Abstract
Mycobacterium tuberculosis, the causative agent of tuberculosis, produces unique sulfated metabolites associated with virulence. One such metabolite from M. tuberculosis lipid extracts, S881, has been shown to negatively regulate the virulence of M. tuberculosis in mouse infection studies, and its cell–surface localization suggests a role in modulating host–pathogen interactions. However, a detailed structural analysis of S881 has remained elusive. Here we use high resolution, high mass accuracy, and tandem mass spectrometry to characterize the structure of S881. Exact mass measurements showed that S881 is highly unsaturated, tandem mass spectrometry indicated a polyisoprene–derived structure, and characterization of synthetic structural analogs confirmed that S881 is a previously–undescribed sulfated derivative of dihydromenaquinone–9, the primary quinol electron carrier in M. tuberculosis. To our knowledge, this is the first example of a sulfated menaquinone produced in any prokaryote. Together with previous studies, these findings suggest that this redox cofactor may play a role in mycobacterial pathogenesis.
Tuberculosis (TB) affects approximately one third of the world’s population and kills approximately two million people a year (1). In order to be an effective pathogen, Mycobacterium tuberculosis, the causative agent of TB, must not only survive the initial onslaught of the host immune response, but also carefully modulate adaptive immunity to allow for bacterial persistence. Sulfated metabolites have been shown to serve as signaling molecules between both symbiotic and pathogenic bacteria and their hosts (2–4), and the sulfate modification is also key to a number of mammalian extracellular signaling events (5). A number of sulfated metabolites have been isolated from the mycobacterial family (6–9), many of which are found in the cell wall (10, 11). While the best–characterized of these molecules is the M. tuberculosis–specific metabolite sulfolipid–1 (SL–1) (9, 12), another sulfated metabolite identified in M. tuberculosis lipid extracts has also been localized to the outer envelope of the cell (8, 10). This previously–uncharacterized metabolite was termed S881 based on its measured mass. Isotopic labeling of S881 with 34SO42− indicated that it contains only one sulfate moiety (8, 10). Despite the identification of this novel metabolite in M. tuberculosis lipid extracts, further structural characterization was hindered by the relatively low abundance of the molecule in M. tuberculosis total lipid extracts.
Although the exact chemical structure of S881 remained elusive, attempts were made to discover the enzymes responsible for the sulfation of the molecule. The M. tuberculosis genome encodes four putative sulfotransferases, named Stf0–3 (13, 14). Stf0 is responsible for the synthesis of SL–1 (14), so it was reasoned that one of the remaining three sulfotransferases would be required for S881 synthesis. M. tuberculosis mutants lacking stf3 (Δstf3) did not produce S881, while complementation with stf3 restored the ability of the Δstf3 mutant strain to produce S881, indicating that Stf3 is necessary for its biosynthesis (10). The S881–deficient Δstf3 mutant was then examined for virulence in a mouse model of TB infection. Interestingly, the Δstf3 mutant shows a hypervirulent phenotype in the mouse model of infection compared to wild–type M. tuberculosis bacteria. From these results, we concluded that S881 is a negative regulator of virulence in the mouse (10).
Enrichment of S881 from M. tuberculosis lipids
Given the hypervirulent phenotype of the S881–deficient Δstf3 mutant and the presence of a sulfate group in the molecule, we resolved to structurally characterize S881. We first partially purified S881 by extracting the total lipids from irradiated H37Rv M. tuberculosis cells, and separating these lipids over an anion exchange resin. The intensity of S881 in the Electrospray Ionization Fourier Transform Ion Cyclotron Resonance (ESI–FT–ICR) mass spectrum of its purest fraction was enriched approximately 200–fold compared to its intensity in crude lipid extracts (Figure S1). This fraction was also free of the contaminating isobar present in M. tuberculosis total lipid extracts (Figure S1B, inset).
Exact Mass and Elemental Composition Analysis
We used the accurate mass capabilities of the FT–ICR MS to measure the exact mass of S881 at 881.5755 Da. The mass spectrum was calibrated internally, and all internal calibrants were measured to <0.7 ppm. The molecular formula generation algorithm from the DataAnalysis software was used to generate putative elemental compositions for the measured mass of S881 (Table S1). Only elemental compositions containing 49 or more carbon atoms were considered based on the intensity of the “M+1” isotope. Only elemental compositions with one sulfur atom were considered in our analysis (10). Elemental compositions with an odd number of nitrogen atoms were not considered, in accordance with the “Nitrogen Rule” (15). Finally, compositions with sulfur and phosphate atoms, but without enough oxygen atoms to support both sulfate and phosphate moieties, were not considered to be possible elemental compositions of S881 because phosphines have not been reported in M. tuberculosis (Table S1, line 2).
Only one such elemental composition that satisfied the above requirements, C56H81O6S1−, was found to be within 0.7 ppm of the measured mass of S881 (Table S1). This elemental composition contains eighteen degrees of unsaturation, including the two on the sulfate, indicating that S881 is highly unsaturated.
Tandem Mass Spectrometry (MSn) of S881
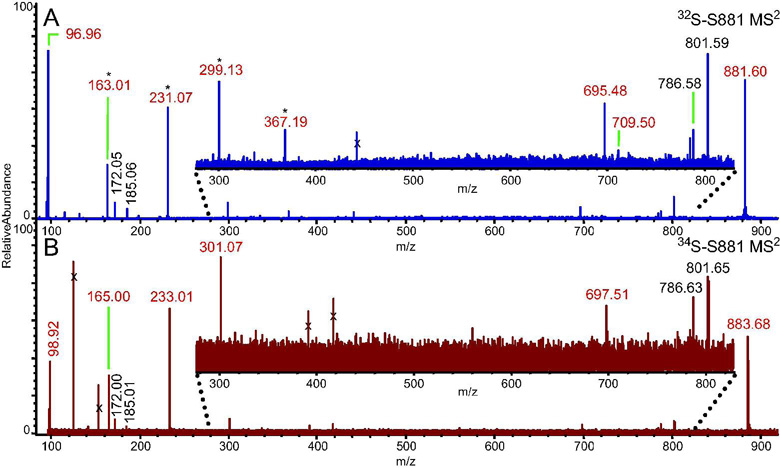
To perform a detailed structural characterization, we analyzed S881 via FT–ICR MSn (Figure 1A). The resulting MS2 spectrum of S881 revealed ions at m/z = 801.59, corresponding to [S881 – SO3]−, and m/z = 96.96, corresponding to HSO4− (Figure 1A). Using the accurate mass capabilities of the FT–ICR MS, we were able to determine the elemental compositions of the dissociation ions (Table S2). We performed MSn on a 34S–labeled sample of S881 (10) to confirm the elemental compositions of the sulfate–containing dissociation ions (Figure 1B). This analysis indicated that the dissociation ions at m/z = 96.96, 163.01, 231.07, 299.13, and 695.48 are indeed sulfated (Figure 1B). This confirms that the dissociation ion at m/z = 96.96 is not H2PO4−, an ion commonly seen in MSn spectra of phosphorylated molecules. Interestingly, we found that many of the sulfate–containing dissociation ions differed in mass by 68 mass units, corresponding to a difference of five carbon and eight hydrogen atoms (Table S2, Figure 1).
Figure 1. MS2 FT–ICR mass spectra of S881.
Ions marked with an “x” were not ejected from the ICR cell prior to activation of the parent ion, and are therefore not dissociation ions of S881. Ions in red are sulfate–containing dissociation ions (Table S2). A) The MS2 FT–ICR mass spectrum of S881. Ions separated by 68 mass units are denoted by an asterisk. B) The MS2 FT–ICR mass spectrum of a 34S–labeled S881. The sulfate–containing dissociation ions are shifted by two mass units compared to the dissociation ions of the 32S–S881.
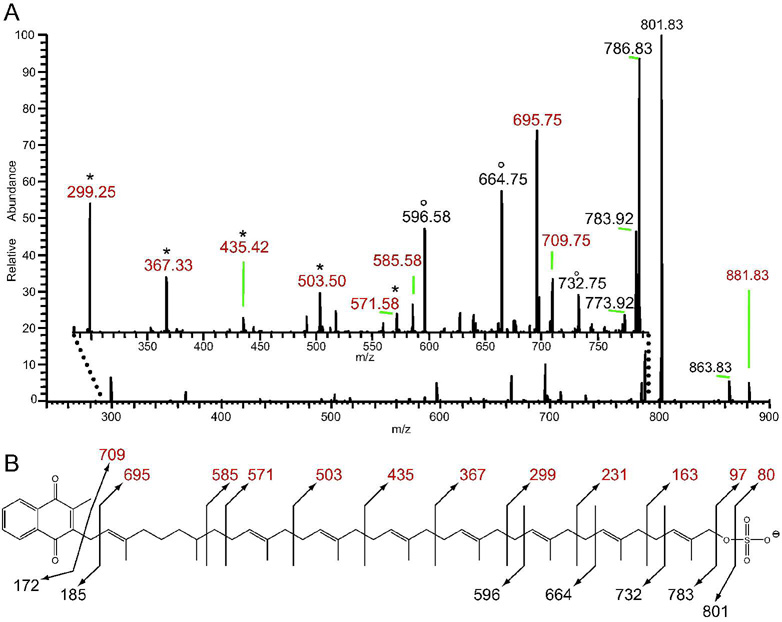
In order to obtain more structural information, we performed MSn of S881 on an LTQ ion trap MS. The resulting MS2 spectrum contained many more dissociation ions compared to those obtained on the FT–ICR MS (Figure 2A). MS3 of the ion at m/z = 801 yielded MS3 dissociation ions without the sulfate residue, and we compared this MS3 spectrum to the MS2 spectrum of S881 to identify ions that contained the sulfate modification. This comparison yielded a series of sulfate–containing ions differing by 68 mass units at m/z = 299, 367, 435, 503, and 571 (Figure 2A). MS3 of the sulfate–containing MS2 dissociation ions yielded [M–68n]− dissociation ions. Taken together with the elemental compositions of the MS2 ions obtained on the FT–ICR MS (Table S2), these data indicate that the sulfate–containing dissociation ions of S881 derive from a structure of a sulfate residue attached to a chain of hydrocarbons with a repeating unit of C5H8.
Figure 2. The LTQ ion–trap MS2 spectrum of S881 indicates that it is a polyisoprenoid–derived molecule.
Values in red denote sulfate–containing dissociation ions. A) A zoom of the m/z = 280–790 region (inset) reveals a number of dissociation ions not obtained via FT–ICR MSn (Figure 1). Two series of dissociation ions separated by 68 mass units are observed; one containing sulfate and denoted by *, and another denoted by °. B) The proposed chemical structure of S881, with observed dissociation ions.
This type of dissociation is consistent with that of a polyisoprenoid chain, which typically dissociates via random allylic cleavage between the individual isoprene units in the chain (16). While mycobacteria contain many polyisoprenoid–derived molecules (17, 18), a key mycobacterial polyisoprene–derived compound with a high degree of unsaturation is dihydromenaquinone–9 (MK–9(H2)) (19). MK–9(H2) consists of a 2–methyl–1,4–napthoquinone moiety alpha–linked to a chain of nine polyisoprene units, however the second isoprene unit from the napthoquinone is saturated. MK–9(H2) is the most abundant quinone in mycobacteria (19). Interestingly, there is no evidence that mycobacteria produce ubiquinone (19) indicating that menaquinone is the sole quinol electron carrier in the mycobacterial respiratory chain.
MSn of S881 Structural Analogs
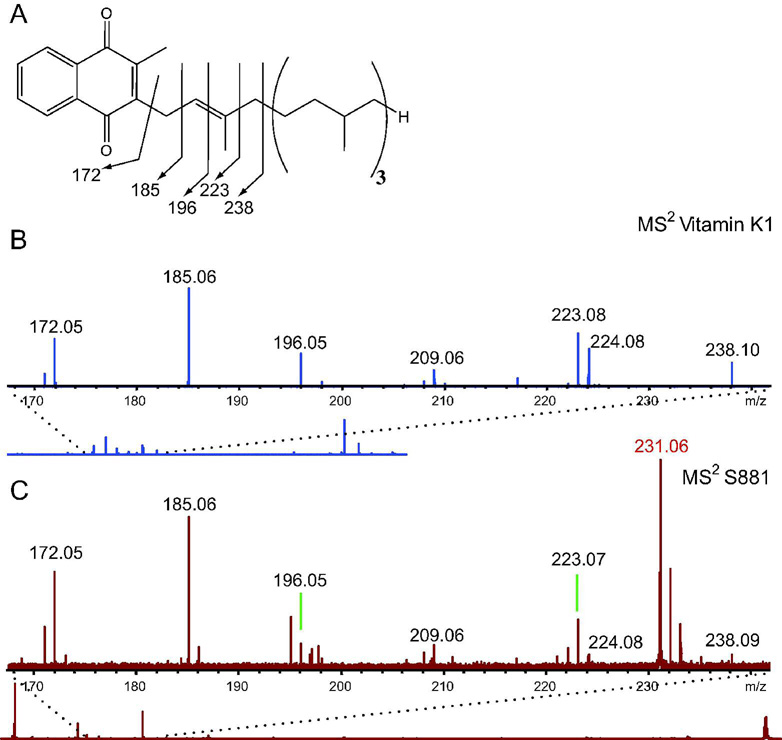
Given the high degree of unsaturation of S881 and its MSn dissociation pattern suggesting a polyisoprenoid structure, we sought to determine if S881 is a sulfated derivative of MK–9(H2) (Figure 2B). FT–ICR MSn was performed on the menaquinone vitamin K1 (Figure 3A) using negative ion mode detection for comparison with the MS2 spectrum of S881. The MS2 spectrum of vitamin K1 revealed ions at m/z = 185 and 223 (Figure 3B), which are consistent with the characteristic dissociation ions at m/z = 187 and 225 observed in the positive mode MS2 spectra of polyisoprenoid menaquinones (19). The vitamin K1 dissociation ions at m/z = 172, 185, 196, 223, and 238 in the MS2 spectrum obtained in the negative ion mode correspond to dissociation of the isoprene unit alpha–linked to the napthoquinone moiety (Figure 3, panel A and B). These ions are also present in the MS2 spectrum of S881 (Figure 3C), clearly supporting a structure containing a napthoquinone moiety.
Figure 3. S881 contains a napthoquinone moiety.
A) The chemical structure of Vitamin K1, and the m/z = 170–240 region of the FT–ICR MS2 spectrum of B) vitamin K1, and C) S881. Dissociation ions at m/z = 172, 185, 196, 223, and 238 are consistent with dissociation of the isoprene unit alpha–linked to the napthoquinone moiety, and are present in both spectra.
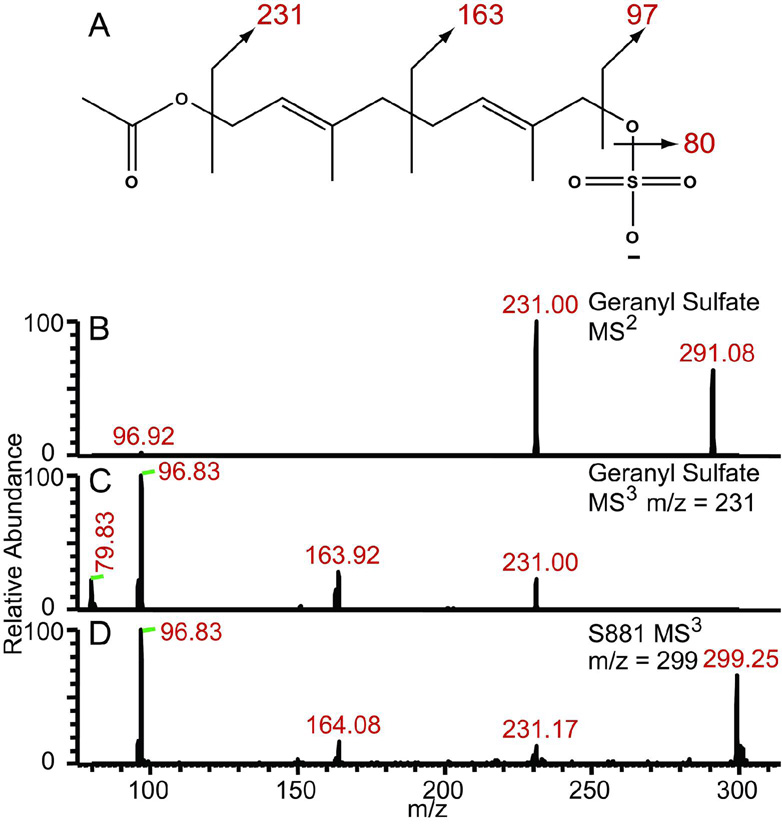
To confirm the proposed position of the sulfate residue on S881 (Figure 2B), we synthesized a geranyl sulfate derivative to compare its dissociation pattern with that of S881 (Figure 4A). The MS2 spectrum of geranyl sulfate yields the expected dissociation ion at m/z = 97 corresponding to HSO4−, as well as an ion at m/z = 231 also present in the MS2 spectrum of S881 (Figure 4B). Conspicuously absent from this spectrum was the expected dissociation ion at m/z = 163 observed in the MS2 spectrum of S881 (Figure 1A). We reasoned that during the MS2 experiment, dissociation is preferred at the ester bond rather than at the carbon–carbon bond between the two isoprene units (Figure 4A). Consistent with this reasoning, MS3 of the m/z = 231 dissociation ion from geranyl sulfate yielded an ion at m/z = 164 (Figure 4C), which is also present in the MS3 spectrum of the m/z = 299 dissociation ion of S881 (Figure 4D). Taken together, these data show that S881 is a sulfated derivative of MK–9(H2) (Figure 2B).
Figure 4. S881 contains an allylic sulfoester.
A) The chemical structure of the synthesized geranyl sulfate, B) The MS2 spectrum of geranyl sulfate, C) the MS3 spectrum of the m/z = 231 dissociation ion from geranyl sulfate, and D) the MS3 spectrum of the m/z = 299 dissociation ion from S881. The MS3 spectra from both molecules yield identical dissociation ions at m/z = 97, 164, and 231, corresponding to loss of sulfate and the dissociation of the polyisoprenoid chain.
To our knowledge, our proposed structure of S881 is unique to M. tuberculosis and represents a novel modification to the common quinone scaffolds. Also, this structure is distinct from previously characterized mycobacterial sulfated metabolites, including the carbohydrate–based SL–1 and its biosynthetic precursors (8, 12, 20, 21) and the sulfated glycopeptidolipids from M. avium and M. fortuitum (6, 7). Sulfur–containing menaquinones have been reported in the thermophilic bacteria Hydrogenobacter thermophilus (22) and Caldariella acidophila (19), however these quinones contain reduced sulfur in the napthoquinone moiety, as opposed to a sulfate modification on the terminal end of the polyisoprenoid chain.
Our elucidation of the chemical structure of S881 will allow us to further probe its biosynthesis. We postulate that S881 is synthesized from MK–9(H2), however this biosynthesis requires at least two steps: oxidation at the terminal position of the polyisoprenoid chain, and sulfation of the resulting hydroxyl moiety. Previously, we provided evidence that the putative sulfotransferase Stf3 is necessary for the production of S881 (10). Interestingly, a putative cytochrome P450 monooxygenase, cyp128, has been annotated in the M. tuberculosis genome directly upstream of stf3 (23). Given the genomic position of this gene in the same putative operon as stf3, we hypothesize that Cyp128 is the enzyme responsible for the terminal oxidation of the polyisoprenoid chain of MK–9(H2), thereby allowing sulfation of the resulting hydroxyl moiety by Stf3. Characterization of Cyp128 in vitro and analysis of the metabolomic profile of a cyp128 mutant will be key to understanding its role in S881 biosynthesis.
The structure of S881, coupled with its localization to the outer cell wall of the bacterium and its role as a negative regulator of virulence, poses many questions regarding a molecular function for the metabolite. The location of S881 in the outer leaflet of M. tuberculosis allows it to interact with host immune cells and modulate the immune response. Disrupting S881 biosynthesis could disrupt the balance M. tuberculosis has with host immune cells, resulting in a lessened immune response. Alternatively, the presence of an allylic sulfate group in S881 suggests that the compound could be an activated intermediate on a pathway to a yet unknown compound, since the sulfate moiety could easily function as a leaving group. Finally, another putative function of S881 is to change the redox balance or availability of menaquinone in the cell. While many recent studies have shed light on the mycobacterial respiratory chain (24, 25), little is known about the regulation of quinone availability in M. tuberculosis. S881 could be an intermediate in a mechanism of non–transcriptional regulation of quinone availability. The conversion of MK–9(H2) to S881 and its subsequent secretion could disrupt the redox balance of the respiratory chain, possibly slowing the respiratory capacity of the cell.
In conclusion, we have successfully characterized the structure of the M. tuberculosis metabolite S881 as a sulfated derivative of MK–9(H2). The structure of S881 represents a novel class of metabolites, and broadens our general understanding of prokaryotic sulfated molecules. Further insight into its mechanism of action will aid our understanding of its contribution to the virulence and life cycle of M. tuberculosis.
METHODS
S881 Extraction From M. tuberculosis Cells
Approximately five grams of M. tuberculosis H37Rv cells (Colorado State University) were extracted in 100 ml CHCl3: MeOH (1:1, v/v) at room temperature for 2 hours. The organic layer was filtered 3 times by vacuum filtration. The filtrate was concentrated to dryness and resuspended in 50 ml CHCl3. The CHCl3 was extracted with water (2 × 25 ml), and the water layer was extracted with 50 ml CHCl3. The CHCl3 layers were combined, concentrated, and the lipid residue was weighed. 40 mg of lipid residue was resuspended in CHCl3: MeOH (4:1, v/v) and passed over an anion exchange resin (AG4–X4, 100–200 mesh, biotechnology grade, free–base form, Bio–RAD) that had been pre–charged with CHCl3: MeOH: AcOH (400:100:0.6, v/v). The void volume was collected, the column was washed with CHCl3: MeOH (4:1, v/v), and the wash was collected. The column was eluted by a gradient of 2–5 mM triethylamine in CHCl3: MeOH (4:1, v/v). Fractions were concentrated and stored at −20 °C until MS analysis.
Synthesis of Geranyl Sulfate Analogue
The synthesis and characterization data for all synthetic analogs is provided in the supporting information section.
Sample Preparation for MS Analysis
The S881 fractions were dried and resuspended in 1 ml of CHCl3: MeOH (2:1) for MS analysis. Vitamin K1 was purchased from Fisher Scientific and was diluted with CHCl3: MeOH (2:1, v/v) to a final concentration of 5 µM for MS analysis. Dry geranyl sulfate solid (Scheme S1, IV) was diluted to a final concentration of 20 µM in CHCl3: MeOH (2:1, v/v).
ESI–FTICR MS
Mass spectra were obtained on an Apex II FT–ICR MS (Bruker Daltonics) equipped with a 7 T actively–shielded superconducting magnet. Samples were introduced into the ion source via direct injection at a rate of 2 µl/min. Ions were generated with an Apollo pneumatically–assisted electrospray ionization source (Bruker Daltonics) operating in the negative ion mode, and were accumulated in an rf–only external hexapole for 0.5–2 s before being transferred to the ICR cell for mass analysis.
For MSn, ions were isolated by a correlated harmonic excitation field isolation sweep, and cleanup shots were used to eject ions not ejected from the initial sweep. Isolated ions were collisionally activated via sustained off–resonance irradiation collision–induced dissociation (SORI–CID) at 1.2 to 1.5 kHz above the cyclotron frequency of the ion of interest, using argon as the collision gas. Dissociation ions were excited for detection after a delay of 2–3 s to allow the residual argon to pump away.
Mass spectra consist of 256k to 1M data points and are an average of 24–32 scans. The spectra were acquired using XMASS version 6.0.0 or 7.0.8 (Bruker Daltonics). For accurate mass measurements, spectra were internally calibrated with four to 12 known compounds. DataAnalysis 3.4 (Bruker Daltonics) was used to determine elemental compositions.
ESI Linear–Ion Trap MS
Mass spectra were obtained on an LTQ ion trap mass spectrometer (ThermoFinnigan) operating in the negative ion mode. Ions were introduced into the ion source via direct injection at a rate of 5 µl/min. For MSn experiments, the precursor ions were isolated with an isolation width of 1–3 Da, the ions were activated with a 25% normalized collision energy for 100 ms, and the qz value was maintained at 0.250. Spectra are an average of 40–100 scans, acquired using Xcalibur, version 1.4 (ThermoFinnigan).
Supplementary Material
This material is available free of charge via the Internet.
Acknowledgments
J.A.L and C.R.B. acknowledge NIH #AI51622 for financial support of this research. The authors would like to thank J. Mougous, and members of the Leary and Bertozzi labs for critical evaluation of this manuscript. H37Rv was kindly provided as part of the NIH, NIAID Contract No. HHSN266200400091C, entitled "Tuberculosis Vaccine Testing and Research Materials," which was awarded to Colorado State University.
REFERENCES
- 1.World Health Organization. Tuberculosis Fact Sheet No. 104. 2007
- 2.Ehrhardt DW, Atkinson EM, Faull KF, Freedberg DI, Sutherlin DP, Armstrong R, Long SR. In vitro sulfotransferase activity of NodH, a nodulation protein of Rhizobium meliloti required for host–specific nodulation. J Bacteriol. 1995;177:6237–6245. doi: 10.1128/jb.177.21.6237-6245.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roche P, Debellé F, Maillet F, Lerouge P, Faucher C, Truchet G, Dénarié J, Promé JC. Molecular basis of symbiotic host specificity in Rhizobium meliloti: nodH and nodPQ genes encode the sulfation of lipo–oligosaccharide signals. Cell. 1991;67:1131–1143. doi: 10.1016/0092-8674(91)90290-f. [DOI] [PubMed] [Google Scholar]
- 4.Shen Y, Sharma P, da Silva FG, Ronald P. The Xanthomonas oryzae pv. lozengeoryzae raxP and raxQ genes encode an ATP sulphurylase and adenosine–5'–phosphosulphate kinase that are required for AvrXa21 avirulence activity. Mol Microbiol. 2002;44:37–48. doi: 10.1046/j.1365-2958.2002.02862.x. [DOI] [PubMed] [Google Scholar]
- 5.Farzan M, Mirzabekov T, Kolchinsky P, Wyatt R, Cayabyab M, Gerard NP, Gerard C, Sodroski J, Choe H. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV–1 entry. Cell. 1999;96:667–676. doi: 10.1016/s0092-8674(00)80577-2. [DOI] [PubMed] [Google Scholar]
- 6.López Marín LM, Lanéelle MA, Promé D, Lanéelle G, Promé JC, Daffé M. Structure of a novel sulfate-containing mycobacterial glycolipid. Biochemistry. 1992;31:11106–11111. doi: 10.1021/bi00160a021. [DOI] [PubMed] [Google Scholar]
- 7.Khoo KH, Jarboe E, Barker A, Torrelles J, Kuo CW, Chatterjee D. Altered expression profile of the surface glycopeptidolipids in drug–resistant clinical isolates of Mycobacterium avium complex. J Biol Chem. 1999;274:9778–9785. doi: 10.1074/jbc.274.14.9778. [DOI] [PubMed] [Google Scholar]
- 8.Mougous JD, Leavell MD, Senaratne RH, Leigh CD, Williams SJ, Riley LW, Leary JA, Bertozzi CR. Discovery of sulfated metabolites in mycobacteria with a genetic and mass spectrometric approach. Proc Natl Acad Sci U S A. 2002;99:17037–17042. doi: 10.1073/pnas.252514899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schelle MW, Bertozzi CR. Sulfate metabolism in mycobacteria. Chembiochem. 2006;7:1516–1524. doi: 10.1002/cbic.200600224. [DOI] [PubMed] [Google Scholar]
- 10.Mougous JD, Senaratne RH, Petzold CJ, Jain M, Lee DH, Schelle MW, Leavell MD, Cox JS, Leary JA, Riley LW, et al. A sulfated metabolite produced by stf3 negatively regulates the virulence of Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:4258–4263. doi: 10.1073/pnas.0510861103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackson M, Stadthagen G, Gicquel B. Long–chain multiple methyl–branched fatty acid–containing lipids of Mycobacterium tuberculosis: biosynthesis, transport, regulation and biological activities. Tuberculosis (Edinb) 2007;87:78–86. doi: 10.1016/j.tube.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 12.Goren M, Brokl O, Das BC. Sulfatides of Mycobacterium tuberculosis: the structure of the principal sulfatide (SL–1) 1976;15:2728–2735. doi: 10.1021/bi00658a003. [DOI] [PubMed] [Google Scholar]
- 13.Mougous JD, Green RE, Williams SJ, Brenner SE, Bertozzi CR. Sulfotransferases and sulfatases in mycobacteria. Chem Biol. 2002;9:767–776. doi: 10.1016/s1074-5521(02)00175-8. [DOI] [PubMed] [Google Scholar]
- 14.Mougous JD, Petzold CJ, Senaratne RH, Lee DH, Akey DL, Lin FL, Munchel SE, Pratt MR, Riley LW, Leary JA, et al. Identification, function and structure of the mycobacterial sulfotransferase that initiates sulfolipid–1 biosynthesis. Nat Struct Mol Biol. 2004;11:721–729. doi: 10.1038/nsmb802. [DOI] [PubMed] [Google Scholar]
- 15.Watson JT. Introduction to Mass Spectrometry. 3rd ed. Philadelphia, PA: Lippincott–Raven; 1997. [Google Scholar]
- 16.Hermansson K, Jansson P, Löw P, Dallner G, Swiezewska E, Chojnacki T. Analysis of Long–chain Polyisoprenoids by Fast Atom Bombardment Mass Spectrometry. Biol Mass Spectrom. 1992;21:548–553. [Google Scholar]
- 17.Kaur D, Brennan PJ, Crick DC. Decaprenyl diphosphate synthesis in Mycobacterium tuberculosis. J Bacteriol. 2004;186:7564–7570. doi: 10.1128/JB.186.22.7564-7570.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahapatra S, Yagi T, Belisle J, Espinosa B, Hill P, McNeil M, Brennan P, Crick D. Mycobacterial lipid II is composed of a complex mixture of modified muramyl and peptide moieties linked to decaprenyl phosphate. J. Bacteriol. 2005;187:2747–2757. doi: 10.1128/JB.187.8.2747-2757.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collins MD, Jones D. Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implication. Microbiol Rev. 1981;45:316–354. doi: 10.1128/mr.45.2.316-354.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Converse SE, Mougous JD, Leavell MD, Leary JA, Bertozzi CR, Cox JS. MmpL8 is required for sulfolipid-1 biosynthesis and Mycobacterium tuberculosis virulence. Proc Natl Acad Sci U S A. 2003;100:6121–6126. doi: 10.1073/pnas.1030024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Domenech P, Reed MB, Dowd CS, Manca C, Kaplan G, Barry CE. The role of MmpL8 in sulfatide biogenesis and virulence of Mycobacterium tuberculosis. J Biol Chem. 2004;279:21257–21265. doi: 10.1074/jbc.M400324200. [DOI] [PubMed] [Google Scholar]
- 22.Ishii M, Kawasumi T, Igarashi Y, Kodama T, Minoda Y. 2–Methylthio–1,4–naphthoquinone, a unique sulfur–containing quinone from a thermophilic hydrogen–oxidizing bacterium, Hydrogenobacter thermophilus. J Bacteriol. 1987;169:2380–2384. doi: 10.1128/jb.169.6.2380-2384.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 24.Matsoso LG, Kana BD, Crellin PK, Lea–Smith DJ, Pelosi A, Powell D, Dawes SS, Rubin H, Coppel RL, Mizrahi V. Function of the cytochrome bc1–aa3 branch of the respiratory network in mycobacteria and network adaptation occurring in response to its disruption. J Bacteriol. 2005;187:6300–6308. doi: 10.1128/JB.187.18.6300-6308.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Voskuil MI, Visconti KC, Schoolnik GK. Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low–oxygen dormancy. Tuberculosis (Edinb) 2004;84:218–227. doi: 10.1016/j.tube.2004.02.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This material is available free of charge via the Internet.