Abstract
The zebrafish has emerged as a powerful genetic model of cancer, but has been limited by the use of stable transgenic approaches to induce disease. Here, a co-injection strategy is described that capitalizes on both the numbers of embryos that can be microinjected and the ability of transgenes to segregate together and exert synergistic effects in forming tumors. Using this mosaic transgenic approach, gene pathways involved in tumor initiation and radiation sensitivity have been identified.
Keywords: Myc, Ras, heat-shock, T-cell acute lymphoblastic leukemia, rhabdomyosarcoma
Cancer models developed in genetically tractable organisms have provided unique insights into the molecular underpinnings of human malignancy. To capitalize on the powerful developmental and genetic tools available in zebrafish, researchers have created numerous zebra-fish tumor models (Langenau et al., 2003, 2007; Yang et al., 2004; Berghmans et al., 2005; Patton et al., 2005; Haramis et al., 2006; Sabaawy et al., 2006; Le et al., 2007). The ultimate goal of the majority of this work is to identify pathways that modify disease states. To achieve this, we sought to develop methodologies to rapidly modify tumor initiation and radiation sensitivity in the zebrafish that do not require the generation of stable transgenic animals.
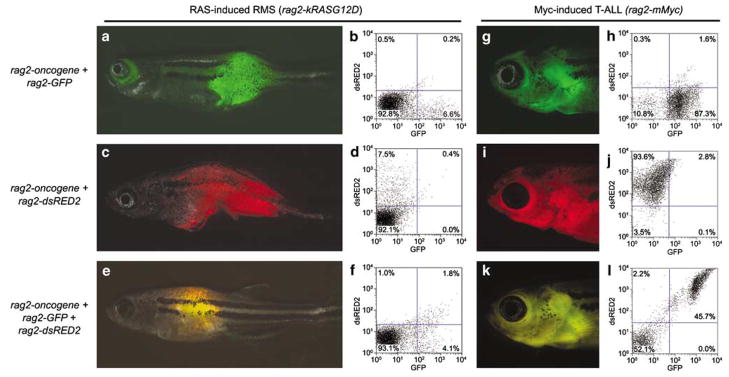
Previously, a mosaic transgenic model of RAS-induced rhabdomyosarcoma (RMS) was developed, in which the rag2 promoter drives expression of activated human kRASG12D within mononuclear muscle cell populations (Langenau et al., 2007). In these studies, co-injection strategies were utilized to label tumor cell populations based on muscle differentiation status. Specifically, rag2-kRASG12D and rag2-dsRED2 were co-injected into alpha-actin-GFP transgenic animals. Cells were isolated from the externally visible portion of the tumor, and a single cell suspension was obtained by mincing RMS in the presence of liberase, a collagenase blend used for tissue dissociation (Langenau et al., 2007). Cells were then filtered (40 μm mesh) and analysed by FACS (fluorescence activated cell sorting). Zebrafish RMS contained mononuclear dsRED2 + cells that express early muscle markers and comprise the tumor-initiating stem cell, whereas alpha-actin-GFP + cells comprised more differentiated tumor cell types. Moreover, the double negative cell population, which comprises a bulk of the RMS tumor mass (75–80% of all mononuclear cells), predominantly comprised blood cells and was non-tumorigenic in transplantation assays (Langenau et al., 2007). In fact, co-injection of rag2-kRASG12D and rag2-dsRED2 or rag2-GFP into one-cell stage embryos leads to highly efficient co-segregation and co-integration of transgenes within a majority of developing RMSs (green fluroscent protein (GFP) expressing tumors, n = 14 of 14; dsRED, n = 67 of 69; Figures 1a–d, Supplementary Figure 1 and Supplementary Methods and Text), providing a novel method to express multiple transgenes within a developing tumor. Taken together, these experiments establish that transgenes co-segregate within RMS and can be used to label tumor cells. Moreover, they suggest that it may be possible to inject two or more transgenes that act synergistically to alter the kinetics of malignant transformation.
Figure 1.
Co-injection of multiple transgenes into one-cell stage zebrafish embryos leads to co-expression in tumors. Rhabdomyosarcoma (RMS) was induced by injection of rag2-kRASG12D (a–f), whereas T-cell acute lymphoblastic leukemia (T-ALL) was induced by injection of rag2-mMyc (g–l). Oncogene-containing transgenes were co-injected with rag2-GFP (a, b, g and h), rag2-dsRED2 (c, d, i and j) or both (e, f, k and l). Fluorescent microscopic images show expression of GFP (a and g), dsRED2 (c and i) or both (e and k). FACS analysis confirms expression patterns of tumor cells from animals shown in the corresponding fluorescent images (b, d, f, h, j and l). The rag2-GFP (Jessen et al., 2001), rag2-mouse cMyc (Langenau et al., 2003), rag2-dsRED2 and rag2-human kRASG12D (Langenau et al., 2007) constructs have been described previously. DNA constructs were linearized with either XhoI or Not1, phenol:chloroform extracted and ethanol precipitated as described previously (Langenau et al., 2003). For rag2-oncogene, rag2-dsRED2 co-injections (a, b, g and h), the oncogene containing construct was linearized with XhoI and the second linearized with NotI. For rag2-oncogene co-injections with rag2-GFP (c, d, i and j), both constructs were digested with the same enzyme, either NotI or XhoI. DNA was quantified by both spectrophotometric analysis and gel quantification. DNA was diluted in 0.5XTE + 100 mM KCl to 100 ng μl−1, with co-injection of two transgenes having 50 ng of each construct per microliter or three transgenes having 33.3 ng of each construct per microliter. FACS, fluorescence activated cell sorting; GFP, green fluorescent protein.
To assess if co-segregation of transgenes is a common feature of mosaic transgenic tumors, transgene co-segregation was also analysed in Myc-induced T-cell acute lymphoblastic leukemia (T-ALL) (Langenau et al., 2003). Specifically, the rag2-mouse cMyc (rag2-mMyc) transgene was co-injected into one-cell stage embryos along with either rag2-GFP or rag2-dsRED2. All T-ALLs that developed in injected animals were fluorescently-labeled (GFP, n = 12 of 12; dsRED2, n = 13 of 13; Figures 1g–j and Supplementary Table 1). Previously, we established that injection of rag2-mMyc into stable transgenic rag2-GFP animals leads to GFP-labeled tumors (Langenau et al., 2003). To determine the efficiency of co-segregation within tumor cell populations, rag2-mMyc and rag2-dsRED2 were co-injected into stable transgenic rag2-GFP animals. T-ALLs that developed were both GFP + and dsRED2 + by microscopic and FACS analysis (n = 3, 42–49 days old, Supplementary Figure 2). In fact, FACS analysis of whole tumor fish revealed that nearly all GFP + cells also express dsRED2 (n = 3, percent co-localization ranged from 98.6–99.6%, Supplementary Figure 2a–d). If the rag2-mMyc transgene integrated independently of rag2-dsRED2 within a portion of tumor cells, then we would have observed a discernable population of tumor cells that are only GFP-labeled; however, this was not observed. Additional morphological analysis showed that blast cells are largely confined to the dsRED2 + sorted cell populations, whereas the dsRED2-negative cells are predominantly red blood cells (Supplementary Figure 2e–h), further demonstrating that T-ALL tumor cells are confined to the dsRED2 + cell population. These data suggest that transgenes segregate together in T-ALL and are coexpressed in established tumors.
Limiting dilution analysis and cell transplantation independently confirmed that dsRED2 + T-ALL cells from animals co-injected with rag2-mMyc and rag2-dsRED2 were tumorigenic ((Tropepe et al., 2000), Supplementary Figure 2i–n and Supplementary Table 2). FACS was used to isolate dsRED2 + and dsRED2-negative cells from T-ALLs that developed in rag2-GFP stable transgenic animals co-injected with rag2-mMyc and rag2-dsRED2 (n = 3, dsRED2 + sort purity 98.2–99.7%, negative sort purity 88.8–96.7%, Supplementary Figure 2e–f and Supplementary Table 2). 1 × 104, 1 × 103, 1 × 102 and 1 × 101 sorted cells were supplemented with red blood cell carrier from non-tumorigeneic AB-strain zebrafish up to 1 × 104 cells per 5-μl volume and introduced into irradiated recipients (n = 11 fish per dilution). Transplant animals were analysed for engraftment at 11 and 21 days post-transplantation under a fluorescent dissecting microscope for both GFP and dsRED2 expression. From this analysis, we found that the numbers of tumor-initiating cells vary widely between Myc-induced T-ALLs, comprising between 1 in 1258 and 1 in 32 046 of the dsRED2 + cells. By contrast, the negative cells contained within these same T-ALLs do not transplant as efficiently (Supplementary Table 2). Tumors that developed in transplant recipients were positive for both GFP and dsRED2, whereas no single positive GFP + tumors were observed (n = 41), again strongly suggesting that rag2-dsRED2 and rag2-mMyc transgenes co-segregate efficiently in established T-ALLs and that transgenes are not globally silenced over time. Finally, integration of both transgenes into developing T-ALL or RMS tumor cells was independent of the enzyme used to linearize the transgenes (either XhoI or NotI; Supplementary Methods and Text). Additional experiments in the mitfa-BRAF transgenic zebrafish models of melanoma established that co-segregation of transgenes is not specific to rag2 promoter-containing constructs (CJC, CB and LIZ, unpublished data).
Co-injection of three constructs (rag2-oncogene, rag2-GFP and rag2-dsRED2) also led to tumors that expressed all three transgenes (RMS, n = 14 of 17; TALL, n = 22 of 23; Figures 1e, f, k, and l). In RMS, two distinct sub-populations of tumor cells were identified in individual triple injected animals, GFP + and GFP +/dsRED2 +. When each cell population was sorted to relative purity and transplanted into irradiated recipient animals at low cell number (n = 1000–1500 tumor cells along with 20 000 red blood cell carrier cells per animal, GFP + sort, >86% pure and 97.8% viable; GFP +/dsRED2 + sort, >81.2% pure and 89% viable), each cell population led to the development of dsRED2 +/GFP + RMS in irradiated recipient animals by 11 days post-transplantation (data not shown). Similar results were also obtained with additional tumors. Moreover, analysis of multiple rag2-kRASG12D/rag2-dsRED2/rag2-GFP tumors revealed that single positive dsRED2 + cell types were never observed by FACS analysis of total tumor (n = 6). If rag2-dsRED2 and rag2-GFP segregated independently into different tumor cell populations or were transcriptionally silenced, one would expect to find that a large subset of RMSs should have single positive dsRED2 + cell types but not single positive GFP + cells. However, this is never seen. Together these data suggest that injection of three constructs can lead to co-segregation in tumors and suggest that GFP and dsRED2 can differentially label cell populations based on the kinetics of the protein folding and maturation when expressed in rapidly dividing cell populations (GFP maturation time is 8–12 h, whereas for dsRED2 it is 24 h; Clontech protocol PT3404-1, clontech.com).
Three transgenes can also efficiently co-segregate into Myc-induced T-ALL (Figures 1i and k). In contrast to RMS, most T-ALL cells coexpress both GFP and dsRED2 when triple injected at the one-cell stage with rag2-mMyc, rag2-dsRED2 and rag2-GFP. Interestingly, unlike RMS cells that are rapidly dividing and undergoing limited differentiation (Langenau et al., 2007), a majority of T-ALL tumor cells comprise non-dividing blasts (Langenau et al., 2005a). These results suggest that coexpression of fluorescent protein containing transgenes will vary based on the tumor type, the kinetics of fluorophore protein folding, the proliferation of the target cell population and the differentiation status of the tumor of interest. In the end, our results show that multiple transgenes can be introduced into one-cell stage embryos very efficiently and eventually become coexpressed in the developing malignancy.
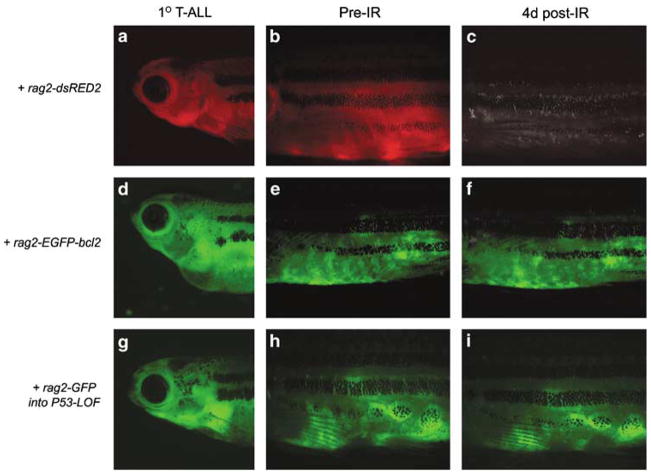
Stable transgenic approaches have shown that Myc-induced T-ALLs regress following radiation treatment, while transgenic overexpression of bcl-2 suppresses cell death within irradiated T-ALLs (Langenau et al., 2005b). To assess whether co-injection strategies can be utilized to modify radiation sensitivity and recapitulate what was observed in stable transgenic animals, rag2-mMyc and rag2-dsRED2 were co-injected into AB-strain zebrafish. Tumor cells were harvested from primary dsRED2-labeled T-ALL (Figure 2a) and introduced by intraperitoneal injection into irradiated recipients (1 × 106 cells). Transplant recipients developed tumors within 10 days after injection (Figure 2b), which were photographed and then again irradiated (23 Gy). By 4 days after irradiation treatment, dsRED2 expressing tumors were absent in transplant animals (Figure 2c). By contrast, transplanted T-ALLs from animals co-injected with rag2-mMyc and rag2-EGFP-bcl-2 were resistant to irradiation-induced cell death (Figures 2d–f, n = 3 tumors analysed per condition).
Figure 2.
Co-injection approaches can be used to modify radiation sensitivity in Myc-induced T-cell acute lymphoblastic leukemias (T-ALLs). Primary T-ALLs were generated by co-injecting rag2-mMyc along with other transgenes denoted to the left of figure (a, d and g). Primary tumor cells were transplanted into irradiated recipients and engrafted cells could be visualized 10 days later (b, e and h). Transplanted animals were irradiated (23 Gy) and assessed for tumor regression at 4 days post-treatment (c, f and i). Control tumors were generated in AB animals injected with rag2-mMyc + rag2-dsRED2 (a–c). T-ALLs from AB fish injected with rag2-mMyc + rag2-EGFP-bcl2 (d–f). p53 loss-of-function animals (p53-LOF) injected with rag2-mMyc + rag2-GFP (g–i). Fish oriented with head toward left and dorsal toward top. The rag2-EGFP-zebrafish bcl2 construct was described previously (Langenau et al., 2005b). Cell transplantation, FACS and irradiation protocols were completed essentially as described (Langenau et al., 2003, 2005b; Traver et al., 2003). 1 × 106 unsorted cells were used in all transplantation experiments. FACS, fluorescence activated cell sorting.
Next, we wanted to establish that co-injection methodologies could be used to rapidly introduce multiple transgenes into diverse genetic backgrounds. Specifically, rag2-mMyc and rag2-GFP were co-injected into p53 loss-of-function animals (p53-LOF) (Berghmans et al., 2005) and assessed as above (Figure 2g–i, n = 3). As may be expected based on findings in P53-deficient T cells (Oda et al., 2000), p53-LOF T-ALLs were insensitive to irradiation. Taken together, these experiments highlight the power of this co-injection approach to identify modulators of radiation sensitivity. Moreover, they illustrate the ability to rapidly perform these analyses without the generation of stable transgenic animals that would require lengthy analysis involving the production and identification of transgenic founders; followed by complex breeding schemes and analysis over multiple generations to assess compound genetic effects on selected cancer phenotypes.
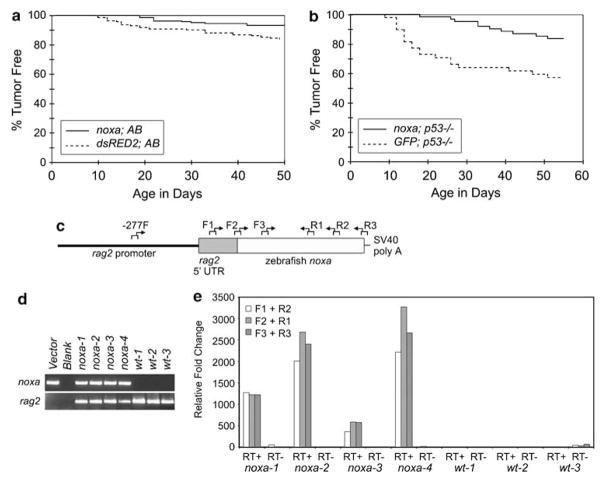
Next, we wanted to determine whether co-injection methodologies could be used to identify genetic modifiers of cancer initiation. Having previously established that p53-LOF collaborates with RAS to induce RMS (Langenau et al., 2007), we hypothesized that reestablishing p53 pathway activation in tumors would alter tumor initiation, ultimately affecting tumor penetrance and latency. Noxa is a potent pro-apoptotic BH3-only protein and is a direct transcriptional target of P53 (Oda et al., 2000). The zebrafish noxa ortholog has recently been identified and functionally tested for its ability to induce apoptosis during embryogenesis (Kratz et al., 2006). Given that RAS-induced RMS can be easily visualized in injected animals (Langenau et al., 2007), tumor onset in wild-type AB-strain animals co-injected with rag2-kRASG12D and rag2-noxa was compared to those injected with rag2-kRASG12D and rag2-dsRED2. In this setting, zebrafish noxa is a potent suppressor of tumor initiation (rag2-kRASG12D + rag2-noxa, n = 12 of 174 vs rag2-kRASG12D + rag2-dsRED2, n = 29 of 179; P = 0.007 using a log-rank analysis, Figure 3a). Tumor onset was also compared in p53-LOF animals co-injected with rag2-kRASG12D and either rag2-noxa or rag2-GFP. Because tumor penetrance is much higher in p53-LOF animals, fewer fish were required to show the effect of noxa expression on tumor initiation (n = 10 of 64 in the noxa cohort and n = 20 of 48 in the GFP cohort, P = 0.001, Figure 3b). To verify that rag2-noxa is expressed in established tumors and is not globally transcriptionally silenced, the presence and expression of the rag2-noxa transgene was confirmed by PCR of either genomic DNA isolated from whole tumor or complementary DNA, respectively (n = 8 tumors, Figures 3d and e, primers available in Supplementary Table 3).
Figure 3.
Noxa is a potent genetic suppressor of RAS-induced rhabdomyosarcoma. Tumor onset in AB wild –type (WT) (a) or p53 loss-of-function animals (b) co-injected with rag2-kRASG12D and either rag2-noxa, rag2-dsRED2 or rag2-GFP. (c) Diagram of rag2-noxa transgene with accompanying primers used in analysis. (d) PCR of genomic DNA isolated from the externally-visible tumor portion of rag2-kRASG12D + rag2-noxa or rag2-kRASG12D (WT) injected AB-strain animals. The upper panel shows presence of the rag2-noxa transgene (primers used in this analysis −277F + R1). The lower panel shows presence of genomic DNA amplification of the endogenous rag2 locus. (e) Quantitative reverse-transcriptase PCR showing that the noxa transgene is expressed in RMS (RT +). No RT (RT −) controls confirm that amplification is not due to contaminating genomic DNA. The rag2-zebrafish noxa transgene was created by cloning a BamHI/NotI fragment that contains both the open reading frame and the SV40 polyadenylation site from the pcs2 + noxa expression vector (CJ and ATL, unpublished data) into the rag2-GFP construct (Jessen et al., 2001). Injection volumes were assessed by injecting DNA samples into oil. One nanoliter of DNA was injected per animal.
P53-deficient tumors that express noxa do not have increased numbers of apoptotic cells when compared to control tumors (n = 6 tumors per group). Specifically, rag2-kRASG12D was co-injected with either rag2-noxa or rag2-GFP into P53-deficient animals at the one-cell stage of life. Animals were scored for tumors at 25 days of life, killed and fixed in 4% PFA. Subsequently, animals were sectioned and stained by TUNEL or hematoxylin/eosin as described previously (Traver et al., 2004). Tumor sections were photographed under ×1000 oil emersion and apoptotic cell number and total cell number were counted per field (n = 3 fields per sample, Supplementary Figure 3). Percentage of apoptotic cells did not differ between the noxa and GFP expressing tumors (0.43 ± 0.31 and 0.94 ± 0.96%, respectively; ± s.d.; P = 0.24; n = 428 ± 134 cells counted per sample), suggesting the existence of a mechanism by which tumor cells evade high levels of noxa transgene expression during tumor development. Taken together, these experiments establish noxa as a potent suppressor of tumor formation in RMS and establish a novel technology to rapidly assess collaborating genetic events in modulating tumor onset.
Heat-shock-inducible transgenic approaches have provided an invaluable tool through which gene expression in development can be modulated (Halloran et al., 2000; Xiao et al., 2003; Burns et al., 2005; Pyati et al., 2005). To establish if co-injection techniques along with heat-shock-inducible transgenesis could be used to elicite gene expression within established tumors, the rag2-kRASG12D, rag2-GFP and hsp70-dsRED2 constructs were co-injected into one-cell stage AB-strain animals. Triple transgenic RMS-affected animals developed GFP-labeled RMS that could induce dsRED2 expression after heat-shock at 37 °C for 45 min (n = 19 of 20, Figure 4). Expression of dsRED2 could be detected in heat-shocked animals from 2–10 days post treatment (Supplementary Figure 4). Importantly, dsRED and GFP expression largely co-localized within tumors that received heat-shock (Figures 4i–k), establishing that the hsp70 promoter is active in most cell types contained in zebrafish RMS (n = 4). In contrast to what is observed in RAS-induced RMS, co-injection of rag2-mMyc, rag2-GFP and hsp70-dsRED2 led to T-ALLs that were both GFP + and strongly dsRED2 + despite never receiving heat-shock treatment (n = 14 of 15, Supplementary Figure 5). Additional confocal analysis established that nearly all T-ALL blast cells expressed high levels of both GFP and dsRED2 in the absence of heat-shock treatment (n = 3, data not shown). In Myc-induced T-ALL, it is likely that the heat-shock response—a stress response that can be affected by other stimuli other than heat—is already active in these malignancies. Although the heat-shock-inducible transgenic approaches will be amenable for use in only a subset of tumor types, our experiments clearly establish that co-injection and inducible approaches can be utilized in zebrafish transgenic models of cancer to selectively turn on genes within established malignancies, providing a novel and rapid methodology to assess gene effects on tumor maintenance.
Figure 4.
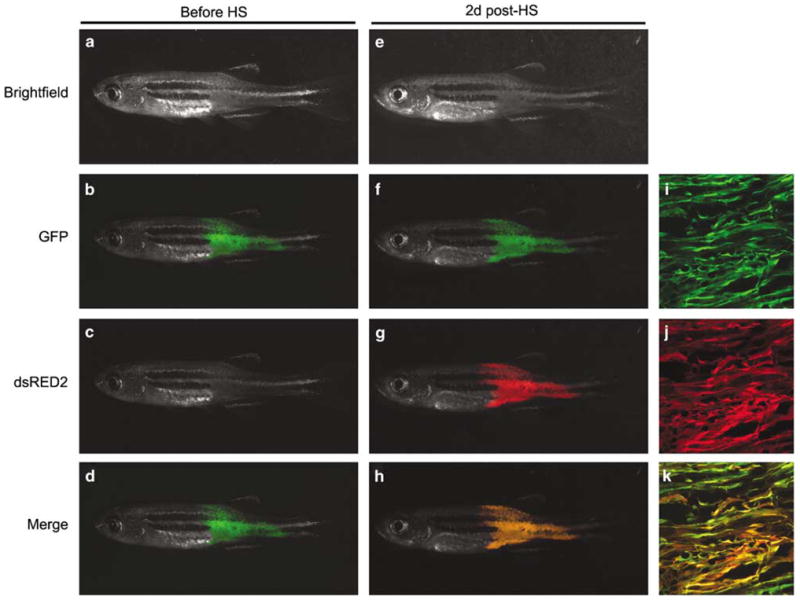
Co-injection approaches can be used to selectively induce gene expression within established RAS-induced rhabdomyosarcomas. The rag2-kRASG12D transgene was co-injected with rag2-GFP and hsp70-dsRED2 into AB strain, one-cell stage embryos. By 30 days of life, co-injected animals that contained GFP-labeled RMS (a–d) were placed into room temperature fish water (40 ml in a 50-ml Falcon tube) and allowed to acclimate for 10–20 min. Next, animals were transferred to 37 °C and incubated for 45 min. Fish were removed from the water bath and allowed to acclimate to room temperature for 10–15 min, after which time they were introduced back into individual tanks and raised at 28.5 °C. Two days post-heat-shock, RMS expressed both GFP and dsRED2 in surviving animals (e–h, n = 19 of 20). Confocal microscopic analysis of the same animal shown in a–h following heat-shock (×10, i–k) was performed essentially as described (Langenau et al., 2007). Brightfield (a and e), GFP (b, f and i), dsRED2 (c, g and j) and merged fluorescent (d, h and k) images. The hsp70-dsRED2 construct was created by amplifying the hsp70-4 promoter from genomic DNA using a forward primer that contains an XhoI digest site and a reverse primer containing a BamHI site (Supplementary Table 3). This PCR fragment was purified, digested and cloned into the rag2-dsRED2 construct, replacing the rag2 promoter sequence. GFP, green fluorescent protein.
Our experiments show that co-injection strategies can be used to modify radiation sensitivity and tumor initiation in transgenic zebrafish models of cancer. Additional experiments in RAS-induced RMS have established that co-injection approaches can also be used to label tumor cell populations based on differentiation status, ultimately allowing for the identification of the cancer stem cell in this disease (Langenau et al., 2007). Although our experiments have focused on these three uses for the co-injection methodology, it is likely that this approach will not be limited to these applications. For example, co-injection methodologies in combination with heat-shock-inducible transgenic approaches can lead to induction of targeted gene expression within established tumors. Such approaches will allow for the rapid assessment of pathways involved in tumor maintenance. Additionally, multigenic diseases, including cancer, can now be easily modeled using the zebrafish. For example, one can inject multiple transgenes into one-cell stage embryos leading to the expression of a combination of factors within a cell type of interest, ultimately allowing for the identification of the key gene combinations required to elicit disease. Taken together, our experiments establish a new methodology for rapidly assessing collaborating gene-modifying events in cancer that does not require the generation of stable transgenic lines. As one investigator can inject 800–1000 surviving embryos in a day, this new co-injection procedure will likely facilitate the rapid discovery of gene pathways that regulate various aspects of cancer biology. Finally, co-injection approaches will most likely be applicable to other model organisms where ex vivo manipulation and DNA injection is possible, including Fugu, medaka and Xenopus.
Acknowledgments
We thank Anna Burrows for expert technical help and David Bellovin and David Adamovich for critical review of this manuscript. DM Langenau is the Edmond J Safra Foundation Fellow from the Irvington Institute and CJ Ceol is supported by a Damon Runyon Fellowship (DRG-1855-05). Funding for this work was provided by NIH Grant 5R01 CA103846-02 (LI Zon), #1RO1 CA119066-01, CA93152-05, CA104605-03, HL-088664 and CA68484-11 (AT Look), #1KO1 DK074555-01 (CA Jette).
Footnotes
Supporting Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- Berghmans S, Murphey RD, Wienholds E, Neuberg D, Kutok JL, Fletcher CD, et al. tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc Natl Acad Sci USA. 2005;102:407–412. doi: 10.1073/pnas.0406252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns CE, Traver D, Mayhall E, Shepard JL, Zon LI. Hematopoietic stem cell fate is established by the Notch–Runx pathway. Genes Dev. 2005;19:2331–2342. doi: 10.1101/gad.1337005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halloran MC, Sato-Maeda M, Warren JT, Su F, Lele Z, Krone PH, et al. Laser-induced gene expression in specific cells of transgenic zebrafish. Development. 2000;127:1953–1960. doi: 10.1242/dev.127.9.1953. [DOI] [PubMed] [Google Scholar]
- Haramis AP, Hurlstone A, van der Velden Y, Begthel H, van den Born M, Offerhaus GJ, et al. Adenomatous polyposis coli-deficient zebrafish are susceptible to digestive tract neoplasia. EMBO Rep. 2006;7:444–449. doi: 10.1038/sj.embor.7400638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen JR, Jessen TN, Voge SS, Lin S. Concurrent expression of recombination activating genes 1 and 2 in zebrafish olfactory sensory neurons. Genesis: the Journal of Genetics and Development. 2001;29:156–162. doi: 10.1002/gene.1019. [DOI] [PubMed] [Google Scholar]
- Kratz E, Eimon PM, Mukhyala K, Stern H, Zha J, Strasser A, et al. Functional characterization of the Bcl-2 gene family in the zebrafish. Cell Death Differ. 2006;13:1631–1640. doi: 10.1038/sj.cdd.4402016. [DOI] [PubMed] [Google Scholar]
- Langenau DM, Feng H, Berghmans S, Kanki JP, Kutok JL, Look AT. Cre/lox-regulated transgenic zebrafish model with conditional myc-induced T cell acute lymphoblastic leukemia. Proc Natl Acad Sci USA. 2005a;102:6068–6073. doi: 10.1073/pnas.0408708102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenau DM, Jette C, Berghmans S, Palomero T, Kanki JP, Kutok JL, et al. Suppression of apoptosis by bcl-2 overexpression in lymphoid cells of transgenic zebrafish. Blood. 2005b;105:3278–3285. doi: 10.1182/blood-2004-08-3073. [DOI] [PubMed] [Google Scholar]
- Langenau DM, Keefe MD, Storer NY, Guyon JR, Kutok JL, Le X, et al. Effects of RAS on the genesis of embryonal rhabdomyosarcoma. Genes Dev. 2007;21:1382–1389. doi: 10.1101/gad.1545007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenau DM, Traver D, Ferrando AA, Kutok JL, Aster JC, Kanki JP, et al. Myc-induced T cell leukemia in transgenic zebrafish. Science. 2003;299:887–890. doi: 10.1126/science.1080280. [DOI] [PubMed] [Google Scholar]
- Le X, Langenau DM, Keefe MD, Kutok JL, Neuberg DS, Zon LI. Heat shock-inducible Cre/Lox approaches to induce diverse types of tumors and hyperplasia in transgenic zebrafish. Proc Natl Acad Sci USA. 2007;104:9410–9415. doi: 10.1073/pnas.0611302104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda E, Ohki R, Murasawa H, Nemoto J, Shibue T, Yamashita T, et al. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science. 2000;288:1053–1058. doi: 10.1126/science.288.5468.1053. [DOI] [PubMed] [Google Scholar]
- Patton EE, Widlund HR, Kutok JL, Kopani KR, Amatruda JF, Murphey RD, et al. BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Curr Biol. 2005;15:249–254. doi: 10.1016/j.cub.2005.01.031. [DOI] [PubMed] [Google Scholar]
- Pyati UJ, Webb AE, Kimelman D. Transgenic zebrafish reveal stage-specific roles for Bmp signaling in ventral and posterior mesoderm development. Development. 2005;132:2333–2343. doi: 10.1242/dev.01806. [DOI] [PubMed] [Google Scholar]
- Sabaawy HE, Azuma M, Embree LJ, Tsai HJ, Starost MF, Hickstein DD. TEL-AML1 transgenic zebrafish model of precursor B cell acute lymphoblastic leukemia. Proc Natl Acad Sci USA. 2006;103:15166–15171. doi: 10.1073/pnas.0603349103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traver D, Paw BH, Poss KD, Penberthy WT, Lin S, Zon LI. Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat Immunol. 2003;4:1238–1246. doi: 10.1038/ni1007. [DOI] [PubMed] [Google Scholar]
- Traver D, Winzeler A, Stern HM, Mayhall EA, Langenau DM, Kutok JL, et al. Effects of lethal irradiation in zebrafish and rescue by hematopoietic cell transplantation. Blood. 2004;104:1298–1305. doi: 10.1182/blood-2004-01-0100. [DOI] [PubMed] [Google Scholar]
- Tropepe V, Coles BL, Chiasson BJ, Horsford DJ, Elia AJ, McInnes RR, et al. Retinal stem cells in the adult mammalian eye. Science. 2000;287:2032–2036. doi: 10.1126/science.287.5460.2032. [DOI] [PubMed] [Google Scholar]
- Xiao T, Shoji W, Zhou W, Su F, Kuwada JY. Transmembrane sema4E guides branchiomotor axons to their targets in zebrafish. J Neurosci. 2003;23:4190–4198. doi: 10.1523/JNEUROSCI.23-10-04190.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HW, Kutok JL, Lee NH, Piao HY, Fletcher CD, Kanki JP, et al. Targeted expression of human MYCN selectively causes pancreatic neuroendocrine tumors in transgenic zebrafish. Cancer Res. 2004;64:7256–7262. doi: 10.1158/0008-5472.CAN-04-0931. [DOI] [PubMed] [Google Scholar]