas.0506345103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Devos D, et al. PLoS Biol. 2004;2:e380. doi: 10.1371/journal.pbio.0020380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alber F, et al. Nature. 2007;450:695. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- 24.Chaudhuri I, Soding J, Lupas AN. Proteins. 2008;71:719. doi: 10.1002/prot.21764. [DOI] [PubMed] [Google Scholar]
- 25.Garcia De La Torre J, Huertas ML, Carrasco B. Biophys. J. 2000;78:719. doi: 10.1016/S0006-3495(00)76630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doye V, Wepf R, Hurt EC. EMBO J. 1994;13:6062. doi: 10.1002/j.1460-2075.1994.tb06953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siniossoglou S, et al. Cell. 1996;84:265. doi: 10.1016/s0092-8674(00)80981-2. [DOI] [PubMed] [Google Scholar]
- 28.Drin G, Casella JF, Gautier R, Boehmer T, Schwartz TU, Antonny B. Nat. Struct. Mol. Biol. 2007;14:138. doi: 10.1038/nsmb1194. [DOI] [PubMed] [Google Scholar]
- 29.Grandi P, Schlaich N, Tekotte H, Hurt EC. EMBO J. 1995;14:76. doi: 10.1002/j.1460-2075.1995.tb06977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fath S, Mancias JD, Bi X, Goldberg J. Cell. 2007;129:1325. doi: 10.1016/j.cell.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 31.Bennett-Lovsey RM, Herbert AD, Sternberg MJ, Kelley LA. Proteins. 2008;70:611. doi: 10.1002/prot.21688. [DOI] [PubMed] [Google Scholar]
- 32.Schrader N, Stelter P, Flemming D, Kunze R, Hurt E, Vetter IR. Mol. Cell. 2008;29:46. doi: 10.1016/j.molcel.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 33.Conti E, Muller CW, Stewart M. Curr. Opin. Struct. Biol. 2006;16:237. doi: 10.1016/j.sbi.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 34.Fotin A, et al. Nature. 2004;432:573. doi: 10.1038/nature03079. [DOI] [PubMed] [Google Scholar]
- 35.Owen DJ, Collins BM, Evans PR. Annu. Rev. Cell Dev. Biol. 2004;20:153. doi: 10.1146/annurev.cellbio.20.010403.104543. [DOI] [PubMed] [Google Scholar]
- 36.Stagg SM, et al. Cell. 2008;134:474. doi: 10.1016/j.cell.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng Y, Boll W, Kirchhausen T, Harrison SC, Walz T. J. Mol. Biol. 2007;365:892. doi: 10.1016/j.jmb.2006.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hinshaw JE, Milligan RA. J. Struct. Biol. 2003;141:259. doi: 10.1016/s1047-8477(02)00626-3. [DOI] [PubMed] [Google Scholar]
- 39.Dujon B. Trends. Genet. 2006;22:375. doi: 10.1016/j.tig.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 40.We thank staff at beamlines 24-ID-C/-E at Argonne National Laboratory and X6A at National Synchrotron Light Source for excellent assistance with data collection, R. Sauer and T. Baker for critically reading the manuscript, G. Wink for her contributions, members of the Schwartz laboratory for discussions, and the Biophysical Instrumentation Facility for the Study of Complex Macromolecular Systems (NSF-0070319 and NIH GM68762) for providing instrumentation. Supported by NIH Grant GM77537 (T.U.S.), a Pew Scholar Award (T.U.S.), a Koch Fellowship Award (S.G.B.) and a Vertex Scholarship (S.G.B.). Coordinates and structure factors of the Nup85•Seh1 crystal structure have been deposited in the Protein Data Bank with accession code 3EWE.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Materials and Methods
Figs S1 to S13
Tables S1, S2
References