Abstract
The pp31 gene of Autographa californica Multicapsid Nucleopolyhedrovirus (AcMNPV) encodes a phosphorylated DNA binding protein that associates with virogenic stroma in the nuclei of infected cells. Prior studies of pp31 by transient late expression assays suggested that pp31 may play an important role in transcription of AcMNPV late genes (Todd et al., 1995. J. Virol. 69, 968–974) although genetic studies of the closely related BmNPV pp31 gene suggested that pp31 may be dispensable (Gomi et al., 1997. Virology 230, 35–47). In the current study, we examined the role of the pp31 gene in the context of the AcMNPV genome during infection. We used a BACmid-based system to generate a pp31 knockout in the AcMNPV genome. The pp31 knockout was subsequently rescued by reinserting the pp31 gene into the polyhedrin locus of the same virus genome. We found that pp31 was not essential for viral replication although the absence of pp31 resulted in a lower viral titer. Analysis of viral DNA replication in the absence of pp31 showed that the kinetics of viral DNA replication were unaffected. An AcMNPV oligonucleotide microarray was used to compare gene expression from all AcMNPV genes in the presence or absence of pp31. In the absence of pp31 a modest reduction in transcripts was detected for many viral genes (99 genes) while no substantial increase or decrease was observed for 43 genes. Transcripts from 6 genes (p6.9, ORF 97, ORF 60, ORF 98, ORF 102 and chitinase) were reduced by 66% or more compared to the levels detected from the control virus. Microarray results were further examined by qPCR analysis of selected genes. In combination, these data show that deletion of the pp31 gene was not lethal and did not appear to affect viral DNA replication, but resulted in an apparent modest down-regulation of a subset of AcMNPV genes that included both early and late genes.
Introduction
The baculovirus Autographa californica Multicapsid Nucleopolyhedrovirus (AcMNPV) is a large double-stranded DNA virus that is highly pathogenic to a number of insect species in the order Lepidoptera. Gene regulation at the transcriptional level can be subdivided into early and late phases. Early genes are recognized and transcribed by host RNA polymerase II whereas late genes are transcribed by a viral-encoded late RNA polymerase (Friesen and Miller, 2001; Guarino et al., 1998). Prior studies of early and late phase transcription revealed that late genes are transcribed from a promoter that contains the core sequence a/g/tTAAG plus approximately 6-20 nucleotides of flanking sequence (Garrity et al., 1997; Morris and Miller, 1994). Transcription from late promoters typically initiates within the core a/g/tTAAG sequence. Unlike early gene transcription, the late RNA polymerase activity is alpha amanitin sensitive, suggesting a polymerase activity distinct from the host RNA polymerase II (Fuchs et al., 1983; Lu and Miller, 1997). To identify AcMNPV genes necessary for late transcription, a transient late expression assay was used to identify 19 early genes that are necessary and sufficient for efficient late transcription from a model AcMNPV late promoter, the vp39 promoter (Lu and Miller, 1995; Passarelli and Miller, 1993; Rapp et al., 1998). The genes identified in the transient late expression assay are referred to as “late expression factor” (lef) genes. The 19 lef genes can be subdivided into two groups: those associated with viral DNA replication, and those more directly related to late transcription. Using a transient origin-dependent DNA replication assay, several laboratories identified lef genes (ie-1, lef-1, lef-2, lef-3, p143, p35, ie-2, lef-7, DNA polymerase, and pe38) that are required for or stimulate DNA replication from a baculovirus origin of replication (Kool et al., 1994; Lu and Miller, 1995). Because viral DNA replication appears to be a necessary prerequisite for late gene transcription in the infection cycle, the role of the above lef genes in late transcription is likely to be indirect and related to their roles in mediating DNA replication. However, direct roles in late transcription cannot be ruled out. Biochemical studies revealed that accurate late transcription can be reconstituted in vitro by four virus-encoded gene products (lef-4, lef-8, lef-9, and p47) (Guarino et al., 1998). Thus, these four viral proteins are believed to comprise the core of the viral RNA polymerase. In addition to these four core proteins of the viral RNA polymerase, expression of six additional gene products are necessary for, or stimulate late transcription in transient late transcription assays. These include AcMNPV genes lef-5, lef-6, lef-10, lef-11, lef-12, and pp31.
pp31 (previously referred to as 39K) was initially studied as a prototype ‘delayed early gene’ that was expressed at high levels in AcMNPV infected cells (Guarino and Smith, 1990; Guarino and Summers, 1986). Delayed early genes are defined as early genes that are dramatically activated by baculovirus early gene products such as IE-1. In transient expression assays performed with plasmid-borne promoters (in the absence of the viral genome), the pp31 early promoter is activated or stimulated by ie-1, ie-2, orf121, and p35 (Gong and Guarino, 1994; Gong et al., 1998). In addition to its early promoter, the pp31 gene is also transcribed from a late promoter (Guarino and Smith, 1992).
The PP31 protein is a phosphorylated DNA binding protein that is localized in the nuclei of infected cells in both early and late phases. PP31 associates with chromatin in the so-called virogenic stroma of infected cells (Guarino et al., 1992) and biochemical studies suggest that PP31 binds to DNA non-specifically, and to both double and single stranded DNA with similar affinities (Guarino et al., 2002). In transient late expression assays using 18 lef genes to reconstitute late transcription, removal of the pp31 gene resulted in an approximately 90–98% reduction in late transcription from the capsid gene (vp39) promoter (Lu and Miller, 1995; Todd et al., 1995). Those data suggested that PP31 plays an important role in late transcription during infection. However, in transient expression assays lef gene products may not be present at physiological levels. Also, because the promoter-reporter construct is supplied on a plasmid in transient assays, and the late promoter is not in the context of the AcMNPV genome, data from such experiments may not precisely reflect the activity of such promoters in a viral infection. Therefore, in the current study we asked whether pp31 was important or essential for late gene transcription in the context of the AcMNPV genome during infection. In a prior study of a related virus, BmNPV, a recombinant BmNPV containing an insertional knockout of pp31 was viable, but showed reduced production of infectious BV. However, in that study the observed phenotype was not rescued to confirm the role of pp31 (Gomi et al., 1997). For the current study of AcMNPV pp31, we used a BACmid-based system to generate a pp31 knockout in the AcMNPV genome. The pp31 knockout was subsequently rescued by reinserting the pp31 gene into the polyhedrin locus of the same virus genome. We then examined the viability of the resulting viruses and used an oligonucleotide microarray representing the AcMNPV genome in combination with quantitative PCR analysis to examine global and specific effects of the pp31 knockout on viral DNA replication and viral gene transcription. We found that pp31 was not essential for AcMNPV replication in Sf9 cells. The absence of pp31 resulted in a decreased viral titer but the kinetics of viral DNA replication were unaffected. In addition, microarray analysis showed that the absence of pp31 resulted in modestly decreased viral transcripts for many viral genes. Microarray results were confirmed by qPCR analysis of selected genes. In combination, these data show that deletion of the pp31 gene was not lethal and resulted in an apparent modest down-regulation of a subset of viral transcripts.
Materials and Methods
Cells and recombinant viruses
Spodoptera frugiperda Sf9 cells were cultured at 27° C in TNMFH medium (Hink, 1970) supplemented with 10% Fetal Bovine Serum. To examine the role of pp31 in the context of an AcMNPV infection, we constructed an AcMNPV BACmid with a deletion of pp31, by homologous recombination in E. coli. To accomplish this, a chloramphenicol resistance gene (chloramphenicol acetyl transferase or cat) flanked by FRT (FLP recognition target) sites was amplified from plasmid pKD3 (Datsenko and Wanner, 2000) using a PCR primer pair in which each primer contained a 5′ terminal addition of a 40 base-pair sequence homologous to the 5′ or 3′ proximal region of the pp31 ORF (5-′ acgggttgtttcaaaggtttacaagaagcaaacatggtaacatatgaatatcctccttag-3′ and 5′-taaaaaccattaaatatacataaaagttttttatttaatcgtgtaggctggagctgcttc-3′). The resulting PCR fragment, which contained a cat gene flanked by FLP recognition target sites and AcMNPV sequences, was treated with Dpn I to eliminate the template plasmid. The PCR product was then transformed into DH10B/bMON14272/pKD46 E. coli cells (containing Bacmid bMON14272) which had been prepared in the following manner. Lambda RED recombinase was induced in DH10B/bMON14272/pKD46 E. coli cells at 30°C by the addition of 1 mM arabinose approximately 4 h prior to preparation of electrocompetent cells. Transformed cells were incubated at 37°C for 4 h in SOC medium and recombinants were selected as resistant colonies on medium containing kanamycin and chloramphenicol as described previously (Lung et al., 2003).
To minimize foreign DNA in the pp31 locus, the cat gene was subsequently removed by FLP recombinase mediated excision. To accomplish this, recombinant BACmid DNA containing the cat gene (described above) was transformed into E. coli EL250 cells which carry a FLP recombinase gene under an arabinose inducible promoter (Lee et al., 2001). Cells were cultured overnight, diluted 50 fold, induced for one hour in the presence of 0.1% arabinose at 30°, then selected at 30° on kanamycin-containing media. The resulting colonies were tested for chloramphenicol sensitivity and cat gene excision was verified by PCR. The resulting BACmid (bMON14272 pp31KO-cat) was used in combination with helper plasmid (pMON7124) to transform DH10B cells to generate DH10Bac-pp31KO cells. Marker genes were inserted into the pp31 knockout BACmid using transposition and a modified pFastBac1 construct that contained two marker genes (lacZ and GUS) and a promoter cassette. For modification of the pFastBac plasmid, the polyhedrin promoter was removed from pFastBac1 and replaced by an expression cassette containing a lacZ gene under the OpMNPV ie-1 promoter, a gus gene under the AcMNPV p6.9 promoter, and an AcMNPV ie-1 promoter (Fig. 1). The resulting pp31 transfer vector containing marker genes and promoter cassette was named pFastBac1-lacZ/gus AcIE-1 Pro. To confirm the phenotype resulting from the pp31 knockout, transfer vectors for genelation of control BACmids were constructed by inserting the pp31 ORF into pFastBac1-lacZ/gus AcIE-1 Pro under either a) the AcMNPV ie1 promoter (pFastBac1-lacZ/gus AcIE-1 pp31) or b) the wild type pp31 promoter (pFastBac1-lacZ/gus pp31 REP) (Fig. 1). DH10Bac-pp31KO cells were transformed with pFastBac1-lacZ/gus AcIE-1 Pro, pFastBac1-lacZ/gus AcIE-1 pp31 and pFastBac1-lacZ/gus pp31 REP for generation of BACmids, bMON14272 pp31KO, bMON14272 pp31KO-REP-ie1P and bMON14272 pp31KO-REP-P, respectively. These BACmids were subsequently used to transfect Sf9 cells to generate recombinant viruses, vAc pp31KO, vAc pp31KO-REP-ie1P and vAc pp31KO-REP-P, respectively, as described previously (Lung et al., 2002). For transfections, each BACmid DNA preparation (approximately 1 μg) was added to 9 × 105 Sf9 cells in a 6-well plate by the calcium phosphate precipitation method (Blissard and Rohrmann, 1991; Graham and Van Der Eb, 1973; Summers and Smith, 1987). At 72 h post transfection, the supernatant was clarified by centrifugation for 10 min at 2,200 × g then used to infect 9 × 105 Sf9 cells for 48 h. Both transfected and infected cells were stained for GUS activity (according to the BAC-to-BAC manual; Invitrogen) to monitor transfection efficiency and to detect infection by virions generated from transfected cells.
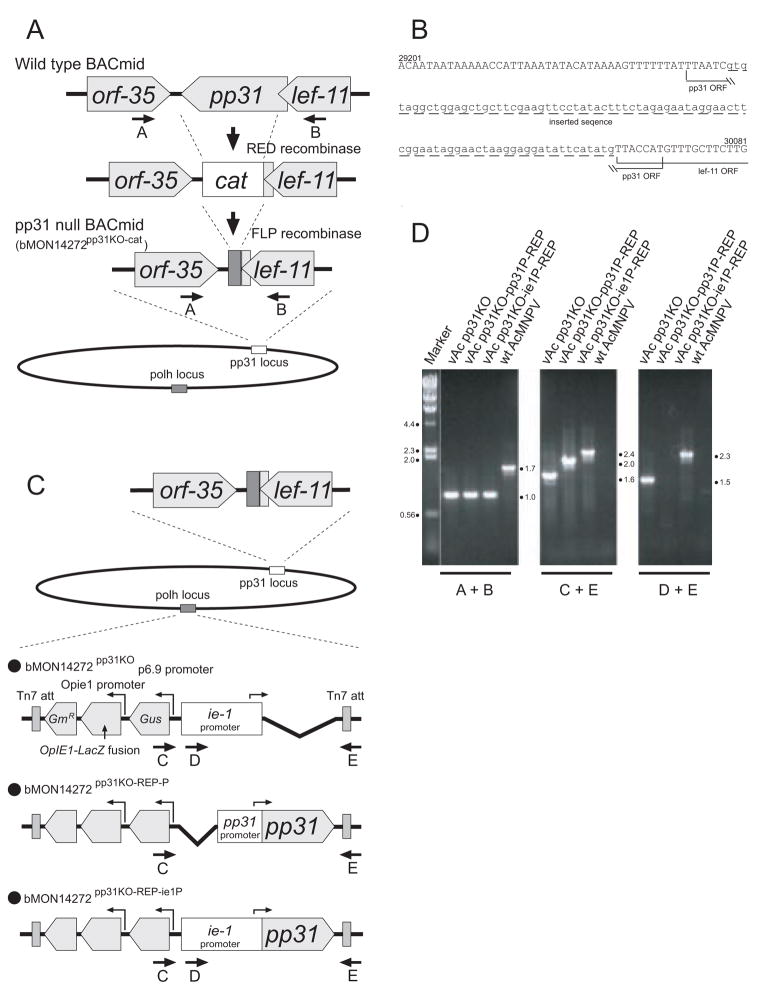
Figure 1.
Construction and analysis of AcMNPV viruses containing a pp31 knockout, and rescue by reinsertion of the pp31 gene.
(A) The strategy for construction of a pp31-null BACmid containing a deletion of the AcMNPV pp31 gene is shown in the diagram. The structure of the pp31 locus is shown in the top line and subsequent modifications are shown below. To eliminate the pp31 ORF, a DNA fragment containing a cat gene flanked by FRT (FLP recognition target) sites and a 40 base-pair sequence homologous to the 5′ or 3′ proximal region of the pp31 ORF, was amplified by PCR and used for homologous recombination with BACmid bMON14272 in KD46 E. coli cells. The structure of the resulting pp31 locus in BACmid (bMON14272 pp31KO+cat) is shown in the second row of the diagram. To minimize foreign DNA in the pp31 locus, the cat gene was subsequently removed by FLP recombinase mediated excision. The structure of the resulting pp31 locus in BACmid bMON14272 pp31KO-cat is shown in the third row of the diagram.
(B) The sequence of the pp31 locus of bMON14272 pp31KO-cat is shown along with the positions of the remaining pp31 ORF sequences and lef-11 ORF sequences. Capital letters indicate wild type AcMNPV sequence and lower case letters represent the location of the pp31 deletion and inserted sequence. Nucleotide position numbers correspond to those reported from the AcMNPV genome sequence (Ayres et al., 1994)(Genbank Accession No. NC_001623).
(C) The strategy for rescue of the pp31 knockout by reinsertion of the pp31 gene is shown. Construction of pp31-repair BACmids by inserting the pp31 gene into the polyhedrin locus is shown in the diagram. One control construct and two pp31 gene constructs were inserted into the polyhedrin locus of the pp31-null BACmid (bMON14272 pp31KO-cat) to generate repair BACmids bMON14272 pp31KO, bMON14272 pp31KO-REP-P and bMON14272 pp31KO-REP-ie1P. The structures of the inserted pp31 constructs are shown and the promoter (wild-type pp31 or wild-type ie-1) used to drive pp31 expression is indicated. Each construct also contains a gentamicin resistance gene (GmR), an OpMNPV ie1 promoter driven OpIE1-LacZ fusion, an AcMNPV p6.9 promoter driven gus gene, and transposon Tn7 attachment sites (Tn7 att).
(D) The results of PCR analysis of the pp31 locus of the pp31 knockout (vAc pp31KO) and the repair (vAc pp31KO-REP-ie1P and vAc pp31KO-REP-P) viruses and wild type AcMNPV are shown on an ethidium bromide-stained agarose gel. Viral DNAs as a template were purified from vAc pp31KO, vAc pp31KO-REP-ie1P, vAc pp31KO-REP-P, and wt AcMNPV infected Sf9 cells. Primer pairs for pp31 knockout locus (A and B) are shown in panel A. Primer pairs for repair locus (C, D and E) are shown in panel C. The sizes of PCR amplification products (in kilobase pairs) are indicated on the right of each gel image.
PCR confirmation of recombinant viruses
To confirm the pp31 knockout, each BACmid was analyzed by PCR with a set of primers, Ac29095 (A: 5′-ttgcgggcaaacaactgg-3′) and Ac30753 (B: 5′-aaagatgacggcgtcacc-3′) that flanked the pp31 ORF (Fig. 1A). To confirm the reinsertion of the pp31 gene into the polyhedrin locus, each BACmid was analyzed by PCR with two pairs of primers. The first primer pair consisted of p6.9 promoter (C: 5′-atccgactgagcgggtacgg-3′) and M13 reverse (E: 5′-ttcacacaggaaacag-3′) corresponding to sequence in the p6.9 promoter and sequence flanking the Tn7-att site, respectively (Fig. 1C). The second primer pair consisted of #689 (D: 5′-ttaatgtttaaatatatcgatgtctttgtgatg-3′, corresponding to ’ sequence in the ie-1 promoter) and an M13 reverse primer which flanks the Tn7-att site (Fig. 1C).
Viral growth curves
To generate viral growth curves, Sf9 cells (1.0 × 106) cultured in 1 ml of TNMFH medium were infected in triplicate with each virus (vAc pp31KO, vAc pp31KO-REP-ie1P, vAc pp31KO-REP-P and wt AcMNPV E2 strain) in six well plates, at a multiplicity of infection (MOI) of 5. After a 1 hour incubation, the inoculum was removed and exchanged with TNMFH. Time zero represents the time point when inoculum was removed after a 1 h incubation. Supernatants were collected at the indicated times post infection and the titers of all supernatants were determined by TCID50 on Sf9 cells (O’Reilly et al., 1992).
Quantitative PCR analysis of viral DNA replication
To analyze viral DNA replication, viral DNAs purified from infected cells were quantified at various times post infection using quantitative real time PCR (qRT-PCR). Sf9 cells (1.0 × 106) were infected in triplicate with each virus (vAc pp31KO, vAc pp31KO-REP-P and vAc pp31KO-REP-ie1P and wt AcMNPV E2 strain) at an MOI of 5 and infected cells were collected at various times post infection. Total DNA was purified from each sample with DNAzol (Invitrogen) according to the manufacturer’s protocol and resuspended in 8 mM NaOH. For qRT-PCR, two primers (AcIE1-84: 5′-ttgtgataaacaacccaacga-3′ and AcIE1-212: 5′-gttaacgagttgacgcttgc-3′) and a 5′ FAM-3′ TAMRA labeled taqman-probe (AcIE1-135-taqmanP: 5′-tcaccgtgtcggctccatcc-3′) were designed within the ie-1 ORF and qRT-PCR reactions were performed using an ABI Prism 7900 and the following conditions: 50 °C for 2 min, 95 °C for 2 min, and 40 cycles of 95 °C for 30 sec and 60° for 30 sec. Each reaction was performed with 500 nM of each flanking primer and 250 nM of the taqman-probe in 1x PCR buffer (Sigma; 10 mM Tris-HCl pH 8.3, 50 mM KCl, 1.5 mM MgCl2, 0.001% gelatin) and 125 μM dNTPs and 0.25 units Taq DNA polymerase (Sigma). As a standard, we used a plasmid (pGEMte-ie1) containing the entire ie-1 ORF in plasmid vector pGEMt-easy. The number of viral DNA genome copies within each sample were calculated using a standard curve generated from a dilution series of the pGEMte-ie1 plasmid.
RNA isolation
For viral transcription analysis using microarray and quantitative reverse transcription PCR analysis, poly(A)+ RNAs were isolated from Sf9 cells (1.5 × 107 cells) infected at an MOI of 5. At each selected time point, cells were lysed in 6 ml of lysis buffer (100 mM Tris-HCl pH 7.5, 500 mM LiCl, 10 mM EDTA, 1% LiDS and 5 mM dithiothreitol) and the lysates were mixed with oligo (dT) cellulose, incubated for 30 min, washed with wash buffer A (10 mM Tris-HCl, pH 7.5, 150 mM LiCl, 1 mM EDTA and 0.1% LiDS), rinsed with wash buffer B (10 mM Tris-HCl, pH 7.5, 150 mM LiCl and 1 mM EDTA) and eluted with 10 mM Tris-HCl pH 7.5. All steps were performed at room temperature and purified mRNAs were quantified by absorbance at 260 nm.
Microarray analysis
For construction of original microarrays designated as BTI-Ac-ver2.1, oligonucleotides of 50 nt were synthesized by MWG-Biotech Inc. Oligonucleotide positions were selected with BioGIST software (MWG-Biotech) in combination with prior results from pilot array experiments. Positioning of the 50mer oligonucleotides representing the AcMNPV genome were biased toward the 3′ end of each ORF, and sequences that contained known overlapping 5′ or 3′ UTRs from adjacent genes were avoided where possible. Oligonucleotides were spotted on UltraGAPSTM Coated Slides (Corning) by the Center for Gene Expression Profiling at the Boyce Thompson Institute. Each array contained 12 replicate spots of each of the oligonucleotides which in total corresponded to: all 155 predicted AcMNPV ORFs (Ayres et al., 1994), 30 cellular genes from Spodoptera frugiperda and Trichoplusia ni, and 10 oligonucleotides corresponding to ‘spike’ RNAs (Table 1). For preparation of each fluorescent labeled probe, 2 μg of poly(A)+ RNA purified from vAc pp31KO or vAc pp31KO-REP-P infected cells was reverse transcribed to generate an aminoallyl-dUTP (aa-dUTP) incorporated cDNA. Reverse transcription reactions were performed under the following conditions: 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 33.3 μM of random hexamer (pd(N)6) primer, 500 μM each of dATP, dCTP, and dGTP, 300 μM of dTTP, 200 μM aa-dUTP, 0.01 M DTT, 10 spike mRNAs at (100 pg each) (Stratagene #252610), 20 units RNase OUT (invitrogen), and 100 units of Super Script II reverse transcriptase (Invitrogen) in a volume of 30 μl. After incubation for 90 min at 42 °, reactions were stopped by the addition of 1 μl of 0.5 M EDTA and 5 μl of 1 N NaOH, followed by incubation for 10 min at 65°. To neutralize each reaction, 25 μl of 1 M HEPES pH 7.0 was added. Unincorporated dNTP and pd(N)6 were removed by filter centrifugation with a Microcon 30 (Millipore) filter unit. cDNA labeled with aa-dUTP was reacted with cy3 or cy5 Mono-Reactive dye (Amersham) in 0.05 M carbonate buffer (pH 9.0) to covalently link incorporated aa-dUTP with cy3 or 5 dye. After a 1 hour incubation, reaction mixtures were stabilized by adding hydroxylamine to a final concentration of 1.5 M. The reaction mixtures were purified with QIAquick PCR Purification kits (Qiagen) and used for hybridization. Parallel hybridizations were carried out using reciprocal dye labelings. The hybridization buffer was comprised of 0.25 M phosphate buffer (pH 7.0), 0.5% SDS, 1 mM EDTA, 1x SSC, and 2x Denhardt’s solution. After overnight incubation at 55 °C, slides were washed in prewarmed 2x SSC and 0.2% SDS at 42 °C for 15 min, 2x SSC at room temperature for 15 min, and 0.2x SSC at room temperature for 15 min.
TABLE 1.
Microarray Oligos
| gene | 5′ a | 3′a | gene | 5′ a | 3′a | gene | 5′ a | 3′a |
|---|---|---|---|---|---|---|---|---|
| PTPase | 955 | 1004 | ORF 54 | 46111 | 46160 | ORF 106 | 93939 | 93988 |
| Ac-Bro | 1090 | 1041 | ORF 55 | 46451 | 46500 | ORF 107 | 94297 | 94346 |
| ctx | 2245 | 2196 | ORF 56 | 46839 | 46888 | ORF 108 | 94486 | 94437 |
| ORF 4 | 2379 | 2330 | ORF 57 | 47509 | 47558 | ORF 109 | 94770 | 94721 |
| ORF 5 | 2963 | 3012 | ORF 58 | 47715 | 47666 | ORF 110 | 96000 | 95951 |
| lef-2 | 3613 | 3662 | ORF 59 | 47931 | 47882 | ORF 111 | 96220 | 96171 |
| ORF 603 | 3808 | 3759 | ORF 60 | 48350 | 48301 | ORF 112 | 96605 | 96654 |
| ph | 5208 | 5257 | fp25K | 48627 | 48578 | ORF 113 | 97244 | 97293 |
| p78/83 | 5681 | 5632 | lef-9 | 50510 | 50559 | ORF 114 | 97935 | 97886 |
| pk1 | 7663 | 7712 | ORF 63 | 51062 | 51111 | ORF 115 | 99282 | 99233 |
| ORF 11 | 7987 | 7938 | gp37 | 51430 | 51381 | ORF 116 | 99853 | 99804 |
| ORF 12 | 9502 | 9551 | DNA pol | 52411 | 52362 | ORF 117 | 100137 | 100186 |
| ORF 13 | 9748 | 9699 | ORF 66 | 57538 | 57587 | ORF 118 | 100303 | 100254 |
| lef-1 | 10932 | 10883 | lef-3 | 58259 | 58210 | ORF 119 | 101935 | 101984 |
| egt | 12818 | 12867 | ORF 68 | 59000 | 59049 | ORF 120 | 102456 | 102505 |
| da18 | 14018 | 14067 | MTase1 | 59981 | 60030 | ORF 121 | 102769 | 102818 |
| da41 | 14521 | 14472 | ORF 70 | 60767 | 60816 | ORF 122 | 102766 | 102717 |
| ORF 19 | 15706 | 15755 | iap2 | 61508 | 61557 | pk2 | 103163 | 103114 |
| arif-1 | 16295 | 16246 | ORF 72 | 61938 | 61987 | ORF 124 | 104458 | 104507 |
| ORF 22 | 18362 | 18411 | ORF 73 | 62120 | 62071 | lef-7 | 104881 | 104832 |
| ORF 23 | 20468 | 20517 | ORF 74 | 62420 | 62371 | chitinase | 106195 | 106146 |
| pkip | 20767 | 20718 | ORF 75 | 63175 | 63126 | v-cath | 107834 | 107883 |
| ORF 25 | 21237 | 21188 | ORF 76 | 63667 | 63618 | gp64 | 108300 | 108251 |
| ORF 26 | 22361 | 22410 | vlf-1 | 63869 | 63820 | p24 | 110352 | 110401 |
| iap1 | 23307 | 23356 | ORF 78 | 65093 | 65044 | gp16 | 110744 | 110793 |
| lef-6 | 23922 | 23971 | ORF 79 | 65353 | 65304 | pp34 | 111536 | 111585 |
| ORF 29 | 24095 | 24046 | gp41 | 65762 | 65713 | ORF 132 | 112356 | 112405 |
| ORF 30 | 24412 | 24363 | ORF 81 | 67073 | 67024 | alk-exo | 113768 | 113817 |
| sod | 26185 | 26234 | tlp | 67698 | 67649 | 94K | 114539 | 114490 |
| fgf | 27095 | 27046 | p95 | 70078 | 70127 | p35 | 117304 | 117353 |
| ORF 33 | 27903 | 27854 | ORF 84 | 71544 | 71593 | p26 | 118707 | 118756 |
| ORF 34 | 28368 | 28319 | ORF 85 | 72046 | 72095 | p10 | 119074 | 119123 |
| v-ubi | 29101 | 29150 | pnk | 72184 | 72135 | p74 | 119401 | 119352 |
| pp31 | 29292 | 29243 | p15 | 74572 | 74621 | me53 | 121351 | 121302 |
| lef-11 | 30247 | 30198 | cg30 | 74786 | 74737 | ORF 140 | 122703 | 122752 |
| ORF 38 | 30647 | 30598 | vp39 | 75934 | 75885 | exon 0 | 123545 | 123594 |
| p43 | 31128 | 31079 | lef-4 | 77871 | 77920 | ie-0 | 122895 | 122944 |
| p47 | 32776 | 32727 | ORF 91 | 78036 | 77987 | 49K | 124643 | 124692 |
| lef-12 | 33872 | 33921 | ORF 92 | 78749 | 78700 | odv-e18 | 125290 | 125339 |
| gta | 35426 | 35475 | ORF 93 | 79658 | 79707 | odv-ec27 | 126051 | 126100 |
| ORF 43 | 35555 | 35604 | p25 | 80608 | 80657 | ORF 145 | 126483 | 126532 |
| ORF 44 | 36099 | 36148 | helicase | 80766 | 80717 | ORF 146 | 126599 | 126550 |
| ORF 45 | 36631 | 36680 | ORF 96 | 84700 | 84749 | ie-1 | 128796 | 128845 |
| odv-e66 | 38719 | 38768 | ORF 97 | 84960 | 85009 | odv-e56 | 129058 | 129009 |
| ets | 39063 | 39014 | ORF 98 | 85135 | 85086 | ORF 149 | 130257 | 130208 |
| etm | 39403 | 39354 | lef-5 | 86451 | 86500 | ORF 150 | 130706 | 130755 |
| pcna-etl | 39742 | 39693 | p6.9 | 86761 | 86712 | ie-2 | 131113 | 131064 |
| lef-8 | 40572 | 40523 | c42 | 86970 | 86921 | ORF 152 | 132227 | 132178 |
| ORF 51 | 44087 | 44136 | ORF 102 | 88350 | 88301 | pe38 | 133403 | 133452 |
| ORF 52 | 44572 | 44523 | p48 | 88507 | 88458 | ORF 154 | 133784 | 133833 |
| ORF 53 | 45068 | 45117 | vp80 | 91544 | 91593 | |||
| lef-10 | 45304 | 45353 | he65 | 91881 | 91832 |
The nucleotide sequence location is indicated for each end of each synthesized oligonucleotide. Numbers correspond to that of the AcMNPV sequence (Ayres et al. 1994), Genbank Acc. No L22858.
Image acquisition was performed using a ScanArray 5000 (Packard BioScience, Meriden, CT, USA) optical scanner. Typically, each slide was scanned twice using high and low gain for the CCD camera. Low gain scans were performed in order to avoid exceeding the dynamic range of the CCD camera for high intensity signals. Signal intensities of the scanned images were quantified with ImaGene software (BioDiscovery, Los Angeles, CA, USA). Fluorescence values with a median signal less than the sum of the local background mean plus two standard deviations were judged indistinguishable from background. Data normalization for differential dye sensitivities was carried out by using the dye signal ratios determined from the spike RNAs to normalize data from each AcMNPV gene after local background subtraction. Infection and synthesis of probes with reciprocal dye labeling were duplicated, resulting 4 hybridization data sets for each gene. The average hybridization ratio and standard deviation were calculated from the data sets (n=4) and a t-test was performed to determine the level of significance for ratios between knockout and repair viruses for each gene. Known early and late genes (viral genes transcribed from TAAG sequence and confirmed by S1 nuclease, primer extention or 5′ RACE assay) are indicated (Table 2 and 3).
TABLE 2.
Microarray analysis of pp31 knockout effects on AcMNPV gene expression
| gene | ratioa | s.d.b | t-testc | p.t.d | gene | ratioa | s.d.b | t-testc | p.t.d | gene | ratioa | s.d.b | t-testc | p.t.d |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ORF 25 | 1.57 | 0.25 | * | ORF 93 | 0.58 | 0.16 | * | ORF 112 | 0.46 | 0.10 | ** | |||
| ORF 115 | 1.36 | 0.20 | * | ORF 30 | 0.57 | 0.17 | * | iap1 | 0.46 | 0.11 | ** | E | ||
| ORF 96 | 1.15 | 0.35 | vp39 | 0.57 | 0.48 | L | ORF 72 | 0.46 | 0.08 | ** | ||||
| egt | 1.10 | 0.35 | E | odv-e66 | 0.57 | 0.11 | ** | L | p95 | 0.46 | 0.03 | ** | ||
| ORF 19 | 1.09 | 0.25 | ORF 13 | 0.56 | 0.16 | * | ORF 45 | 0.46 | 0.07 | ** | ||||
| me53 | 1.08 | 0.10 | Ee | alk-exo | 0.56 | 0.11 | ** | DNA pol | 0.46 | 0.09 | ** | E | ||
|
| ||||||||||||||
| PTPase | 0.98 | 0.31 | L | lef-3 | 0.55 | 0.19 | * | E | lef-10 | 0.45 | 0.04 | ** | ||
| lef-1 | 0.96 | 0.10 | E | gta | 0.55 | 0.10 | * | ORF 66 | 0.44 | 0.09 | ** | |||
| ORF 33 | 0.93 | 0.06 | ORF 81 | 0.55 | 0.16 | * | odv-e18 | 0.44 | 0.17 | ** | L | |||
| ORF 4 | 0.89 | 0.17 | ORF 92 | 0.55 | 0.04 | ** | p74 | 0.43 | 0.13 | ** | ||||
| ORF 34 | 0.89 | 0.14 | p43 | 0.55 | 0.10 | ** | p47 | 0.42 | 0.04 | ** | ||||
| ORF 26 | 0.89 | 0.11 | L | ORF 43 | 0.54 | 0.21 | * | ORF 109 | 0.42 | 0.05 | ** | |||
| pk1 | 0.88 | 0.08 | L | p10 | 0.54 | 0.15 | ** | L | ORF 116 | 0.42 | 0.14 | ** | ||
| da41 | 0.83 | 0.10 | * | p24 | 0.54 | 0.11 | ** | L | ORF 59 | 0.42 | 0.14 | ** | ||
| ORF 68 | 0.83 | 0.35 | he65 | 0.53 | 0.15 | ** | E | ORF 57 | 0.41 | 0.11 | ** | |||
| ORF 85 | 0.82 | 0.16 | exon 0 | 0.53 | 0.10 | ** | L | ORF 114 | 0.41 | 0.13 | ** | |||
| ORF 108 | 0.81 | 0.12 | ORF 140 | 0.53 | 0.10 | ** | ORF 110 | 0.41 | 0.08 | ** | ||||
| ORF 122 | 0.79 | 0.10 | * | gp41 | 0.53 | 0.20 | * | L | lef-7 | 0.40 | 0.12 | ** | ||
| tlp | 0.79 | 0.12 | * | ORF 603 | 0.53 | 0.11 | ** | cg30 | 0.40 | 0.05 | ** | E | ||
| ph | 0.75 | 0.05 | ** | L | ctx | 0.52 | 0.09 | ** | 49K | 0.40 | 0.11 | ** | ||
| arif-1 | 0.75 | 0.18 | etm | 0.52 | 0.09 | ** | ORF 106 | 0.40 | 0.12 | ** | ||||
| ORF 152 | 0.74 | 0.11 | * | MTase1 | 0.52 | 0.04 | ** | L | gp16 | 0.40 | 0.10 | ** | ||
| iap2 | 0.73 | 0.08 | ** | L | ORF 150 | 0.52 | 0.09 | ** | ORF 154 | 0.39 | 0.04 | ** | L | |
| v-ubi | 0.72 | 0.11 | * | L | ORF 132 | 0.52 | 0.13 | ** | ORF 76 | 0.39 | 0.13 | ** | ||
| helicase | 0.71 | 0.08 | ** | E/L | ORF 145 | 0.52 | 0.09 | ** | ORF 121 | 0.39 | 0.06 | ** | ||
| ORF 55 | 0.71 | 0.11 | * | da18 | 0.52 | 0.16 | ** | L | p25 | 0.38 | 0.08 | ** | ||
| lef-6 | 0.70 | 0.06 | ** | E | p35 | 0.52 | 0.12 | ** | E/L | lef-8 | 0.38 | 0.07 | ** | |
| pk2 | 0.70 | 0.14 | * | E | lef-4 | 0.51 | 0.07 | ** | E | ORF 29 | 0.38 | 0.10 | ** | |
| ORF 12 | 0.69 | 0.05 | ** | p78/83 | 0.51 | 0.04 | ** | L | ORF 117 | 0.38 | 0.08 | ** | ||
| ORF 53 | 0.69 | 0.12 | * | ORF 84 | 0.51 | 0.09 | ** | ORF 58 | 0.37 | 0.04 | ** | |||
| ie-0 | 0.69 | 0.19 | * | E/L | ORF 56 | 0.50 | 0.22 | * | ORF 78 | 0.37 | 0.06 | ** | ||
| pcna-etl | 0.67 | 0.12 | * | E | ORF 51 | 0.50 | 0.03 | ** | f | lef-12 | 0.37 | 0.12 | ** | |
|
|
||||||||||||||
| ORF 22 | 0.67 | 0.16 | * | odv-e56 | 0.50 | 0.10 | ** | L | gp37 | 0.37 | 0.07 | ** | L | |
| p26 | 0.66 | 0.18 | * | E | ie-1 | 0.50 | 0.11 | ** | E | odv-ec27 | 0.37 | 0.15 | ** | L |
| ORF 120 | 0.65 | 0.08 | ** | ORF 52 | 0.50 | 0.07 | ** | ORF 119 | 0.37 | 0.06 | ** | |||
| lef-5 | 0.64 | 0.13 | * | vlf-1 | 0.50 | 0.09 | ** | L | lef-11 | 0.36 | 0.01 | ** | E | |
| ie-2 | 0.63 | 0.09 | ** | E | ORF 124 | 0.49 | 0.08 | ** | ORF 107 | 0.36 | 0.08 | ** | ||
| ORF 54 | 0.63 | 0.11 | ** | pe38 | 0.49 | 0.05 | ** | E | c42 | 0.35 | 0.10 | ** | L | |
| lef-9 | 0.63 | 0.13 | * | pkip | 0.48 | 0.12 | ** | ORF 73 | 0.35 | 0.05 | ** | |||
| vp80 | 0.62 | 0.11 | ** | L | pnk | 0.48 | 0.10 | ** | E | p48 | 0.35 | 0.03 | ** | L |
| ORF 23 | 0.62 | 0.07 | ** | ORF 79 | 0.48 | 0.05 | ** | ORF 75 | 0.35 | 0.12 | ** | |||
| fgf | 0.61 | 0.16 | E | Ac-Bro | 0.48 | 0.07 | ** | ORF 74 | 0.35 | 0.06 | ** | g | ||
|
|
||||||||||||||
| ORF 91 | 0.61 | 0.03 | ** | lef-2 | 0.48 | 0.07 | ** | chitinase | 0.31 | 0.05 | ** | |||
| ORF 70 | 0.61 | 0.11 | ** | fp25K | 0.48 | 0.13 | ** | ORF 102 | 0.30 | 0.08 | ** | |||
| ets | 0.61 | 0.10 | ** | E | ORF 5 | 0.47 | 0.13 | ** | ORF 98 | 0.30 | 0.05 | ** | L | |
| ORF 38 | 0.60 | 0.11 | ** | 94K | 0.47 | 0.06 | ** | E | ORF 60 | 0.28 | 0.03 | ** | ||
| ORF 111 | 0.59 | 0.04 | ** | pp34 | 0.47 | 0.05 | ** | ORF 97 | 0.24 | 0.10 | ** | |||
| v-cath | 0.58 | 0.17 | * | gp64 | 0.47 | 0.02 | ** | E/L | p6.9 | 0.24 | 0.10 | ** | L | |
| ORF 118 | 0.58 | 0.09 | ** | ORF 113 | 0.47 | 0.09 | ** | pp31 | 0.09 | 0.03 | ** | E/L | ||
| sod | 0.58 | 0.07 | ** | L | ORF 44 | 0.47 | 0.10 | ** | ||||||
Ratio of the microarray signal intensity from mRNA isolated from cells infected with pp31 knockout (vAcpp31KO) vs pp31 repair (vAcpp31KO-REP-P) viruses.
Standard deviation (n=4).
* and ** represent p values of less than 0.05. and 0.01, respectively. The number of genes with a p value of less than 0.01 is 106.
Phase of Transcription.
“E” represents viral early genes defined by prior studies of either alpha amanitine sensitivity, aphidicolin resistance, cycloheximid resistance, or by analysis of temporal transcription and start site sequence. Viral genes transcribed from a TAAG start site sequence are classified as late and indicated by an “L”. References for early and late genes are summarized in Table 3. Dashed lines represent the positions of ratios of 1.0 (e), 0.5 (f) and 0.33 (g).
Table 3.
References for phase of transcription
Quantitative reverse transcription PCR analysis of viral transcripts
To confirm the trends identified by microarray analysis, we used quantitative reverse transcription PCR to analyze transcripts of several viral genes. As target genes, we selected ie-1 and me53 as representative early genes; p6.9, vp39, p78/83, odv-e18 and odv-e56 as representative late genes; and chitinase which showed a dramatic difference in the microarray analysis. Sf9 cells were infected with the pp31 knockout (vAc pp31KO), repaired viruses (vAc pp31KO-REP-P and vAc pp31KO-REP-ie1P), or wild type AcMNPV at an MOI of 5. Poly(A)+ RNAs were isolated from infected cells at 12, 18, 24 and 36 hours post infection as described above. To generate cDNA, 1 μg of each purified poly(A)+ RNA preparation was used for reverse transcription under the following conditions: 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 5 μM of oligo dT, 500 μM of each dNTP, 0.01 M DTT, 10 units of RNase OUT (Invitrogen) and 50 units of super script II reverse transcriptase (Invitrogen). Reactions were incubated at 42 °C for 90 min and terminated by heating at 70 °C for 10 min.
For qPCR analysis of transcripts from each of the genes below, each cDNA template (described above) was used with the indicated primer sets and Taqman-probes: ie-1 gene (AcIE1-84: 5′-ttgtgataaacaacccaacga-3′, AcIE1-212: 5′-gttaacgagttgacgcttgc-3′ and AcIE1-135-taqmanP: 5′-tcaccgtgtcggctccatcc-3′, 5′ FAM-3′ TAMRA labeled), me53 gene (AcME53-263: 5′-ccgagtttgggaacaagttt-3′, AcME53-379: 5′-ctttcatgatgtcgcgttct-3′ and AcME53-285-taqmqnP: 5′-cgctgtacgcgggcaaacct-3′, 5′ FAM-3′ TAMRA labeled), p6.9 gene (p6.9-37: 5′-accacatatggttcgacacg-3′, p6.9-154: 5′-ttctgtaacttcggcgacct-3′ and p6.9-57 taqmanP: 5′-aacccgagcttctgcgcctg-3′, 5′ FAM-3′ TAMRA labeled), vp39 gene (vp39_75647: 5′-ttgcgcaacgactttatacc-3′, vp39_75535: 5′-tagacggctattcctccacc-3′ and vp39_75566 taqmanP: 5′-caacaccaggcgcaggacct-3′, 5′ FAM-3′ TAMRA labeled), p78/83 gene (p78-1070: 5′-tgatcgcggacgtgttagcc-3′, p78-1197: 5′-cgtgttagctttattaggcc-3′ and p78-1103 taqman-p: 5′-tgccatagccacacgacgcc-3′, 5′ FAM-3′ TAMRA labeled), odv-e18 gene (odvE18-26: 5′- gcgctacgactagcacagac -3′, odvE18-84: 5′- ccgagctgtttccattactg-3′ and odvE18-81 taqmanP: 5′-ccaaacatgttcttgaccatcttggc-3′, 5′ FAM-3′ TAMRA labeled), odv-e56 gene (odvE56-36: 5′- attgtatcctaatcaggccag-3′, odvE56-143: 5′-ccaatgttgcgtacactggg-3′ and odvE56-88: 5′-agcactcccgccggtttcac-3′, 5′ FAM-3′ TAMRA labeled), and chitinase gene (chiti_105422: 5′-agcgtcgactctgtgttagg-3′, chiti_105316: 5′-atcgcgttgagcaagtcgcc-3′, and chiti_105351 taqmanP: 5′-ctcccaagcaaacaggccgc-3′, 5′ FAM-3′ TAMRA labeled). In each case, the amplified sequences were designed so that they were included within each ORF. As a standard for qPCR analysis of each gene transcript, we also performed qPCR analysis using a dilution series of a template plasmid that contained the target ORF sequence. For the standard plasmids, we cloned a PCR amplified fragment from the ORF of each target gene into a pGEMt-easy vector. The copy numbers of standard plasmids used for qPCR analysis were calculated from DNA concentrations of purified plasmids. Poly(A)+ RNA was isolated from 3 independent infections for each time point as described above and qPCR was performed with 1 μg of the each poly(A)+ RNA resulting in 3 data sets for calculations of average, standard deviation and significance (t-test).
Results and Discussion
Generation of pp31 knockout and the repair viruses
In the current study, we examined the role of AcMNPV PP31 in the context of the infection cycle. The pp31 gene was deleted from an AcMNPV genome propagated in E. coli (bMON14272), using a long-primer PCR, RED recombinase system that was described previously (Lung et al., 2003). Rescued or repaired viruses were subsequently generated by reinserting the pp31 gene into the polyhedrin locus, and the phenotypes of knockout and rescued viruses were directly compared.
To generate an AcMNPV virus with a pp31 gene knockout, the pp31 gene was first replaced with an antibiotic resistance gene (cat) cassette by inserting the cat cassette between AcMNPV nt 29,238 and 30,071 in AcMNPV BACmid (bMON14272 pp31KO+cat) (Fig. 1A). After confirming the replacement of the pp31 ORF by the cat gene, the cat gene was subsequently excised by recombination with FLP recombinase (Datsenko and Wanner, 2000) resulting in BACmid bMON14272 pp31KO-cat. Because the pp31 ORF overlaps the lef-11 ORF, a small portion of the 5′ end of the pp31 ORF (7 nt) was retained in the final pp31 null construct in order to preserve the intact lef-11 ORF. The majority of the pp31 gene was replaced with a 84 nt “scar” sequence (Fig. 1A and 1B). Next, a repaired viral genome was generated by reinserting the pp31 gene into the polyhedrin locus of BACmid bMON14272 pp31KO-cat by transposition (Fig. 1C). Two ‘repair’ BACmids were generated: one in which the pp31 gene was under the control of the native pp31 promoter (bMON14272 pp31KO-REP-P), and another in which the pp31 gene was under the transcriptional control of the AcMNPV ie-1 promoter (bMON14272 pp31KO-REP-ie1P). As a control, the ie-1 promoter alone was also inserted into the polyhedrin locus (bMON14272 pp31KO) (Fig. 1C).
To generate recombinant viruses, the resulting bacmids were transfected into Sf9 cells. The pp31 knockout (vAc pp31KO) and the rescued viruses (vAc pp31KO-REP-P and vAc pp31KO-REP-ie1P) were examined to determine if they were able to replicate in Sf9 cells. To accomplish this, we performed a transfection-infection assay, as described previously (Lung et al., 2002). Sf9 cells were first transfected with each bacmid and incubated for 72 hours, then supernatants were removed from the transfected cells and transferred to a second culture of Sf9 cells to monitor the infectivity of the supernatant. Transfections and infections were confirmed by GUS activity screening as all constructs also carried a GUS marker gene under the control of the AcMNPV p6.9 promoter. For both pp31 knockout (vAc pp31KO) and rescued viruses (vAc pp31KO-REP-P and vAc pp31KO-REP-ie1P), replication in Sf9 cells was detected (data not shown).
The pp31 locus of the knockout viruses was confirmed by PCR with a primer set that flanks the pp31 ORF (Fig. 1A, primers A and B). In the pp31 knockout and repaired viruses (Fig. 1C; vAc pp31KO, vAc pp31KO-REP-P and vAc pp31KO-REP-ie1P), the replacement of the pp31 gene by the “scar” sequence resulted in a 1.0 kb PCR product, whereas a 1.7 kb PCR product resulted from a similar amplification of the wild type pp31 locus in wt AcMNPV (Fig. 1D, A+B). Rescue or repair of the virus by insertion of the pp31 gene into the polyhedrin locus of the BACmid was also confirmed by PCR but with 2 different primer pairs as indicated in Figure 1D (primers C+E). The expected amplification products of 1.6, 2.0 and 2.4 kb were detected from the viruses, vAc pp31KO, vAc pp31KO-REP-P and vAc pp31KO-REP-ie1P, respectively. In addition, constructs with the ie-1 promoter were also confirmed by PCR with an ie-1 specific primer and primer E (Fig. 1D, D+E). Using primers D and E, we detected amplification products of 1.5 and 2.3 kb from constructs vAc pp31KO and vAc pp31KO-REP-ie1P.
Virus replication
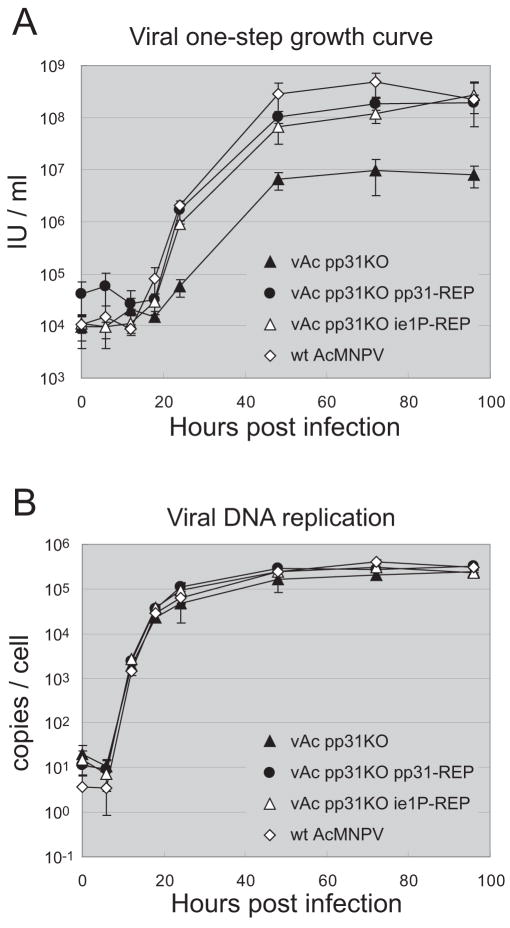
To measure the effect of the pp31 knockout on viral replication, a one-step growth curve analysis was performed (Fig. 2A). By 24 h pi, the titers of the repair (vAc pp31KO-REP-P and vAc pp31KO-REP-ie1P) and wild type viruses were substantially higher than that from the pp31 knockout virus (vAc pp31KO) and the difference (approximately 10–100 fold) was observed throughout the remainder of the timecourse (24 – 96 h pi) of the infection.
Figure 2.
Analysis of the effects of a pp31 knockout on infectious AcMNPV budded virion production and viral DNA replication in Sf9 cells.
(A) Virus one-step growth curves were generated from Sf9 cells infected with either the pp31 knockout virus (vAc pp31KO), the repair viruses (vAc pp31KO-REP-ie1P and vAc pp31KO-REP-P), or wild-type AcMNPV. Infections were performed in triplicate at an MOI of 5, and supernatants were collected and assayed for production of infectious virus by TCID50. Error bars represent the standard deviation from the mean.
(B) To analyze AcMNPV viral DNA replication, Sf9 cells were infected with either the pp31 knockout virus (vAc pp31KO), the repair viruses (vAc pp31KO-REP-ie1P and vAc pp31KO-REP-P), or wild-type AcMNPV at an MOI of 5, and the accumulation of viral DNA in infected Sf9 cells was monitored by qRT-PCR at various time points post infection. Infections were performed in triplicate. The vertical axis represents average viral genome copies per Sf9 cell. Error bars represent the standard deviation from the mean.
Viral DNA replication
To determine whether viral DNA replication in AcMNPV infected Sf9 cells was affected by the pp31 knockout, we used quantitative PCR to measure the accumulation of viral genomic DNA in infected cells (Fig. 2B). Cells were infected at an MOI of 5 with either pp31 knockout (vAc pp31KO), rescued (vAc pp31KO-REP-P and vAc pp31KO-REP-ie1P), or wild type AcMNPV virus and viral genomic DNA was quantified by qPCR at 0, 6, 12, 18, 24, 48, 72, and 96 h pi, using a taqman probe (see Materials and Methods). At 0 and 6 h pi, we detected app. 3 – 20 viral genomes per cell, consistent with the detection of infectious DNA prior to DNA replication. By 12 h pi, we detected the initiation of viral genome replication in all viruses, with approximately 1,000 – 3,000 genomes/cell detected in all cases. By 48 h pi, intracellular viral genome numbers reached approximately 150,000–300,000 genomes/cell and the accumulation of viral genomic DNA appears to have plateaued in all viruses. Thus, the pattern of accumulation of viral genomic DNA in infected cells appears to be similar and the pp31 knockout did not appear to affect viral DNA replication in infected cells. Although these estimates of genomes per cell appear high, they are consistent with prior measurements (Lin and Blissard, 2002; Gomi et al., 1997; Rosinski et al., 2002).
Analysis of global AcMNPV transcription
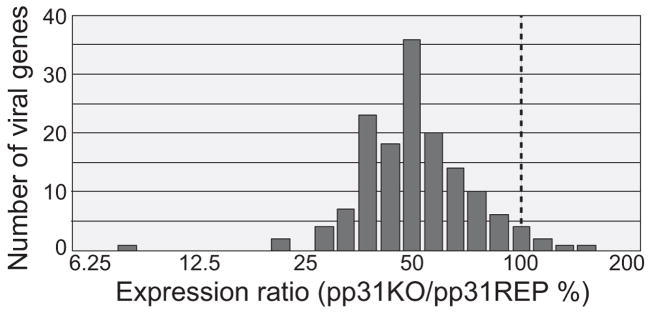
To determine the effect of PP31 on transcriptional regulation in the AcMNPV infection cycle, and to identify specific genes that may be regulated by pp31, an AcMNPV oligonucleotide microarray was used to examine infected cells for effects of the pp31 knockout on accumulation of specific mRNAs. An oligonucleotide microarray consisting of 50mer oligonucleotides and representing 154 described genes of the AcMNPV genome plus a series of control genes (see Materials and Methods) was spotted (Table 1). Each oligonucleotide was spotted 12 times and hybridized using reciprocal labeling of cDNA samples. For analysis at 24 h pi, the study represents 2 technical replicates and 2 biological replicates (see Materials and Methods). The results of the comparison of infected cell mRNAs from vAc pp31KO (pp31 knockout) and vAc pp31KO-REP-P (rescued) virus infected cells are shown in Table 2 and Figure 3. The effects of the pp31 knockout on accumulation of viral mRNAs in general, appeared to be subtle. There was no viral gene in which expression in the pp31 knockout (vAc pp31KO) was increased more than 2 times that in cells infected with the control virus (vAc pp31KO-REP-P). We detected a small (between 1–2x) increase in mRNA in the absence of pp31 for only 6 of the 154 AcMNPV genes (Table 2 and Fig. 3). In contrast, we detected a modest decrease in transcription from many genes when pp31 was deleted (Fig. 3). For 76 AcMNPV genes, we detected a very modest decrease, with mRNAs from the pp31 knockout present at levels between 50 and 100% of that from cells infected with the control virus (vAc pp31KO-REP-P). For 60 AcMNPV genes, mRNA levels were reduced to 33–50 % of the level from the repaired virus. Six genes (p6.9, ORF 97, ORF 60, ORF 98, ORF 102 and chitinase) showed the most dramatic effects with mRNA levels reduced to 20 -33 % of that from the control virus. Of all genes for which expression was detected in control virus infections (vAc pp31KO-REP-P), none appeared to be completely shut down in cells infected with the pp31 knockout virus (vAc pp31KO). A very low level signal was detected from the pp31 gene in the pp31 knockout virus, presumably resulting from non-specific background hybridization to the pp31 oligo. mRNAs for 5 AcMNPV genes (ORF 11, ORF 63, p15, ORF 146 and ORF 149) were not detected in either the pp31 knockout or the repaired virus.
Figure 3.
Effects of an AcMNPV pp31 knockout on transcript levels from AcMNPV genes as determined by AcMNPV microarray analysis. Sf9 cells were infected with either pp31 knockout or repair viruses (vAc pp31KO or vAc pp31KO-REP-P). Cells were infected at an MOI of 5 and mRNAs were isolated at 24 h pi and used to synthesize fluorescent probes for hybridization to an AcMNPV oligonucleotide microarray (see Materials and Methods section). Experiments were performed in duplicate (biological replicates). Reciprocal labeling and hybridizations were performed for each data set and each array contained 12 replica spots of each oligonucleotide. Data points for each gene were averaged from 48 hybridization data points (spots). The horizontal axis represents the average transcript levels of genes from the pp31 knockout virus (vAc pp31KO) infection expressed as a percentage of that from the control or repair virus (vAc pp31KO-REP-P). The vertical axis represents number of viral genes that correspond to each expression percentage. For reference, the dashed line shows genes from which expression was equal (100%) to that of the control or repaired virus.
Analysis of AcMNPV transcription by quantitative RT-PCR
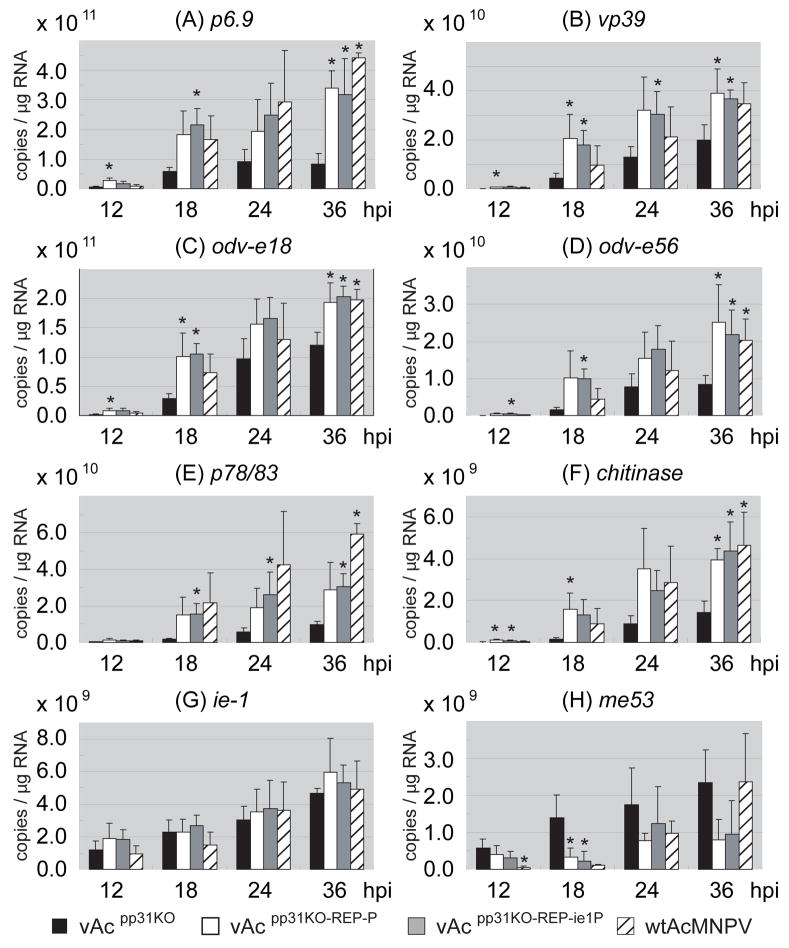
Microarrays are not well suited for measuring small quantitative differences, and large differences were not detected in the microarray analysis of the pp31 knockout. Therefore, to further examine the effects of the pp31 knockout, we examined mRNA accumulation from several well-characterized genes (p6.9, vp39, odv-e56, oed-e18, p78/83, me53, chitinase, and ie1) by quantitative RT-PCR (qRT-PCR). The microarray analysis suggested a general low level decrease in transcription from most AcMNPV genes. Because the chitinase and p6.9 gene transcripts were among those genes from which transcription appeared more severely reduced in the absence of pp31, those genes were included in the analysis of mRNAs by qRT-PCR. Analysis by qRT-PCR showed that p6.9 and chitinase transcripts were substantially reduced when pp31 was absent (Fig. 4A and F, vAcpp31KO vs. pp31repair or control wt AcMNPV). For genes vp39, odv-e18, odv-e56, and p78/83, transcripts were also decreased in the absence of pp31 (Fig. 4). In the later cases, transcription was typically reduced to approximately 20–50% of that from the pp31 repair virus. In the case of two early genes, ie1 and me53, transcripts were either unaffected (ie-1) or even perhaps somewhat increased (me53) in the absence of pp31. These results differed slightly from microarray data since ie-1 showed a very modest decrease (50%) and me53 levels were not substantially different in the absence of pp31. Thus, overall we could not detect any substantial change in the expression of the early genes (ie-1 and me53) in absence of pp31.
Figure 4.
Quantification of transcripts from selected viral genes [p6.9 (A), vp39 (B), odv-e18 (C), odv-e56 (D), p78 (E), chitinase (F), ie1 (G) and me53 (H)] by qRT-PCR. For each graph, the vertical axis indicates the number of transcripts estimated from 1 μg poly(A)+RNA purified from infected Sf9 cells. Infections were performed in triplicate at an MOI of 5 and poly(A)+RNAs were purified and assayed in triplicate. Average qRT-PCR data was derived from 9 data points for each time point post infection (n=9). Error bars represent the standard deviation from the mean. Statistically significant differences between the pp31 knock out and control viruses are indicated by asterisks (* P <0.05).
In several previous studies, the AcMNPV pp31 gene was identified as important for transcription from a late promoter in transient late expression factor assays (Berretta and Passarelli, 2006; Lu and Miller, 1995; Todd et al., 1995). However, in another study (Gomi et al., 1997) the BmNPV pp31 gene was inactivated by insertional mutagenesis and the resulting BmNPV virus was viable. The BmNPV pp31 knockout virus was characterized as having reduced production of infectious BV while not apparently affecting viral DNA replication. However, in that study the pp31 knockout virus was not rescued by reinserting the pp31 gene into the BmNPV genome. Thus, it was unclear whether the observed effects resulted from inactivation of pp31 or whether unapparent mutations may have contributed to the presumed pp31 knockout phenotype. Indeed, the dramatic results from transient late expression assays with AcMNPV lef genes appeared inconsistent with the minor phenotype observed in the BmNPV pp31 knockout virus. In the current study, we generated a pp31 knockout in an AcMNPV BACmid genome and examined the effects of that knockout in detail. Similar to the results of Gomi and colleagues in studies of BmNPV pp31 (Gomi et al., 1997), the AcMNPV pp31 gene knockout was not lethal and resulted in a reduction in the production of infectious virus (Fig. 2A). This reduction in infectious virus production was rescued by reinserting the pp31 gene into the knockout background, confirming the phenotype of the AcMNPV pp31 knockout. Analysis of DNA replication by the AcMNPV pp31 knockout virus indicated no substantial effect of pp31 on AcMNPV DNA replication. In addition, genomic analysis of viral transcripts using an AcMNPV oligonucleotide microarray suggested a general but very modest down regulation of viral transcripts in the absence of pp31.
Of the 154 AcMNPV genes examined by microarray analysis in this study, we detected 149 positive signals indicating that potential changes could be measured for these genes. Comparisons of the signal ratio (pp31knockout vs control virus) for each gene and statistical analysis of the data revealed that for the149 genes, we could not detect significan differences in the expression of 43 genes when pp31 was absent (for these, p values were less than 0.01). In contrast, 99 genes showed modest but statistically significant decreases in transcript levels. The remaining 6 genes (p6.9, ORF 97, ORF 60, ORF 98, ORF 102 and chitinase) showed relatively dramatic and statistically significant reductions in transcript levels (less than 33% of that from control viruses expressing PP31). The most dramatic effects observed from the AcMNPV pp31 knockout was the reduction of transcripts from two genes (orf97 and p6.9) to approximately 22% of their levels in the control (repair) virus.
PP31 is a phosphorylated DNA binding protein that is associated with the virogenic stroma late in infection and prior studies indicate that pp31 is abundantly expressed both early and late in infection. The observation that this highly abundant DNA binding protein was an important component of transient late transcription assays was therefore not surprising. However, the report that the very closely related BmNPV pp31 appeared to be dispensable (Gomi et al., 1997) appeared to conflict with data suggesting a critically important function for PP31 in the AcMNPV infection cycle. In the current study, we found that pp31 was also dispensable in the AcMNPV genome and the absence of PP31 resulted in reduced viral titers, a general but modest down regulation of viral transcripts, but no apparent effect on viral DNA replication. Perhaps most surprising was the observation that the effect of the pp31 knockout on viral transcripts was not limited to or more pronounced in late gene transcripts, but rather included both early and late genes. Because only a fraction of the AcMNPV genes have been transcriptionally mapped and classified (as early, late, or very late, see Table 2 and 3), it is not yet possible to fully understand the implications of the current data. However, when more detailed transcriptional data become available it will be interesting to further examine pp31 knockout data with regard to transcriptional classification.
Because most effects of the pp31 knockout on transcript levels represented either low level or undetectable changes, caution must be exercised in interpreting these data. The combination of biochemical data indicating that pp31 non-specifically binds to DNA (Guarino et al., 2002), and data showing that pp31 is important in transient late promoter assays (Todd et al., 1995), suggests the hypothesis that PP31 may stabilize viral transcription units or otherwise enhance efficiency of viral transcription. The observation that inactivation of pp31 is not lethal in either AcMNPV or BmNPV viruses suggests that either a) PP31 plays a dispensable accessory role in the context of the viral infection, or b) PP31 may play a critical role but its function is redundant with that of another viral or cellular protein.
In this study, we examined the effect of a pp31 knockout on infectious virus production, DNA replication, and global transcription in AcMNPV. We observed a modest reduction or defect in production of infectious budded virus, and data on viral DNA replication suggests that the defect was not related to production of viral genomic DNA. Growth curve data also suggest that the reduction in infectious virus did not result simply from a delay in the infection cycle, but rather an overall limit in production of infectious progeny BV. Combined, these data suggest that the general reduction in budded virion production may have resulted from either reduced production of a limiting protein factor (due to generally reduced transcript levels) or perhaps from a defect in DNA packaging or virus assembly. More detailed biochemical and genetic analyses will be necessary to understand the precise function of the abundant PP31 protein in the baculovirus infection cycle. The pp31 knockout system developed for the current study should be of value in examining PP31 mutants and testing hypotheses regarding the function of PP31.
Acknowledgments
The authors thank Warren Lamboy for discussions on microarray analysis and Takayuki Miyazawa for help in design of taqman probes. This work was supported by USDA grant 2002-35302-12342 and project 1255 of the Boyce Thompson Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- Ayres MD, Howard SC, Kuzio J, Lopez-Ferber M, Possee RD. The complete DNA sequence of Autographa californica Nuclear Polyhedrosis Virus. Virology. 1994;202:586–605. doi: 10.1006/viro.1994.1380. [DOI] [PubMed] [Google Scholar]
- Becker D, Knebel-Morsdorf D. Sequence and temporal appearance of the early transcribed baculovirus gene HE65. J Virol. 1993;67(10):5867–72. doi: 10.1128/jvi.67.10.5867-5872.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berretta MF, Passarelli AL. Function of Spodoptera exigua nucleopolyhedrovirus late gene expression factors in the insect cell line SF-21. Virology. 2006 doi: 10.1016/j.virol.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Blissard GW, Rohrmann GF. Baculovirus gp64 gene expression: Analysis of sequences modulating early transcription and transactivation by IE1. J Virol. 1991;65:5820–5827. doi: 10.1128/jvi.65.11.5820-5827.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunagel SC, Elton DM, Ma H, Summers MD. Identification and analysis of an Autographa californica nuclear polyhedrosis virus structural protein of the occlusion-derived virus envelope: ODV-E56. Virology. 1996a;217(1):97–110. doi: 10.1006/viro.1996.0097. [DOI] [PubMed] [Google Scholar]
- Braunagel SC, Guidry PA, Rosas-Acosta G, Engelking L, Summers MD. Identification of BV/ODV-C42, an Autographa californica nucleopolyhedrovirus orf101-encoded structural protein detected in infected-cell complexes with ODV-EC27 and p78/83. J Virol. 2001;75(24):12331–12338. doi: 10.1128/JVI.75.24.12331-12338.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunagel SC, He H, Ramamurthy P, Summers MD. Transcription, translation, and cellular localization of three Autographa californica nuclear polyhedrosis virus structural proteins: ODV-E18, ODV-E35, and ODV-EC27. Virology. 1996b;222(1):100–114. doi: 10.1006/viro.1996.0401. [DOI] [PubMed] [Google Scholar]
- Carson DD, Summers MD, Guarino LA. Transient expression of the Autographa californica nuclear polyhedrosis virus immediate-early gene, IE-N, is regulated by three viral elements. J Virol. 1991;65(2):945–51. doi: 10.1128/jvi.65.2.945-951.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm GE, Henner DJ. Multiple early transcripts and splicing of the Autographa californica nuclear polyhedrosis virus IE-1 gene. J Virol. 1988;62(9):3193–3200. doi: 10.1128/jvi.62.9.3193-3200.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford AM, Miller LK. Characterization of an early gene accelerating expression of late genes of the baculovirus Autographa californica nuclear polyhedrosis virus. J Virol. 1988;62(8):2773–2781. doi: 10.1128/jvi.62.8.2773-2781.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Stewart TM, Pathakamuri JA, Li Q, Theilmann DA. Autographa californica multiple nucleopolyhedrovirus exon0 (orf141), which encodes a RING finger protein, is required for efficient production of budded virus. J Virol. 2004;78(18):9633–9644. doi: 10.1128/JVI.78.18.9633-9644.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detvisitsakun C, Berretta MF, Lehiy C, Passarelli AL. Stimulation of cell motility by a viral fibroblast growth factor homolog: proposal for a role in viral pathogenesis. Virology. 2005;336(2):308–317. doi: 10.1016/j.virol.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Durantel D, Croizier G, Ravallec M, Lopez-Ferber M. Temporal expression of the AcMNPV lef-4 gene and subcellular localization of the protein. Virology. 1998a;241(2):276–284. doi: 10.1006/viro.1997.8971. [DOI] [PubMed] [Google Scholar]
- Durantel D, Croizier L, Ayres MD, Croizier G, Possee RD, Lopez-Ferber M. The pnk/pnl gene (ORF 86) of Autographa californica nucleopolyhedrovirus is a non-essential, immediate early gene. J Gen Virol. 1998b;79 (Pt 3):629–637. doi: 10.1099/0022-1317-79-3-629. [DOI] [PubMed] [Google Scholar]
- Friesen PD, Miller LK. Divergent transcription of early 35- and 94-kilodalton protein genes encoded by the HindIII K genome fragment of the baculovirus Autographa californica nuclear polyhedrosis virus. J Virol. 1987;61(7):2264–2272. doi: 10.1128/jvi.61.7.2264-2272.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen PD, Miller LK. Insect Viruses. In: Knipe DM, Howley PM, editors. Fields Virology. 4. Vol. 1. Lippincott Williams & Wilkins; Philadelphia: 2001. pp. 599–628. [Google Scholar]
- Fuchs LY, Woods MS, Weaver RF. Viral transcription during Autographa californica nuclear polyhedrosis virus infection: A novel RNA polymerase induced in infected Spodoptera frugiperda cells. J Virol. 1983;48(3):641–646. doi: 10.1128/jvi.48.3.641-646.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrity DB, Chang MJ, Blissard GW. Late promoter selection in the baculovirus gp64 envelope fusion protein gene. Virology. 1997;231(2):167–81. doi: 10.1006/viro.1997.8540. [DOI] [PubMed] [Google Scholar]
- Glocker B, Hoopes RR, Jr, Hodges L, Rohrmann GF. In vitro transcription from baculovirus late gene promoters: accurate mRNA initiation by nuclear extracts prepared from infected Spodoptera frugiperda cells. J Virol. 1993;67(7):3771–3776. doi: 10.1128/jvi.67.7.3771-3776.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomi S, Zhou CE, Yih W, Majima K, Maeda S. Deletion analysis of four of eighteen late gene expression factor gene homologues of the baculovirus, BmNPV. Virology. 1997;230(1):35–47. doi: 10.1006/viro.1997.8457. [DOI] [PubMed] [Google Scholar]
- Gong M, Jin J, Guarino LA. Mapping of ORF121, a factor that activates baculovirus early gene expression. Virology. 1998;244(2):495–503. doi: 10.1006/viro.1998.9116. [DOI] [PubMed] [Google Scholar]
- Gong M, Guarino LA. Expression of the 39k Promoter of Autographa californica Nuclear Polyhedrosis Virus Is Increased by the Apoptotic Suppressor P35. Virology. 1994;204(1):38–44. doi: 10.1006/viro.1994.1508. [DOI] [PubMed] [Google Scholar]
- Graham FL, Van Der Eb AJ. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Guarino LA. Identification of a viral gene encoding a ubiquitin-like protein. Proc Natl Acad Sci U S A. 1990;87(1):409–413. doi: 10.1073/pnas.87.1.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarino LA, Dong W, Xu B, Broussard DR, Davis RW, Jarvis DL. Baculovirus phosphoprotein pp31 is associated with virogenic stroma. J Virol. 1992;66(12):7113–7120. doi: 10.1128/jvi.66.12.7113-7120.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarino LA, Mistretta TA, Dong W. DNA binding activity of the baculovirus late expression factor PP31. Virus Res. 2002;90(1–2):187–195. doi: 10.1016/s0168-1702(02)00152-1. [DOI] [PubMed] [Google Scholar]
- Guarino LA, Xu B, Jin J, Dong W. A virus-encoded RNA polymerase purified from baculovirus-infected cells. J Virol. 1998;72(10):7985–7991. doi: 10.1128/jvi.72.10.7985-7991.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarino LA, Smith M. Regulation of delayed-early gene transcription by dual TATA boxes. J Virol. 1992;66:3733–3739. doi: 10.1128/jvi.66.6.3733-3739.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarino LA, Smith MW. Nucleotide sequence and characterization of the 39k gene region of Autographa californica nuclear polyhedrosis virus. Virology. 1990;179(1):1–8. doi: 10.1016/0042-6822(90)90266-t. [DOI] [PubMed] [Google Scholar]
- Guarino LA, Summers MD. Functional mapping of a trans-activating gene required for expression of a baculovirus delayed-early gene. J Virol. 1986;57(2):563–571. doi: 10.1128/jvi.57.2.563-571.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarino LA, Summers MD. Nucleotide Sequence and Temporal Expression of a Baculovirus Regulatory Gene. J Virol. 1987;61(7):2091–2099. doi: 10.1128/jvi.61.7.2091-2099.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hink WF. Established insect cell line from the cabbage looper, Trichoplusia ni. Nature. 1970;226:466–467. doi: 10.1038/226466b0. [DOI] [PubMed] [Google Scholar]
- Hong T, Braunagel SC, Summers MD. Transcription, translation, and cellular localization of PDV-E66: a structural protein of the PDV envelope of Autographa californica nuclear polyhedrosis virus. Virology. 1994;204(1):210–22. doi: 10.1006/viro.1994.1525. [DOI] [PubMed] [Google Scholar]
- Hoopes RR, Jr, Rohrmann GF. In vitro transcription of baculovirus immediate early genes: accurate mRNA initiation by nuclear extracts from both insect and human cells. Proc Natl Acad Sci U S A. 1991;88(10):4513–4517. doi: 10.1073/pnas.88.10.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard SC, Ayres MD, Possee RD. Mapping the 5′ and 3′ ends of Autographa californica nuclear polyhedrosis virus polyhedrin mRNA. Virus Research. 1986;5:109–119. [Google Scholar]
- Huh NE, Weaver RF. Identifying the RNA polymerases that synthesize specific transcripts of the Autographa californica nuclear polyhedrosis virus. J Gen Virol. 1990a;71:195–202. doi: 10.1099/0022-1317-71-1-195. [DOI] [PubMed] [Google Scholar]
- Huh NE, Weaver RF. Categorizing some early and late transcripts directed by the Autographa californica nuclear polyhedrosis virus. J Gen Virol. 1990b;71:2195–2200. doi: 10.1099/0022-1317-71-9-2195. [DOI] [PubMed] [Google Scholar]
- Kim D, Weaver RF. Transcription mapping and functional analysis of the protein tyrosine/serine phosphatase (PTPase) gene of the Autographa californica nuclear polyhedrosis virus. Virology. 1993;195(2):587–595. doi: 10.1006/viro.1993.1410. [DOI] [PubMed] [Google Scholar]
- Knebel-Morsdorf D, Kremer A, Jahnel F. Baculovirus gene ME53, which contains a putative zinc finger motif, is one of the major early-transcribed genes. J Virol. 1993;67(2):753–758. doi: 10.1128/jvi.67.2.753-758.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kool M, Ahrens CH, Goldbach RW, Rohrmann GF, Vlak JM. Identification of genes involved in DNA replication of the Autographa californica baculovirus. Proc Natl Acad Sci U S A. 1994;91(23):11212–11216. doi: 10.1073/pnas.91.23.11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs GR, Guarino LA, Graham BL, Summers MD. Identification of spliced baculovirus RNAs expressed late in infection. Virology. 1991a;185(2):633–643. doi: 10.1016/0042-6822(91)90534-i. [DOI] [PubMed] [Google Scholar]
- Kovacs GR, Guarino LA, Summers MD. Novel regulatory properties of the IE1 and IE0 transactivators encoded by the baculovirus Autographa californica multicapsid nuclear polyhedrosis virus. J Virol. 1991b;65(10):5281–5288. doi: 10.1128/jvi.65.10.5281-5288.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krappa R, Knebel-Morsdorf D. Identification of the very early transcribed baculovirus gene PE-38. J Virol. 1991;65(2):805–812. doi: 10.1128/jvi.65.2.805-812.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer A, Knebel-Morsdorf D. The early baculovirus he65 promoter: On the mechanism of transcriptional activation by IE1. Virology. 1998;249(2):336–51. doi: 10.1006/viro.1998.9288. [DOI] [PubMed] [Google Scholar]
- Kuzio J, Jaques R, Faulkner P. Identification of p74, a gene essential for virulence of baculovirus occlusion bodies. Virology. 1989;173(2):759–763. doi: 10.1016/0042-6822(89)90593-x. [DOI] [PubMed] [Google Scholar]
- Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73(1):56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- Li Y, Passarelli AL, Miller LK. Identification, sequence, and transcriptional mapping of lef-3, a baculovirus gene involved in late and very late gene expression. J Virol. 1993;67(9):5260–5268. doi: 10.1128/jvi.67.9.5260-5268.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Miller LK. Expression and functional analysis of a baculovirus gene encoding a truncated protein kinase homolog. Virology. 1995;206(1):314–323. doi: 10.1016/s0042-6822(95)80047-6. [DOI] [PubMed] [Google Scholar]
- Lin G, Slack JM, Blissard GW. Expression and localization of LEF-11 in Autographa californica nucleopolyhedrovirus-infected Sf9 cells. J Gen Virol. 2001;82(Pt 9):2289–2294. doi: 10.1099/0022-1317-82-9-2289. [DOI] [PubMed] [Google Scholar]
- Lin G, Blissard GW. Analysis of an Autographa californica multicapsid nucleopolyhedrovirus lef-6-null virus: LEF-6 is not essential for viral replication but appears to accelerate late gene transcription. J Virol. 2002;76(11):5503–5514. doi: 10.1128/JVI.76.11.5503-5514.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Carstens EB. Nucleotide sequence and transcriptional analysis of the p80 gene of Autographa californica nuclear polyhedrosis virus: a homologue of the Orgyia pseudotsugata nuclear polyhedrosis virus capsid-associated gene. Virology. 1992a;190(1):201–209. doi: 10.1016/0042-6822(92)91206-a. [DOI] [PubMed] [Google Scholar]
- Lu A, Carstens EB. Transcription analysis of the EcoRI D region of the baculovirus Autographa californica nuclear polyhedrosis virus identifies an early 4-kilobase RNA encoding the essential p143 gene. J Virol. 1992b;66(2):655–663. doi: 10.1128/jvi.66.2.655-663.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Miller LK. The roles of eighteen baculovirus late expression factor genes in transcription and DNA replication. J Virol. 1995;69:975–982. doi: 10.1128/jvi.69.2.975-982.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Miller LK. Regulation of baculovirus late and very late gene expression. In: Miller LK, editor. The Baculoviruses. Plenum Press; New York: 1997. pp. 193–216. [Google Scholar]
- Lung OY, Cruz-Alvarez M, Blissard GW. Ac23, an envelope fusion protein homolog in the baculovirus Autographa californica multicapsid nucleopolyhedrovirus, is a viral pathogenicity factor. J Virol. 2003;77(1):328–339. doi: 10.1128/JVI.77.1.328-339.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung O, Westenberg M, Vlak JM, Zuidema D, Blissard GW. Pseudotyping Autographa californica multicapsid Nucleopolyhedrovirus (AcMNPV): F proteins from Group II NPVs are functionally analogous to AcMNPV GP64. J Virol. 2002;76:5729–5736. doi: 10.1128/JVI.76.11.5729-5736.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mans RM, Knebel-Morsdorf D. In vitro transcription of pe38/polyhedrin hybrid promoters reveals sequences essential for recognition by the baculovirus-induced RNA polymerase and for the strength of very late viral promoters. J Virol. 1998;72(4):2991–2998. doi: 10.1128/jvi.72.4.2991-2998.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris TD, Miller LK. Mutational analysis of a baculovirus major late promoter. Gene. 1994;140(2):147–153. doi: 10.1016/0378-1119(94)90538-x. [DOI] [PubMed] [Google Scholar]
- Nissen MS, Friesen PD. Molecular analysis of the transcriptional regulatory region of an early baculovirus gene. J Virol. 1989;63(2):493–503. doi: 10.1128/jvi.63.2.493-503.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly DR, Miller LK, Luckow VA. Baculovirus expression vectors, a laboratory manual. W. H. Freeman and Co.; New York: 1992. [Google Scholar]
- O’Reilly DR, Passarelli AL, Goldman IF, Miller LK. Characterization of the DA26 gene in a hypervariable region of the Autographa californica nuclear polyhedrosis virus genome. J Gen Virol. 1990;71 (Pt 5):1029–1037. doi: 10.1099/0022-1317-71-5-1029. [DOI] [PubMed] [Google Scholar]
- O’Reilly DR, Miller LK. Regulation of expression of a baculovirus ecdysteroid UDPglucosyltransferase gene. J Virol. 1990;64(3):1321–1328. doi: 10.1128/jvi.64.3.1321-1328.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oellig C, Happ B, Muller T, Doerfler W. Overlapping sets of viral RNAs reflect the array of polypeptides in the EcoRI J and N fragments (map positions 81.2 to 85.0) of the Autographa californica nuclear polyhedrosis virus genome. J Virol. 1987;61(10):3048–3057. doi: 10.1128/jvi.61.10.3048-3057.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passarelli AL, Miller LK. Identification and characterization of lef-1 a baculovirus gene involved in late and very late gene expression. J Virol. 1993;67(6):3481–3488. doi: 10.1128/jvi.67.6.3481-3488.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passarelli AL, Miller LK. Identification and transcriptional regulation of the baculovirus lef-6 gene. J Virol. 1994;68(7):4458–4467. doi: 10.1128/jvi.68.7.4458-4467.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possee RD, Sun TP, Howard SC, Ayres MD, Hill-Perkins M, Gearing KL. Nucleotide sequence of the Autographa californica nuclear polyhedrosis 9.4 kbp EcoRI-I and -R (polyhedrin gene) region. Virology. 1991;185(1):229–241. doi: 10.1016/0042-6822(91)90770-c. [DOI] [PubMed] [Google Scholar]
- Rankin C, Ladin BF, Weaver RF. Physical mapping of temporally regulated, overlapping transcripts in the region of the 10K protein gene in Autographa californica nuclear polyhedrosis virus. J Virol. 1986;57(1):18–27. doi: 10.1128/jvi.57.1.18-27.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp JC, Wilson JA, Miller LK. Nineteen baculovirus open reading frames, including LEF-12, support late gene expression. J Virol. 1998;72:10197–10206. doi: 10.1128/jvi.72.12.10197-10206.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly LM, Guarino LA. The pk-1 gene of Autographa californica multinucleocapsid nuclear polyhedrosis virus encodes a protein kinase. J Gen Virol. 1994;75 (Pt 11):2999–3006. doi: 10.1099/0022-1317-75-11-2999. [DOI] [PubMed] [Google Scholar]
- Rosinski M, Reid S, Nielsen LK. Kinetics of baculovirus replication and release using real-time quantitative polymerase chain reaction. Biotechnol Bioeng. 2002;77(4):476–480. [PubMed] [Google Scholar]
- Summers MD, Smith GE. Texas Agricultural Experiment Station Bulletin No. 1555. 1987. A manual of methods for baculovirus vectors and insect cell culture procedures. [Google Scholar]
- Thiem SM, Miller LK. A baculovirus gene with a novel transcription pattern encodes a polypeptide with a zinc finger and a leucine zipper. J Virol. 1989a;63(11):4489–4497. doi: 10.1128/jvi.63.11.4489-4497.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiem SM, Miller LK. Identification, sequence, and transcriptional mapping of the major capsid protein gene of the baculovirus Autographa californica nuclear polyhedrosis virus. J Virol. 1989b;63(5):2008–2018. doi: 10.1128/jvi.63.5.2008-2018.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd JW, Passarelli AL, Miller LK. Eighteen baculovirus genes, including lef-11, p35, 39K, and p47, support late gene expression. J Virol. 1995;69:968–974. doi: 10.1128/jvi.69.2.968-974.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomalski MD, Eldridge R, Miller LK. A baculovirus homolog of a Cu/Zn superoxide dismutase gene. Virology. 1991;184(1):149–161. doi: 10.1016/0042-6822(91)90831-u. [DOI] [PubMed] [Google Scholar]
- Tomalski MD, Wu JG, Miller LK. The location, sequence, transcription, and regulation of a baculovirus DNA polymerase gene. Virology. 1988;167(2):591–600. [PubMed] [Google Scholar]
- Whitford M, Faulkner P. Nucleotide sequence and transcriptional analysis of a gene encoding gp41, a structural glycoprotein of the baculovirus Autographa californica nuclear polyhedrosis virus. J Virol. 1992;66(8):4763–4768. doi: 10.1128/jvi.66.8.4763-4768.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson ME, Mainprize TH, Friesen PD, Miller LK. Location, transcription, and sequence of a baculovirus gene encoding a small arginine-rich polypeptide. J Virol. 1987;61(3):661–666. doi: 10.1128/jvi.61.3.661-666.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JG, Miller LK. Sequence, transcription and translation of a late gene of the Autographa californica nuclear polyhedrosis virus encoding a 34.8K polypeptide. J Gen Virol. 1989;70 (Pt 9):2449–2459. doi: 10.1099/0022-1317-70-9-2449. [DOI] [PubMed] [Google Scholar]
- Wu X, Guarino LA. Autographa californica nucleopolyhedrovirus orf69 encodes an RNA cap (nucleoside-2′-O)-methyltransferase. J Virol. 2003;77(6):3430–3440. doi: 10.1128/JVI.77.6.3430-3440.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Miller LK. Expression and mutational analysis of the baculovirus very late factor 1 (vlf-1) gene. Virology. 1998;245(1):99–109. doi: 10.1006/viro.1998.9152. [DOI] [PubMed] [Google Scholar]