Summary
Models of macromolecular assemblies are essential for a mechanistic description of cellular processes. Such models are increasingly obtained by fitting atomic-resolution structures of components into a density map of the whole assembly. Yet, current density-fitting techniques are frequently insufficient for an unambiguous determination of the positions and orientations of all components. Here, we describe MultiFit, a method for simultaneously fitting atomic structures of components into their assembly density map at resolutions as low as 25 Å. The component positions and orientations are optimized with respect to a scoring function that includes the quality-of-fit of components in the map, the protrusion of components from the map envelope, as well as the shape complementarity between pairs of components. The scoring function is optimized by our exact inference optimizer DOMINO that efficiently finds the global minimum in a discrete sampling space. MultiFit was benchmarked on 7 assemblies of known structure, consisting of up to 7 proteins each. The input atomic structures of the components were obtained from the Protein Data Bank as well as by comparative modeling based on 16 – 99% sequence identity to a template structure. A near-native configuration was usually found as the top-scoring model. Therefore, MultiFit can provide initial configurations for further refinement of many multi-component assembly structures described by electron microscopy.
Keywords: electron microscopy, protein structure modeling, docking, optimization, macromolecular assemblies
Introduction
Structural description of macromolecular assemblies is essential for a mechanistic understanding of the cell1. The scope of the problem is revealed by protein interaction studies: The yeast cell contains approximately 800 distinct core complexes of 4.9 proteins on average2, most of which have not yet been structurally characterized3. The human proteome is likely to have an order of magnitude more distinct assemblies than the yeast cell. Therefore, there are thousands of biologically relevant assemblies whose structures still need to be determined.
Structural determination of macromolecular assemblies is a major challenge in structural biology. X-ray crystallography can provide structures of stable assemblies at atomic resolution4. However, there are many other assemblies that are refractory to crystallographic determination. A low-resolution structure of these assemblies can be determined by cryo-electron microscopy (cryoEM)5. The resolution usually ranges from 4 Å, where the backbone of the protein can be traced, to 30 Å, where only the outer envelope of the assembly is visible6.
The increasing numbers of the atomic and cryoEM datasets7 have stimulated the development of computational techniques for fitting atomic structures of assembly components into a cryoEM density map of the whole assembly. The result is a pseudo-atomic model of the assembly that can reveal significant insights into its structure, dynamics, function, and evolution8–12.
Here, we focus on determining the positions and orientations (i.e., placements) of multiple atomic component models within the assembly density. When the structure of a homologous assembly (template) is available, the placements of the components can be computed by fitting the template into the target assembly density, superposing the target component models on the corresponding template components, and refining the model13; 14. Alternatively, the component positions can be determined experimentally by a number of protein labeling methods, relying for example on gold-labeled antibodies15. However, when only a cryoEM map and component structures are available, a general method for solving the configuration problem is not yet available.
A sequential method for fitting multiple components into an assembly map has been described16. The method starts by fitting the largest component into the map, followed by an iterative fitting of the largest remaining component into the unoccupied density, until all components are fitted. The fitting of a component into a given map can be performed manually using interactive visualization tools17. More desirably, automated fitting methods that assess the placement of a component by a fit between the component and a segmented6 or complete density of the assembly can also be used; the fit is optimized over the translational and rotational degrees of freedom of a rigid component relative to the map18. The sequential method is applicable if the components to be fitted dominate the unoccupied densities. Unfortunately, this condition is generally not satisfied, especially when the resolution is low, the number of components is large, and component models are inaccurate19. For example, sequential fitting is not expected to work for the 19S proteasome with 18 component proteins20, the mammalian ribosome for which 30 out of 80 proteins are not present in the known archaeal or bacterial ribosomes13, nor the ryanodine receptor isoform 1 (RyR1) for which some domains are poorly modeled while for others no template is available21.
Here, we describe a method named MultiFit for determining the configuration of multiple high-resolution component structures based on the quality-of-fit of each component into the density map, the protrusion of each component from the map envelope, and the shape complementarity between pairs of components. The combination of these terms reduces the ambiguity of the final solution, compared to using any individual term on its own.
The task of sampling the configuration space is challenging because the placement of a component depends on the placements of other components. MultiFit tackles this combinatorial challenge by reformulating the problem as an inferential optimization over a discrete sampling space. In outline, a discrete set of possible placements for each component is first generated independently of other components. Next, the globally optimal combination of placements with respect to a scoring function is found by a combination of branch-and-bound search and the DOMINO (Discrete Optimization of Multiple INteracting Objects) inferential optimizer. The relative translations and orientations of pairs of components in the best ranking configurations are then refined; specifically, a refined discrete sampling space is generated by pairwise geometrical docking between interacting components, and the optimal refined combination of placements is again found using DOMINO. We successfully validated the method on a simulated benchmark of 6 assemblies, consisting of up to 7 proteins each. In addition, for a more realistic test, we determined the configuration of 4 domains in the subunit of GroES-ADP7-GroEL-ATP7 chaperonin from Echerichia coli based on an experimentally determined map at the resolution of 23.5 Å22. A near-native configuration scored best in 4 test cases, 3rd best in 2 cases, and 4th best in the remaining case.
Below, we begin with a detailed description of general combinatorial optimization by DOMINO, followed by a formal definition of the component configuration problem and the MultiFit algorithm to solve it using DOMINO (Theory). We then demonstrate the performance of MultiFit on the benchmark cases (Results). Finally, we discuss the implications of MultiFit and DOMINO for structural characterization of large assemblies (Discussion).
Theory
Combinatorial optimization by DOMINO
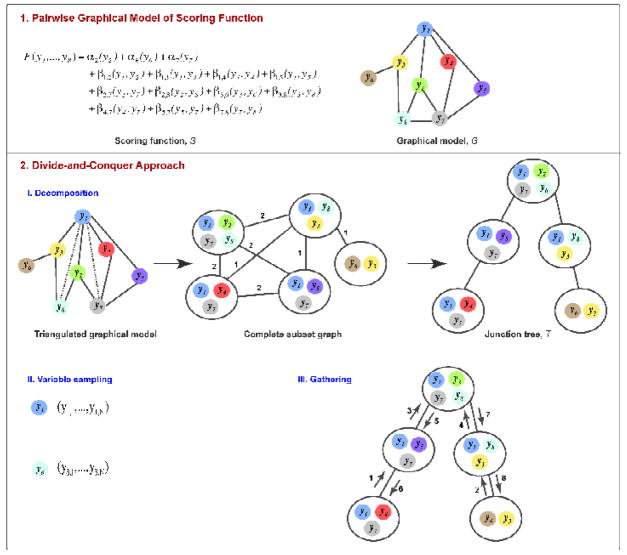
DOMINO applies a divide-and-conquer approach to efficiently find solutions with the globally optimal score within a discrete sampling space (Fig. 1)23; 24. The idea is to decompose the set of variables into relatively uncoupled but potentially overlapping subsets that can be sampled independently form each other, followed by efficiently gathering the subset solutions into the global minimum. The strength of this approach derives from the decomposition procedure that helps reduce the size of the search space from exponential in the number of components in the whole system to exponential in the number of components in the largest subset. Next, we describe DOMINO’s application to the minimization of a scoring function F corresponding to a sum of single-body terms {αi} and pairwise terms {βi,j}:
where {yi} are the variables being optimized; for example, in MultiFit, these variables are the positions and orientations of the components. The scoring function F is represented by a graphical model G=(V, E). The graphical model G of the scoring function F is a graph whose nodes V correspond to the variables {yi} and edges E connect all pairs of nodes. The weight of a node corresponding to yi is αi and the weight of an edge between nodes corresponding to yi and yj is βi,j. Thus, the scoring function F is the sum of all node and edge weights.
Figure 1. DOMINO outline.
(1) The DOMINO optimizer is illustrated with the scoring function F of 8 variables {yi} composed of a sum between 3 single body terms {αi} and 11 pairwise terms {βi, j}. The scoring function is encoded in the graphical model G. (2) (I) Decomposition of the graphical model results into a junction tree T 23; 24. The graphical model is first triangulated; a graph is triangulated if there are no cycles with more than three edges without a chord (a chord is an edge connecting two non-adjacent nodes in a cycle). The triangulation procedure adds edges (dotted lines) to the graphical model until no cycle is chordless. The triangulated graphical model is then converted into a complete subset graph. The nodes of the complete subset graph are maximum cliques in the triangulated graphical model (gray circles); a maximum clique is a sub-graph whose nodes are connected directly to each other and are not all part of another clique. The weight of an edge in the complete subset graph is the number of the shared variables between the adjacent subsets, as indicated; edges of weight zero are not shown. Next, the junction tree is the maximum spanning tree of the complete subset graph; a maximal spanning tree of a graph spans all of the nodes without cycles, using a subset of the original edges with the maximal sum of their weights. (II) The sampling space of each variable is discretized. (III) Finally, the globally optimal solution of F is gathered from enumerated subset states by passing messages between subset nodes. The numbers on the edges indicate a valid sequence of message passing.
The problem of finding the minimum of the scoring function F is equivalent to the maximum a posteriori problem in a graphical model. This problem is known to be NP-hard (nondeterministic polynomial-time hard) for an arbitrary graph G25; NP-hard is a class of decision, search, and optimization problems whose computing time increases at least exponentially with the number of optimized variables. When a graphical model has at most one path between any two given nodes (i.e., it does not contain cycles and thus is a singly connected graphical model or a tree), it can be efficiently optimized by the belief-propagation algorithm26.
Unfortunately, the belief-propagation algorithm is not guaranteed to converge to the globally optimal solution for graphs with cycles, such as the graphical models used for the MultiFit application. Therefore, to ensure finding the global minimum of G efficiently, we apply a divide-and-conquer approach. First, the variables to be optimized are decomposed into smaller relatively uncoupled but potentially overlapping subsets, using a junction tree construction algorithm (the decomposition step). Second, each variable is discretized (the variable sampling step); for example, by uniform sampling. Third, the discrete states of the individual subsets are constructed and gathered into the globally optimal solution, using the belief-propagation algorithm (the gathering step). Graph theory provides efficient algorithms for decomposition (i.e., junction tree construction) and gathering (i.e., belief-propagation). Next, we elaborate on each of the three steps.
In the decomposition step, the graphical model G is converted into a tree T whose nodes U are potentially overlapping subsets of variables {yi} (Fig. 1). Importantly, for any two non-adjacent subsets in T that share some variables, the subsets that connect them must also contain these variables (i.e., T is a junction tree). In such a case, it is possible to gather the discrete states of individual subsets into the globally optimal solution using the belief-propagation algorithm. For maximum efficiency, we aim toward decomposing the graphical model into the junction tree such that the size of the largest subset is minimal, which is an NP-hard problem. We use the minimum-degree method that was shown empirically to result in smallest subsets for sparse graphical models27.
In the variable sampling step, a discrete set of values for each variable is created. The details of this discretization may depend on the scoring function F. Most generally, uniform sampling over a relevant range of values can be used. A potentially better possibility is to use the union of the local minima of scoring functions spanned by the variables in the subsets containing the discretized variable.
In the final, gathering step, the HUGIN version of the belief-propagation algorithm28 is applied to the junction tree T to find the global minimum of F (Fig. 1). The computational complexity of the HUGIN algorithm is O(|U| ·Ls), where s is the size of the largest subset of T and L is the number of values of a node in the graphical model G.
The belief-propagation algorithm is based on passing messages between the nodes (i.e., subsets of variables) of the junction tree. A subset is allowed to send a message to a neighbor subset if it has received messages from all of its remaining neighbor subsets. Thus, propagation of messages is initiated in subsets connected only to a single subset (i.e., the leaf subsets) and proceeds to the neighboring subsets until some subset receives messages from all of its neighbors (i.e., the root subset). The content of a message to a target subset is a vector of the minimal values of the partial scoring function F over the variables in all previously visited subsets and the target subset, for each combination of values of the remaining variables in the target subset; a partial scoring function over a subset of variables includes only those terms of F that involve these variables. Messages from the root subset are then sent back to the other subsets, completing the message passing process when the leave subsets receive back the messages from the root subset. For messages from the root subset, the partial scoring function is the scoring function F (because all subsets were already visited), and thus each subset that received a message from all other subsets can infer the values of its variables in the global minimum. The efficiency of message passing derives from enumerating combinations of values for only those variables that are shared between different subsets.
MultiFit: Simultaneous fitting of multiple components into a density map of their assembly
The goal is to find the positions and orientations (i.e., placements) of components (e.g., sub-complexes, proteins, domains, secondary structure segments), represented at atomic resolution, within a cryoEM density map of their assembly. We express this structure characterization challenge as a combinatorial optimization problem. Next, we outline a representation of the modeled system, a scoring function, and an optimization algorithm.
Representation
The assembly density map is represented by a three-dimensional (3D) grid, in which every voxel is assigned an estimated density value. The components are represented by their atoms and remain rigid throughout the entire optimization process (Fig. 2).
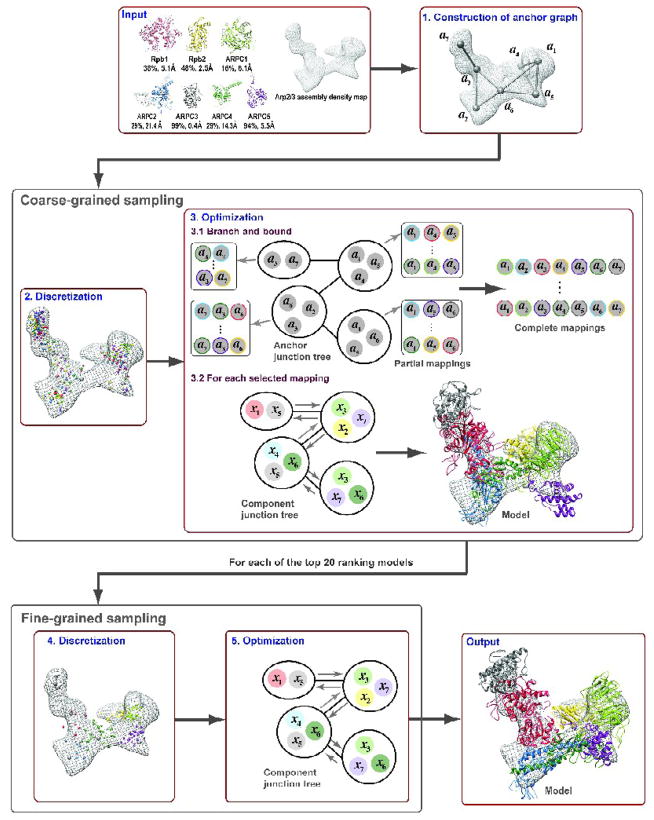
Figure 2. MultiFit outline.
The MultiFit algorithm is illustrated using an assembly between models of Rpb1 (red), Rpb2 (yellow), ARPC1 (light green), ARPC2 (blue), ARPC3 (gray), ARPC4 (dark green), and ARPC5 (purple) (PDB entry 1tyq). The component – template sequence identities and Cα RMSDs are indicated. The input to MultiFit is the assembly density map (grey mesh) and the atomic structures of the individual components (top left). The output is a ranked list of assembly models that optimize the MultiFit scoring function (one model is shown on bottom right). (1) The anchor points (the 7 labeled nodes) are constructed for the input density map by QVOL; the 9 grey edges indicate pairs of anchor points that are sufficiently close to allow components placed in their vicinity to interact with each other. (2) The sampling space of component placements is discretized by fitting each of the 7 components around each of the 7 anchor points (regions), and selecting a number of top-ranking placements for each component in each region; the small colored spheres indicate placement centroids. (3) The optimal combination of component placements is found by optimizing the scoring function S for each mapping of components to anchor points using DOMINO. (3.1) For efficiency, we replace the enumeration by a branch-and-bound procedure that eliminates some of the mappings and makes use of partial results. In the branch stage, we first decompose the anchor graph into an anchor junction tree using DOMINO’s decomposition algorithm (Fig. 1). The top 60% mappings of components to anchor points for each subset of anchor points (partial mappings) are found and stored by iterating over all possible partial mappings; the color of the circle indicates which component is mapped to the anchor point. The partial mappings are scored by partial scoring function S including only the terms involving the mapped components. Complete mappings consistent with the stored partial mappings are generated efficiently with a hashing procedure (not described). (3.2) Next, for each of these complete mappings, the optimal combination of placements for the 7 components is found by DOMINO; the color of the solid circles in the component junction tree indicates the component mapped to the corresponding anchor point in the anchor junction tree. The molecular model shown has a mapping score of 0 and the rank of 10. (4) The top 20 scoring coarse models are further refined. A refined sampling space is generated for each coarse configuration by docking pairs of its interacting components and selecting only those placements that are approximately consistent with the initial coarse configuration. (5) DOMINO is applied again to find the optimal combination of placements for the 7 components; the molecular model shown has a mapping score of 0 and the rank of 4.
Scoring
We evaluate potential configurations based on the quality-of-fit of individual components in the density map, the protrusion of each component from the map envelope, as well as the shape complementarity between pairs of components.
Optimization
The component configuration that optimizes the scoring function is identified by a combinatorial optimization protocol, consisting of three stages: (i) anchor graph construction, (ii) coarse-grained sampling, and (iii) fine-grained sampling (Fig. 2). In anchor graph construction, the density map is discretized into regions and the connectivity between them is calculated. In coarse-grained sampling, the sampling space is first discretized by fitting each of the components into each of the map regions and selecting a number of top-ranking placements for each component in each region. Next, a branch-and-bound search through all mappings of components to regions combined with DOMINO finds top 20 scoring configurations. In fine-grained sampling, each of these top configurations is refined by DOMINO; a refined sampling space is generated for each coarse configuration by docking pairs of its interacting components and selecting only those placements that are approximately consistent with the initial coarse configuration.
Scoring function for MultiFit
The score of placements of N components 29 in an assembly density map is:
ϕ1(xi) is the quality-of-fit of xi into the assembly density map D. In the extreme case, the configuration that optimizes may occupy only the highest density region in the assembly density map. To overcome this problem, we add two geometric terms (ϕ2 and ϕ3) to the scoring function. The component protrusion term ϕ2 (xi) scores how well xi is placed inside the density envelope. The interaction term ϕ3 (xi, xj) scores the pairwise shape complementarity between the structures xi and xj, and also accounts for their excluded volume.
Quality-of-fit term
The fit of a given structure xi into the assembly density map D is usually assessed by a cross-correlation measure between the densities of xi and the assembly5; 19. Here, we use the “normalized fitting score” C as implemented in Mod-EM (Eq. 2 in ref.30); the density of xi is simulated at the same resolution as the assembly density map D, using the uniform-sphere model. However, C is insufficient for comparing placements of different components because small domains have a better chance of higher cross-correlation with the map31. Thus, we calculate the quality-of-fit of a component into a map by expressing C as a Z-score, (C − m)/s where m and s are respectively the mean and standard deviation of a reference distribution of C. The reference distribution is generated by optimally fitting randomly selected, similarly-sized protein structures into simulated maps of randomly selected, similarly-sized protein structures (F. Davis, M.S. Madhusudan, N. Eswar, A. Sali, and M. Topf, unpublished results).
Interaction term
The pairwise shape complementarity score between structures xi and xj is calculated as a weighted sum of a reward for interaction areas and a penalty for steric clashes between the components32; 33. Specifically, the reward is the total number of surface atom pairs of xi and xj within a distance cutoff and the penalty is a weighted sum of all clashing pairs of atoms of xi and xj. To speed up the calculation of the reward, we first classify atoms as buried or exposed, by placing each atom on a grid and dividing the grid into a surface and 4 core shells according to the closest distance from the molecular surface (the surface shell contains all grid points that are at most half of the map resolution away from the surface)32. The reward is calculated by indexing the surface atoms of xi in a geometric hash table34;35, querying the hash table for each surface atom of xj, and summing the number of hits to get the reward. To calculate the steric clash penalty, we determine the accessibility of each atom of xi (and xj) using the grid of xj (xi). If an atom in xi (xj) is located within the surface (k=0) or the k-th core shell of xj (xi), we add (k+1)·27 to the penalty. The sum of the penalty score of xi with respect to xj and the penalty score of xj with respect to xi is divided by 2 to obtain the steric clash penalty. Due to fitting errors, the correct configuration of components might include some minor clashes between interacting components. These clashes are not significantly penalized because of the thickness of the surface shell and because of the evaluation of the favorable and penalty terms using only mainchain atoms. The choice of the shell thickness and the weight of the penalty score were chosen by trial-and-error.
Component protrusion
The protrusion of a component from the assembly envelope is defined to be the negative value of the shape complementarity score between the component surface and the assembly envelope. The assembly envelope is calculated by representing each density voxel above a threshold as an atom and calculating the Connolly surface36 of this collection of atoms.
Optimization for MultiFit
Construction of anchor graph
The centroids of L approximately equally-sized regions of density voxels are calculated from the density map D using the QVOL procedure of SITUS37; a density voxel belongs to the region with the closest centroid. When L equals N and the components are of similar size, the centroids of the regions correspond approximately to the centroids of the N assembly components. These points are the nodes of the anchor graph. We then calculate connectivity between the anchor points (i.e., the edges of the anchor graph); a pair of anchor points (ai, aj) are connected if (i) the distance between ai and aj is below a predefined threshold (by default, 1.5 the sum of radii of the two largest components in the system); and (ii) the variance of the gradient of the density along a line of voxels that connects ai with aj is below a predefined threshold (by default, two times the variance of the assembly density).
Discretization step in coarse-grained sampling
We construct a discrete sampling space of component placements, represented by a set of M′ placements (by default, 50) for each of the N components in each of the L regions. Thus, each set of placements for all components in region i (Ai) contains M=M′·N “local” placements around an anchor point ai. Here, we set L to N, although L can also be larger than N.
In detail, for each component j, the discrete sampling space is constructed as follows. Placements around each anchor point ai are sampled by optimizing C in a cube surrounding the anchor point (the edge length of the cube is half the resolution of the map). This optimization is performed by Mod-EM30, starting with a random starting orientation of the component centered at the anchor point. Next, the sampled placements for all anchor points are clustered based on their pairwise Cα RMSD values: The highest scored placement (by C) initiates the first cluster and is its pivot. The closest remaining placement is either joined with the first cluster for which its Cα RMSD with the cluster’s pivot is less than the threshold (half the resolution of the map) or initiates its own cluster otherwise. The process is repeated with the best scoring non-clustered placement until all placements are clustered. The best scoring placement from each cluster is assigned to the set of placements Ai,j corresponding to the closest anchor point ai; each anchor point is assigned at most M′ placements.
Optimization step in coarse-grained sampling
We find the optimal combination of placements of components by optimizing the scoring function S within the discrete sampling space constructed in the previous step. The global minimum of S is the minimum of the optimal solutions for each of the mappings of components to anchor points Π={πk}, where πk is a function that maps a component j to an anchor point i (i = πk (j)); formally, we solve , where xj are placements of component j in the set Aπk(j),j, as constrained by mapping πk.
Naively, this optimization could be achieved by a nested double loop in which the outer loop consists of enumerating the mappings and the inner loop consists of applying DOMINO to the scoring function S constrained by the given mapping. However, enumerating over all possible mappings becomes computationally expensive as the number of components increases. To improve the efficiency of MultiFit, we replace the enumeration by a branch-and-bound procedure that eliminates some of the mappings and makes use of partial results (Fig. 2).
The scoring function F optimized by DOMINO for each mapping ( ) is a simplified S that does not contain uninformative interaction terms ϕ3 corresponding to physically non-interacting components (Fig. 2); specifically, we eliminate interaction terms between pairs of components that are mapped to unconnected anchor points. Importantly, it is this simplification that results in a relatively “sparse” graphical model G, thus allowing it to be optimized efficiently by DOMINO.
Discretization step in fine-grained sampling
We construct a refined discrete sampling space for a coarse configuration found in coarse-grained sampling, ( ). The refined set of placements of component j is first initialized with the placements in Aπ(j),j, as found in coarse-grained sampling. We then enrich this set of placements by sampling binding of component j to neighboring components {w} with PATCHDOCK32. A PATCHDOCK-produced binding mode of component j to component w (xj) is added to the refined set of placements of component j if (i) the distance between the centroid of xj and the centroid of is below half the resolution of the map and (ii) xj is consistent with the density map boundaries (i.e., if ϕ2(xj) is below a predefined threshold). Finally, the refined set of placements of component j is re-ranked by the quality-of-fit score and clustered according to Cα RMSD (described above).
Optimization step in fine-grained sampling
The optimal combination of component placements is found by DOMINO, through optimizing the scoring function S within the refined discrete sampling space.
Results
Benchmark with simulated maps
Benchmark
We tested MultiFit on a benchmark of 6 simulated test cases. The assembly density maps were simulated at 20 Å resolution using the PDB2VOL program of SITUS38 with voxel size of 3 Å. The input atomic structures of the components included native structures from the Protein Data Bank (PDB39) as well as models calculated by comparative modeling using MODELLER-9v3 (http://salilab.org/modeller)40 based on related template structures with sequence identity ranging from 16% to 99%. The accuracy of the individual comparative models is assessed using Cα RMSD and native overlap to the corresponding native structure. Native overlap (NO3.5) measures the percentage of Cα atoms of the model that are within 3.5 Å to the corresponding Cα atoms in the native structure. The native overlap was calculated by superposing the model on the corresponding native structure using a rigid-body least-squares minimization, as implemented in the model. superpose command of MODELLER-9v3.
We use three scores to assess the accuracy of modeled configurations at different levels of resolution: First, the mapping score is the number of substitutions needed to convert the assessed mapping of components to anchor points to the native mapping of components to anchor points (the Hamming distance); the native mapping has a mapping score of 0. Second, the configuration score is the fraction of the components positioned correctly; we define a component as positioned correctly, if the distance between its centroid and the corresponding reference centroid is smaller than half of the map resolution. Third, the assembly placement score is the average of its components placement scores, each of which is composed of a distance and an angle to the reference placement; the distance is calculated between the centroids of the placements and the angle is the axis angle of the rotation matrix between the two placements41. Because the components are kept rigid throughout the optimization process, the reference components used in the assessment of an assembly model are the component models superposed on the corresponding components in the native assembly (i.e., the reference placement). We chose not to use the Cα RMSD measure to assess assembly models because the significance of Cα RMSD values depends strongly on the number of assembly components and their sizes42.
Determining the configuration of Arp2/3
To illustrate MultiFit, we first describe in detail a challenging application to Arp2/3 (Table 1, Figs. 2 and 3). The Arp2/3 complex of seven proteins is crucial for regulating the initiation of actin polymerization and the organization of the resulting filaments43. A density map was simulated from the Arp2/3 crystal structure with ATP and Ca2+ (PDB entry 1TYQ44). The atomic structures of the Arp2/3 components (proteins) were modeled using templates with sequence identity ranging from 16% to 99%; the Cα RMSD error for these models varied from 0.4 Å to 21.4 Å and their native overlap varied between 38% and 100%; we intentionally used inaccurate comparative models to benchmark the robustness of our method with respect to errors in the component conformations.
Table 1.
Determining the configuration of the Arp2/3 assembly.
| Component(name, chain, residue range) | Component modeling | Coarse-grained sampling | Fine-grained sampling | |||||
|---|---|---|---|---|---|---|---|---|
| Template(PDB entry, residue range) | % Seq. Id.a | Cα RMSD (Å)b | NO3.5 (%)b | Discretization (best placement score, fitting rank)c | Optimization (best- scoring placement score)d | Discretizatio n(best placement score, fitting rank)c | Optimizati on(best- scoring placement score) d | |
| Rpb1 A, 4-408 | 2q1nB, 4-370 | 40 | 5.1 | 74 | (4.4,12), 22 | (7.3, 179) | (4.4, 12), 4 | (4.4, 12) |
| Rpb2 B, 143-349 | 1nwkA, 140- 334 | 48 | 2.5 | 93 | (2.4,30), 32 | (2.5, 30) | (2.4, 30), 39 | (9.2, 14) |
| ARPC1 C, 5-361 | 1erjC, 342- 708 | 16 | 6.1 | 52 | (3.6, 20), 42 | (12.1, 115) | (3.6, 20), 101 | (1.4, 44) |
| ARPC2 D, 1-274 | 1u2vF, 3-168 | 29 | 21.4 | 42 | (14.9, 52), 38 | (19.9, 172) | (9.1, 25), 5 | (9.1, 25) |
| ARPC3 E, 1-169 | 2p9nE, 2-173 | 99 | 0.4 | 100 | (1.2, 23), 31 | (3.9, 163) | (1.2, 23), 5 | (1.2, 23) |
| ARPC4 F, 3-186 | 1u2vD, 137-279 | 29 | 14.3 | 38 | (21.1, 84), 1 | (23.0, 177) | (11.8, 46), 36 | (11.8, 46) |
| ARPC5 G, 11-150 | 2p9nG, 11- 150 | 94 | 5.5 | 88 | (6.7, 117), 50 | (6.7, 117) | (6.7, 117), 61 | (12.6, 9) |
The percentage of sequence identity between the template and the component, as calculated from their alignment used for comparative modeling.
The Cα RMSD and native overlap NO3.5 between the modeled component superposed on its native structure.
The placement score and rank of the best placement, calculated by Cα RMSD to the reference. The placements were ranked by the normalized fitting score C.
The placement score of the placement found in the top ranking assembly configuration.
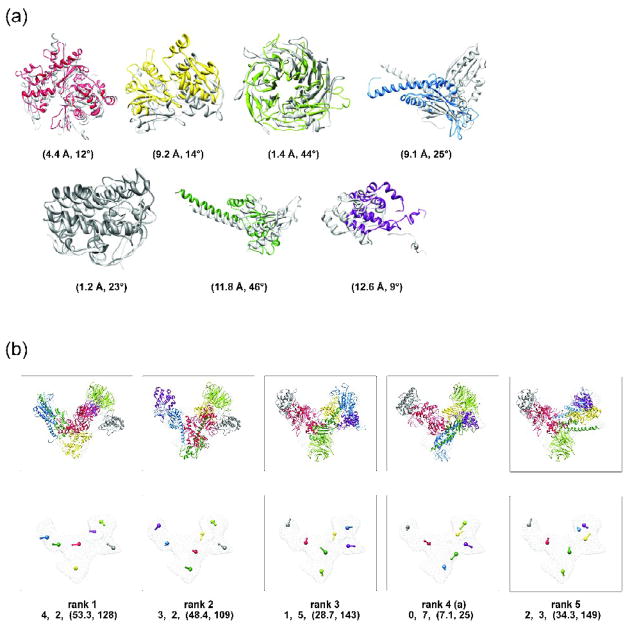
Figure 3. MultiFit results for Arp2/3.
(a) An assessment of the final model with the mapping score of 0. The model has the 4th smallest value of the scoring function S (the 4th model in (b)). The modeled and reference placements of the individual components are compared (Results); the corresponding placement scores are indicated below each comparison. (b) Five top ranked models for Arp2/3. The atomic representations of the models are displayed in the top row. The bottom row shows the centroid and the rotation axis for each component; the corresponding rank, the mapping score, the configuration score, and the assembly placement score are indicated below each model.
In the final output of MultiFit, the near-native model with an assembly placement score of (7.1 Å, 25°) was ranked 4th among all the sampled configurations. In coarse-grained sampling, this model was ranked 10th, with a configuration score of 4/7 and an assembly placement score of (10.8 Å, 136°). The centroids of the individual components were positioned in the vicinity of their native centroids; however, the orientations of some components were incorrect, resulting in steric clashes between components. In fine-grained sampling, the top 20 scored models were refined. The refinement procedure was able to resolve many of the clashes in the model, which in turn improved its global score, resulting into the final rank of 4. Next, we elaborate on the individual steps of the optimization protocol.
In anchor graph construction, we computed 7 anchor points from the density map using the QVOL program of Situs38. The average distance between the anchor points and the centroids of the corresponding reference components was 7.2 Å. We then identified pairs of anchor points that are sufficiently close to allow components placed in their vicinity to interact with each other. The procedure pruned 12 of the possible 21 pairs (i.e., 7 · 6/2). The remaining 9 pairs allowed identification of 9 of the 12 native contacts between the 7 components.
In the discretization step of coarse-grained sampling, we fitted by Mod-EM each component in the neighborhood of each anchor point. We assessed the accuracy of the discretization by the placement score of the best placement of each component (i.e., the placement with the lowest Cα RMSD to the corresponding reference). These best placement scores ranged from (2.5 Å, 30°) to (23.0 Å, 177°). As expected, as the model accuracy measured by Cα RMSD and native overlap decreases, so does the rank of the best placement. The most accurate placement was ranked within the top 50 solutions for each component by the normalized fitting score C.
In the optimization step of coarse-grained sampling, we first represented the scoring function as a graphical model. The globally optimal component configuration was then found by a branch-and-bound search in conjunction with the DOMINO optimizer. We utilized DOMINO for decomposing the simplified graphical model into an anchor junction tree of subsets of anchor points. The anchor junction tree contained 4 subsets of 2, 3, 3, and 3 anchor points. The branch-and-bound procedure resulted in 486 complete mappings for the 7 components (out of 7! = 5040 possible mappings). For each of these 486 mappings, the optimal placements of the 7 components were inferred by the gathering algorithm of DOMINO. A configuration with a mapping score of 0, a configuration score of 4/7, and an assembly placement score of (10.8 Å, 136°) was ranked 10th. The total running time with pre-computed scoring terms was approximately 70 minutes on a single CPU; it takes approximately 2 hours to pre-compute the scoring function terms.
This prediction demonstrates some of the benefits and problems with coarse-grained sampling. For example, an accurate placement of Rpb2 and ARPC5 could not have been obtained solely based on the quality-of-fit due to non-native conformations of their models (Table 1). Nevertheless, global optimization of the scoring function for the entire assembly did result in the correct placement for these two components. However, global optimization can also make a prediction less accurate. For example, ARPC4 was placed inaccurately, because of the need for shape complementarity with inaccurately modeled neighbors Rpb1, ARPC1, ARPC2, and ARPC5. Such problems can be partly resolved by finer discretization of the sampling space (i.e., the fine-grained sampling, below), in addition to flexible fitting (not attempted here).
In fine-grained sampling of a given model, we repopulated the sampling space for the corresponding complete mapping with pairwise docking solutions between the interacting components. Specifically, we enriched the set of placements by sampling binding modes of a component to the corresponding placed components of its neighboring anchor points using PATCHDOCK32. We then ran DOMINO again to find the optimally refined configuration. The assembly placement score of the refined configuration is (7.1 Å, 25°), which clearly demonstrates the improvement in the accuracy of the relative orientations. For example, the placement accuracy of ARPC4 improved from (23.0 Å, 177°) to (11.8 Å, 46°). The improved placement was ranked only 499 in the pairwise docking between ARPC4 and ARPC1. However, global optimization relying on restraints derived from coarse-grained sampling (i.e., shape complementarity between interacting components and protrusion from the map envelope) resulted in this placement occurring in the best-scoring assembly configuration.
To validate the contribution of the shape complementarity score, we optimized a scoring function lacking this term (ϕ3 in the scoring function S, Theory). The top-ranking configuration had a mapping score of 3, a configuration score of 3/7, and an assembly placement score of (42.5 Å, 94°). A model with a mapping score of 0 was not found in the top 50 solutions. This comparison demonstrates the positive contribution of the shape complementarity score to the accuracy of the generated assembly models.
Benchmark
To assess MultiFit more comprehensively than is possible by a single example, we also applied it to a benchmark that included 5 additional simulated test cases. In all 6 simulated tests, a model with the mapping score of 0 was found within the top 4 solutions (Table 2); in fact, a model with the mapping score of 0 was the best scoring model in all cases for which the structures of the individual components were modeled based on templates with sequence identities higher than 60%. The assembly placement score of the model with the mapping score of 0 ranged between (2.6 Å, 4°) and (7.1 Å, 25°). These results demonstrate the utility of MultiFit in predicting the configuration of atomic components in a low-resolution density map of their assembly. Next, we report the benchmark results at each of the 5 steps of the algorithm.
Table 2.
Benchmark.
| Assembly | Component modeling | Coarse-grained sampling | Fine-grained sampling | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Assembly (name, PDB entry) | # component s | % Seq. Id. average rangea | Cα RMSD (Å) average rangeb | NO3.5 (%) average rangeb | Rankc | Optimization (assembly placement score) | Rankc | Optimization (assembly placement score) | |||
| Chaperonin GroEL, 1oel | 3 domains | 65 | 60–72 | 0.9 | 0.2–1.0 | 96 | 92–100 | 1 | (2.6, 13) | 1 | (2.6, 13) |
| SUMO-RanGAP1-Ubc9-Nup358/RanBP 2 complex, 1z5s | 4 proteins | 100 | 100– 100 | 0.0 | 0.0–0.0 | 10 0 |
100–100 | 1 | (5.9, 113) | 1 | (5.0, 67) |
| SUMO-RanGAP1-Ubc9-Nup358/ArnBP2 complex, 1z5s | 4 proteins | 37 | 18–56 | 7.9 | 1.2–15.0 | 52 | 13–98 | 3 | (7.7, 92) | 3 | (6.4, 62) |
| Dihydropyrimid ine Dehydrogenase, 1gte | 5 domains | 100 | 100– 100 | 0.0 | 0.0–0.0 | 10 0 |
100–100 | 1 | (2.6,4) | 1 | (2.6, 4) |
| Archaeon Methanopyrus kandleri,1e6v | 6 proteins | 61 | 57–68 | 0.0 | 0.0–0.0 | 10 0 |
100–100 | 1 | (2.5, 8) | 1 | (2.5, 8) |
| Arp2/3 complex, 1tyq | 7 proteins | 51 | 16–99 | 7.9 | 0.4–21.4 | 70 | 38–100 | 10 | (10.8, 136) | 4 | (7.1, 25) |
The average, minimum, and maximum percentage of sequence identity between the assembly components and their templates.
The average, minimum, and maximum Cα RMSD and native overlap NO3.5 between the modeled components superposed on their corresponding component in the native assembly.
The rank of a model with mapping score 0.
In anchor graph construction, the average distance between the predicted anchor point and the centroid of the corresponding reference component in the near-native configuration was between 4 Å and 7 Å.
In the discretization step of coarse-grained sampling, a near native configuration was sampled within the discrete sampling space in all test cases. However, this configuration was not necessarily ranked highly according to our scoring function, due to steric clashes between interacting components.
In the optimization step of coarse-grained sampling, a model with the mapping score of 0 was found in the top 10 solutions in all test cases; and in 4 of the 6 cases it was the best-scoring solution. The assembly placement score of the model with the mapping score of 0 ranged from (2.6 Å, 4°) to (10.8 Å, 136°). The prediction accuracy depended on the component accuracy (Table 2). As the accuracy of the component models is decreased, the rank of the correct configuration as well as its placement score also become worse. The benchmark shows that coarse-grained sampling is able to determine component positions quite accurately, but frequently fails to result in accurate relative orientations. The main reason is the coarseness of the discrete sampling space, as demonstrated by the Arp2/3 and 1z5s examples. In the latter case, we obtained the near-native assembly (i.e., (5.9 Å, 113°)) with the native components and a less accurate configuration (i.e., (7.7 Å, 92°)) with distorted components.
In the discretization step of fine-grained sampling, the PATCHDOCK docking program32 was able to sample near-native interaction modes between pairs of components. However, these interactions were generally not ranked highly by PATCHDOCK. For example, in the 1z5s case with distorted components, the most accurate docking prediction of chains C and D against chain A ranked 405 and 138, respectively.
In the optimization step of fine-grained sampling, the refined models were at least as accurate as the most accurate models generated in coarse-grained sampling, sometimes much more so. In particular, the accuracy of the relative orientations between pairs of interacting components improved. For example, in the 1z5s case with distorted components, the assembly placement score improved from (7.7 Å, 92°) to (6.4 Å, 62°). The refined model contained placements derived from the docking prediction of chains C and D against chain A. These placements were ranked 405 and 138 by PATCHDOCK; reweighing the placements by the normalized fitting score C increased their ranks to 78 and 43, respectively. DOMINO finally correctly selected these placements for the final best-scoring configuration.
Benchmark with an experimentally determined map
To test the method in a realistic setting, we benchmarked it again by modeling the component configuration for an assembly with an experimentally determined cryo-EM map.
GroEl-GroES domains
GroEL/GroES is a chaperonin that aids protein folding in E. coli. GroEL consists of two back-to-back rings of 7 identical subunits, each of which contains three domains (i.e., the equatorial, apical, and intermediate domain). GroES is a ring of seven identical single-domain proteins that cap GroEL. We applied MultiFit to model the configuration of the four domains in an interacting pair of the GroEL and GroES subunits. Atomic coordinates for the four domains were obtained from a crystal structure of the GroEL-ADP-GroES complex (ADP-state; PDB entry 1AON45). The corresponding density was segmented from a cryoEM map of the bacterial GroES-ADP7-GroEL-ATP7 chaperonin determined at 23.5 Å resolution (ATP-state; EMDB ID 104622). The crystal structure of the ADP-state was fitted to the density (as one rigid body) and used as a reference for assessment. The main structural differences between the ATP- and ADP-states are the downward rotation of the intermediate domain and the counterclockwise twist of the apical domain22.
The configuration with the mapping score 0 was ranked third, with an assembly placement score of (13.9 Å, 160°). A sampling space of approximately 14 million combinations was searched in 16 minutes of CPU time. The fine-grained sampling was able to generate a more accurate model with an assembly placement score of (11.0 Å, 84°). We note in passing that fitting all 49 domains (i.e., 3 × 7 × 2+7) into the density of both rings would presumably benefit from the added information in the subunit-subunit interactions within and across rings; however, to test MultiFit in a more challenging setting, we deliberately modeled only a single symmetry unit consisting of 3 GroEL domains and 1 GroES domain.
Discussion
We described MultiFit, a computational method for determining the positions and orientations (i.e., placements) of multiple atomic components in a cryoEM density map of their assembly. The problem is formulated in terms of combinatorial optimization, solved by our inferential optimizer DOMINO that guarantees finding the global minimum within a given discrete sampling space. The input is a density map and a set of atomic components, which are kept rigid throughout the optimization process. For a given configuration of components, the scoring function measures the quality-of-fit of the atomic structures in the map, the protrusion from the map envelope, as well as the shape complementarity between pairs of components. The optimization process consists of the coarse- and fine-grained sampling. Each sampling stage starts with a discretization step, achieved respectively by fitting and docking, followed by an optimization step that relies on DOMINO. Both DOMINO and MultiFit are available as part of Integrative Modeling Platform (IMP) (http://salilab.org/imp46; 47).
Accurate MultiFit’s predictions for 7 test cases demonstrated its utility (Table 2). Specifically, our benchmark demonstrated the utility of MultiFit in predicting the configuration of components with known folds within density map at resolutions between 20 Å to 23.5 Å; the average assembly placement score for the near native configurations was (5.3 Å, 38°). MultiFit was able to determine the assembly configuration even in cases where the fitting scores were ambiguous. Examples include Arp2/3 (Table 1) and the 1z5s test case with distorted components (Table 2).
Next, we discuss (i) the benefits of simultaneous multiple component fitting, (ii) inaccuracies resulting from the discrete sampling space, and (iii) broad utility of combinatorial optimization.
Benefits of simultaneous fitting
Most methods for modeling assemblies in the context of a cryoEM map rely on a segmented assembly map and/or a model of the whole assembly. In the absence of such information, sampling the configuration space is computationally challenging, as the placement of each component may depend on the placements of other components. For example, the configuration of the Arp2/3 assembly with modeled components could not have been solved by iteratively fitting the largest remaining component in the unoccupied region using Mod-EM30. The configuration also cannot be modeled accurately without the component protrusion and the interaction terms in the scoring function used by MultiFit. However, by considering the placements of all components simultaneously, the protrusion of a component from the assembly envelope, and the shape complementary between the interacting components, we were able to determine the assembly configuration with an assembly placement score of (7.1 Å, 25°).
Inaccuracies resulting from the discrete sampling space
MultiFit prediction will be accurate when a near-native configuration exists in the discrete sampling space and corresponds to the global minimum of the scoring function. These two conditions depend, in turn, on the accuracy of the atomic models of the individual components and the choice of anchor points. Next, we elaborate on these two dependencies.
Accuracy of component models
The atomic models of the individual components might be inaccurate due to modeling errors and/or induced fit. As the accuracy of the component models decreases, the discretized sampling space (either by fitting or docking) is less likely to contain near-native placements (i.e., the sampling problem) and the global minimum is less likely to correspond to the most accurate sampled configuration (i.e., the scoring problem). In other words, these errors may affect the accuracy of the predicted assembly configuration due to scoring and sampling inaccuracies. One such example is the pair of 1z5s test cases (Table 2): The inputs to the first test case were the native components and the assembly density. The discretization steps of coarse- and fine-grained sampling resulted in near-native placements and the top ranked configuration detected by DOMINO had a relatively accurate assembly placement score of (5.0 Å, 67°). The inputs to the second test case were models with average Cα RMSD error of 6.3 Å. The discrete sampling spaces generated in the coarse- and fine-grained sampling contained less accurate placements. As a result, the utility of the scoring terms (especially the protrusion from the map envelope and the shape complementarity) decreased. The assembly placement score of the final assembly model with distorted component models was significantly worse (6.4 Å, 62°) than the assembly placement score of the assembly model with the native components. More accurate assembly models may be obtained by using a shape complementarity score that is less sensitive to component model errors and/or by an explicit treatment of the component conformations. To this end, techniques might be adopted from flexible fitting of a component into a density map41; 48 as well as from flexible molecular docking49;50.
Accuracy of anchor points
Given the QVOL algorithm, the utility of the anchor points is affected by the variances in the size and shape of the components (data not shown). The utility of the anchor points is also affected by the resolution of the map (data not shown). To obtain a discrete sampling space that contains a near-native configuration, we sample candidate placements of each component in a neighborhood of each anchor point. However, there are many assemblies for which the variation in component sizes is too large for reasonable neighborhood sizes. We intend to improve the utility of anchor point calculation by considering component sizes and density map segmentation51; 52.
Combinatorial optimization in structural biology
Modeling challenges in structural biology can generally be expressed as optimization problems46. These optimization problems often fall into a general class of NP-complete problems (Theory)53. Combinatorial optimization is a type of optimization in which the set of feasible solutions is discrete, and the goal is to find the best possible solution within this discrete set. Combinatorial optimizers have been suggested for various modeling tasks, such as sidechain packing54–56, threading27, ab initio RNA folding57, and prediction of quaternary structures of multi-protein complexes58. These methods can in principle be re-formulated as a combinatorial optimization of a scoring function represented by a graphical model, benefiting from graph theory techniques23; 24. Such a formulation has already been proposed for the sidechain packing problem56.
Our DOMINO method can in principle be applied to many problems in structural modeling, from low-resolution assembly modeling to sidechain refinement. Its strength derives from the junction tree algorithm that helps reduce the size of the search space from exponential in the number of components in the whole system to exponential in the number of components in the largest subset. More specifically, the computational complexity is O(| U | · Ls) where |U| is the number of subsets in the junction tree, L is the size of the largest subset, and s is the number of discrete values of a single variable in the graphical model. Fortunately, at the granularity level used in MultiFit’s application to protein assemblies in our benchmark, the theoretical complexity of the junction tree algorithm has not been a limiting factor. Nevertheless, in other applications that involve a dense graphical model of the scoring function and extensively sampled variable values, incomplete sampling of a discrete space may have to be accepted.
In conclusion, MultiFit and DOMINO can help to bridge the gap between the atomic structures of the individual proteins and the cryoEM maps of their assemblies. In particular, they can provide initial configurations for further refinement of many multi-component assembly structures described by electron microscopy41; 48; 59; 60.
Acknowledgments
We thank Frank Alber for stimulating discussions, Ben Webb for help with the IMP software, and Dina Schneidman-Duhovny for help with the PATCHDOCK software. K.L. is supported in part by a fellowship from the Edmond J. Safra Bioinformatics Program at Tel-Aviv University. M.T. is funded by an MRC Career Development Award (G0600084). A.S. is supported by the Sandler Family Supporting Foundation, NIH (R01 GM54762, U54 RR022220, PN2 EY016525, and R01 GM083960), NSF (IIS-0705196), Hewlett-Packard, NetApp, IBM, and Intel. H.J.W. acknowledges support by the Binational U.S.-Israel Science Foundation, Israel Science Foundation (281/05), NIAID, and the Hermann Minkowski-Minerva Center for Geometry at TAU.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Robinson CV, Sali A, Baumeister W. Molecular sociology of the cell. Nature. 2007;450:973–82. doi: 10.1038/nature06523. [DOI] [PubMed] [Google Scholar]
- 2.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, LJJ, Bastuck S, Dümpelfeld B, Edelmann A, Heurtier MA, Hoffman V, Hoefert C, Klein K, Hudak M, Michon AM, Schelder M, Schirle M, Remor M, Rudi T, Hooper S, Bauer A, Bouwmeester T, Casari G, Drewes G, Neubauer G, Rick JM, Kuster B, Bork P, Russell RB, Superti-Furga G. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–6. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 3.Abbott A. Proteomics: the society of proteins. Nature. 2002;417:894–6. doi: 10.1038/417894a. [DOI] [PubMed] [Google Scholar]
- 4.Drenth J. Principles of Protein X-ray Crystallography. Springer; 1999. [Google Scholar]
- 5.Frank J. Three-Dimensional Electron Microscopy of Macromolecular Assemblies: Visualization of Biological Molecules in Their Native State. Oxford University Press; 2006. [Google Scholar]
- 6.Chiu W, Baker M, Jiang W, Dougherty M, Schmid M. Electron cryomicroscopy of biological machines at subnanometer resolution. Structure. 2005;13:363–72. doi: 10.1016/j.str.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 7.Berman HM. The Protein Data Bank: a historical perspective. Acta Crystallogr A. 2008;64:88–95. doi: 10.1107/S0108767307035623. [DOI] [PubMed] [Google Scholar]
- 8.Davis JA, Takagi Y, Kornberg RD, Asturias FA. Structure of the yeast RNA polymerase II holoenzyme: mediator conformation and polymerase interaction. Mol Cell. 2002;20:409–15. doi: 10.1016/s1097-2765(02)00598-1. [DOI] [PubMed] [Google Scholar]
- 9.Marlovits TC, Kubori T, Lara-Tejero M, Thomas D, Unger VM, Galan JE. Assembly of the inner rod determines needle length in the type III secretion injectisome. Nature. 2006;441:637–40. doi: 10.1038/nature04822. [DOI] [PubMed] [Google Scholar]
- 10.Mitra K, Schaffitzel C, Shaikh T, Tama F, Jenni S, Brooks CL, Ban N, Frank J. Structure of the E. coli protein-conducting channel bound to a translating ribosome. Nature. 2005;438:318–24. doi: 10.1038/nature04133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schaffitzel C, Oswald M, Berger I, Ishikawa T, Abrahams JP, Koerten HK, Koning RI, Ban N. Structure of the E. coli signal recognition particle bound to a translating ribosome. Nature. 2006;444:503–6. doi: 10.1038/nature05182. [DOI] [PubMed] [Google Scholar]
- 12.Schmid MF, Sherman MB, Matsudaira P, Chiu W. Structure of the acrosomal bundle. Nature. 2004;431:104–7. doi: 10.1038/nature02881. [DOI] [PubMed] [Google Scholar]
- 13.Chandramouli P, Topf M, Ménétret J, Eswar N, Cannone J, Gutell R, Sali A, Akey C. Structure of the mammalian 80S ribosome at 8.7 A resolution. Structure. 2008;16:535–48. doi: 10.1016/j.str.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kostek S, Grob P, De Carlo S, Lipscomb JS, Garczarek F, Nogales E. Molecular architecture and conformational flexibility of human RNA polymerase II. Structure. 2006;14:1691–700. doi: 10.1016/j.str.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 15.Hainfeld J, Powell R. New Frontiers in Gold Labeling. J Histochem Cytochem. 2000;48:471–80. doi: 10.1177/002215540004800404. [DOI] [PubMed] [Google Scholar]
- 16.Rossmann MG, Bernal R, Pletnev SV. Combining electron microscopic with X ray crystallographic structures. J Struct Biol. 2001;136:190–200. doi: 10.1006/jsbi.2002.4435. [DOI] [PubMed] [Google Scholar]
- 17.Goddard T, Huang C, Ferrin T. Visualizing density maps with UCSF Chimera. J Struct Biol. 2007;157:281–7. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 18.Ceulemans H, RB R. Fast fitting of atomic structures to low-resolution electron density maps by surface overlap maximization. J Mol Biol. 2004;338:783–93. doi: 10.1016/j.jmb.2004.02.066. [DOI] [PubMed] [Google Scholar]
- 19.Fabiola F, Chapman MS. Fitting of high-resolution structures into electron microscopy reconstruction images. Structure. 2005;13:389–400. doi: 10.1016/j.str.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 20.Baumeister W, Walz J, Zühl F, Seemüller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;93:367–80. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 21.Serysheva I, Ludtke S, Baker M, Cong Y, Topf M, Eramian D, Sali A, Hamilton S, Chiu W. Subnanometer-resolution electron cryomicroscopy-based domain models for the cytoplasmic region of skeletal muscle RyR channel. Proc Natl Acad Sci USA. 2008;105:9610–5. doi: 10.1073/pnas.0803189105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ranson NA, Farr GW, Roseman AM, Gowen B, Fenton WA, Horwich AL, Saibil HR. ATP-bound states of GroEL captured by cryo-electron microscopy. Cell. 2001;107:869–79. doi: 10.1016/s0092-8674(01)00617-1. [DOI] [PubMed] [Google Scholar]
- 23.Jordan MI. Graphical models. Statistical Science. 2004;19:140–155. [Google Scholar]
- 24.Lauritzen S. Graphical Models. Oxford University Press; New York, NY: 1996. [Google Scholar]
- 25.Shimony SE. Finding MAPs for belief networks is NP-hard. Artif Intell. 1994;68:399–410. [Google Scholar]
- 26.Pearl J. Probabilistic Reasoning in Intelligent Systems: Networks of Plausible Inference. Morgan Kaufmann; 1998. [Google Scholar]
- 27.Xu J, Jiao F, Berger B. A tree-decomposition approach to protein structure prediction. Proc IEEE Comput Syst Bioinform Conf. 2005:247–56. doi: 10.1109/csb.2005.9. [DOI] [PubMed] [Google Scholar]
- 28.Andersen S, Olesen K, Jensen F. Readings in uncertain reasoning. Morgan Kaufmann Publishers Inc; 1990. HUGIN—a shell for building Bayesian belief universes for expert systems; pp. 332–7. [Google Scholar]
- 29.Krogan N, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis A, Punna T, Peregrin-Alvarez J, Shales M, Zhang X, Davey M, Robinson M, Paccanaro A, Bray J, Sheung A, Beattie B, Richards D, Canadien V, Lalev A, Mena F, Wong P, Starostine A, Canete M, Vlasblom J, Wu S, Orsi C, Collins S, Chandran S, Haw R, Rilstone J, Gandi K, Thompson N, Musso G, St Onge P, Ghanny S, Lam M, Butland G, Altaf-Ul A, Kanaya S, Shilatifard A, O’Shea E, Weissman J, Ingles J, Hughes T, Parkinson J, Gerstein M, Wodak S, Emili A, Greenblatt J. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–43. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 30.Topf M, Baker ML, John B, Chiu W, Sali A. Structural characterization of components of protein assemblies by comparative modeling and electron cryo-microscopy. J Struct Biol. 2005;149:191–203. doi: 10.1016/j.jsb.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 31.Lasker K, Dror O, Shatsky M, Nussinov R, Wolfson HJ. EMatch: discovery of high resolution structural homologues of protein domains in intermediate resolution cryo-EM maps. IEEE/ACM Trans Comput Biol Bioinform. 2007;4:28–39. doi: 10.1109/TCBB.2007.1003. [DOI] [PubMed] [Google Scholar]
- 32.Duhovny D, Nussinov R, Wolfson HJ. Second International Workshop on Algorithms in Bioinformatics Italy.2002. [Google Scholar]
- 33.Chen R, Weng Z. A Novel Shape Complementarirty Scoring Function for Protein-Protein Docking. Proteins. 2003;51:397–408. doi: 10.1002/prot.10334. [DOI] [PubMed] [Google Scholar]
- 34.Wolfson H, Rigoutsos I. Geometric hashing: An overview. IEEE Computational Science and Eng. 1997;11:263–78. [Google Scholar]
- 35.Lamdan Y, Wolfson HJ. Geometric Hashing: A General And Efficient Model-based Recognition Scheme. Proc Intl Conf on Computer Vision, IEEE Computer Society Press. 1988:238–49. [Google Scholar]
- 36.Connolly M. Analytical molecular surface calculation. J Appl Cryst. 1983;16:548–58. [Google Scholar]
- 37.Wriggers W, Milligan RA, Schulten K, McCammon JA. Self-organizing neural networks bridge the biomolecular resolution gap. J Mol Biol. 1998;284:1247–54. doi: 10.1006/jmbi.1998.2232. [DOI] [PubMed] [Google Scholar]
- 38.Wriggers W, Milligan RA, McCammon JA. Situs: A Package for Docking Crystal Structures into Low-Resolution Maps from Electron Microscopy. J Struct Biol. 1999;125:185–95. doi: 10.1006/jsbi.1998.4080. [DOI] [PubMed] [Google Scholar]
- 39.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28:235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 41.Topf M, Lasker K, Webb B, Wolfson HJ, Chiu W, Sali A. Protein Structure Fitting and Refinement Guided by cryoEM Density. Structure. 2008;16:295–307. doi: 10.1016/j.str.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cohen F, Sternberg M. On the prediction of protein structure: The significance of the root-mean-square deviation. J Mol Biol. 1980;138:321–33. doi: 10.1016/0022-2836(80)90289-2. [DOI] [PubMed] [Google Scholar]
- 43.Goley ED, Welch MD. The ARP2/3 complex: an actin nucleator comes of age. Nat Rev Mol Cell Biol. 2006;7:713–26. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- 44.Nolen BJ, Littlefield RS, Pollard TD. Crystal structures of actin-related protein 2/3 complex with bound ATP or ADP. Proc Natl Acad Sci USA. 2004;101:15627–32. doi: 10.1073/pnas.0407149101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu Z, Horwich AL, Sigler PB. The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex. Nature. 1997;388:741–50. doi: 10.1038/41944. [DOI] [PubMed] [Google Scholar]
- 46.Alber F, Forster F, Korkin D, Topf M, Sali A. Integrating Diverse Data for Structure Determination of Macromolecular Assemblies. Annu Rev Biochem. 2008:77. doi: 10.1146/annurev.biochem.77.060407.135530. [DOI] [PubMed] [Google Scholar]
- 47.Alber F, Dokudovskaya S, Veenhoff L, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait B, Rout M, Sali A. Determining the architectures of macromolecular assemblies. Nature. 2007;450:683–94. doi: 10.1038/nature06404. [DOI] [PubMed] [Google Scholar]
- 48.Schröder G, Brunger A, Levitt M. Combining efficient conformational sampling with a deformable elastic network model facilitates structure refinement at low resolution. Structure. 2007;15:1630–41. doi: 10.1016/j.str.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bahar I, Rader A. Coarse-grained normal mode analysis in structural biology. Curr Opin Struct Biol. 2005;15:586–92. doi: 10.1016/j.sbi.2005.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bonvin A. Flexible protein-protein docking. Curr Opin Struct Biol. 2006;16:194–200. doi: 10.1016/j.sbi.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 51.Birmanns S, Wriggers W. Multi-Resolution Anchor-Point Registration of Biomolecular Assemblies and Their Components. J Struct Biol. 2007;157:271–80. doi: 10.1016/j.jsb.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 52.Kawabata T. Multiple subunit fitting into a low-resolution density map of a macromolecular complex using a gaussian mixture model. Biophys J. 2008;95:4643–58. doi: 10.1529/biophysj.108.137125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wales D, Scheraga H. Global Optimization of Clusters, Crystals, and Biomolecules. Science. 1999;285:1368–72. doi: 10.1126/science.285.5432.1368. [DOI] [PubMed] [Google Scholar]
- 54.Canutescu A, Shelenkov A, Dunbrack RJ. A graph-theory algorithm for rapid protein side-chain prediction. Protein Sci. 2003;12:2001–14. doi: 10.1110/ps.03154503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu J, Berger B. Fast and Accurate Algorithms for Protein Side-Chain Packing. JACM. 2006;53:533–57. [Google Scholar]
- 56.Yanover C, Schueler-Furman O, Weiss Y. Minimizing and Learning Energy Functions for Side-Chain Prediction. RECOMB 2007. 2007 doi: 10.1089/cmb.2007.0158. [DOI] [PubMed] [Google Scholar]
- 57.Zhao J, Malmberg RL, Cai L. Rapid ab initio RNA folding including pseudoknots via graph tree decomposition. The 6th Workshop on Algorithms in Bioinformatics (WABI 2006); Zurich, Switzerland. 2006. [Google Scholar]
- 58.Inbar Y, Benyamini H, Nussinov R, Wolfson HJ. Prediction of multimolecular assemblies by multiple dockin. J Mol Biol. 2005;349:435–47. doi: 10.1016/j.jmb.2005.03.039. [DOI] [PubMed] [Google Scholar]
- 59.Trabuco L, Villa E, Mitra K, Frank J, Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure. 2008;16:673–83. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Orzechowski M, Tama F. Flexible fitting of high-resolution x-ray structures into cryoelectron microscopy maps using biased molecular dynamics simulations. Biophys J. 2008;95:5692–705. doi: 10.1529/biophysj.108.139451. [DOI] [PMC free article] [PubMed] [Google Scholar]