Abstract
Cocaine induced neuroplasticity changes in the mesocorticolimbic dopamine systems are thought to be involved in the pathophysiology of cocaine dependence. Since neurotrophic factors have been observed to prevent/reverse and mimic cocaine-induced neurobiological changes in the brain, related genes are plausible candidates for susceptibility to cocaine dependence. The novel conserved dopamine neurotrophic factor protein (CDNF) promotes the survival, growth, and function of dopamine-specific neurons and is expressed in brain regions that undergo cocaine-induced neuroplasticity. In this study, we hypothesize that polymorphisms in the CDNF gene (CDNF/ARMETL1) contribute to increased risk for cocaine dependence. Cocaine dependent individuals (n=351) and unaffected controls (n=257) of African descent were genotyped for four single nucleotide polymorphisms (SNPs) in the CDNF gene (rs11259365, rs7094179, rs7900873, rs2278871). We observed no significant differences in allele, genotype, or haplotype frequencies between cases and controls for any of the tested SNPs. Our study suggests that there is no association between variants in the CDNF gene and cocaine dependence. However, additional studies using larger sample sizes, comprehensive SNP coverage, and clinically homogenous populations are necessary before confidently excluding CDNF as a significant genetic risk factor for cocaine dependence.
Keywords: Addiction, Association study, Cocaine, Neurotrophic factor, Genetics, Substance abuse, ARMETL1
INTRODUCTION
Cocaine dependence is characterized by compulsive cocaine seeking and continued cocaine use despite adverse consequences. The profound loss of behavioral control is the hallmark of cocaine addiction and contributes to the high risk of relapse. Today, there are no FDA approved pharmacological treatments for cocaine dependence available. While the interplay between genetic and environmental factors underlying cocaine dependence is not fully understood, studies have estimated that approximately two-thirds of an individual’s risk for developing cocaine dependence is heritable [23, 24]. Identifying genetic susceptibility factors for cocaine dependence may provide important insights into pathophysiology and eventually may lead to better and more effective therapies.
Research over the last several decades indicates that neuronal activity in the mesocorticolimbic dopamine system is responsible for cocaine reward [39, 50] and contributes to relapse [21, 42]. Current hypotheses of cocaine dependence invoke drug induced neuroadaptations in reward-related learning and memory processes in the mesocorticolimbic dopamine system and glutamatergic corticolimbic circuitry [12, 22, 32, 51]. Cocaine induced neuroplasticity changes in certain brain regions, including the striatum and ventral tegmental area (VTA), may underlie mechanisms of cocaine dependence [5, 6, 34, 43, 47, 48, 52]. Recent evidence suggests that neurotrophins, such as brain-derived neurotrophic factor (BDNF) and its intracellular signaling pathways, are involved in neuroadaptive changes in the dopaminergic or glutamate systems [7, 41, 45]. Several experiments in rodents for example show that acute and chronic administration of BDNF has effects on behavioral paradigms, including cocaine self-administration, cocaine seeking and conditioned responses to cocaine. Taken together, these studies suggest that neurotrophic factors play an important role in the pathophysiology of cocaine addiction or addictive behavior in general.
Recently, a novel neurotrophic factor has been discovered. The conserved dopamine neurotrophic factor (CDNF) is a trophic factor for dopamine neurons that promotes the survival, growth, and function for dopamine-specific neurons. CDNF is present in various brain regions, including both the striatum and VTA. A recent study showed that CDNF protein injections restored dopaminergic function and prevented degeneration of dopaminergic neurons in mouse models of Parkinson’s disease [26]. No further studies have investigated whether CDNF plays a role in other neurological, psychiatric or substance use disorders. Given the biological rationale for studying this gene, we tested the hypothesis that genetic variation in the conserved dopamine neurotrophic factor gene (CDNF/ARMETL1) might contribute to neuroplasticity changes in mesocorticolimbic dopamine system and confer susceptibility to cocaine dependence.
MATERIALS AND METHODS
Sample collection
Blood samples for DNA isolation were collected from cocaine-dependent individuals of African-American decent (n=351; 72% male, mean age: 43) during clinical studies of cocaine dependence at the University of Pennsylvania Treatment Research Center. Subjects were at least 18 years of age. All were assessed with the Structured Clinical Interview for DSM Disorders (SCID) and urine drug screens were obtained. All patients had a clinical diagnosis of cocaine dependence as defined by DSM-IV. Patients with drug use that did not fulfill abuse or dependence criteria were included in the study. Family history was not obtained and ethnicity was determined by self-report. All psychiatric axis I disorders except alcohol dependence/abuse and nicotine dependence were used as exclusion criteria. In addition, participants were excluded if they had a history of a seizure disorder (except cocaine-induced seizures) or a severe medical illness, including a history of AIDS (but not merely of HIV+ status). Individuals currently being treated with psychotropic medications or with psychiatric symptoms, including psychosis, dementia, suicidal or homicidal ideation, mania or depression requiring antidepressant therapy were also excluded [27]. Blood samples from control persons of African-American descent (n=257; 29% male, mean age: 40) were collected at the University of Pennsylvania, Thomas Jefferson University, and the National Institute of Mental Health Genetics Initiative (www.nimhgenetics.org). Control individuals were screened for history of substance use disorders or other psychiatric illness. They were not assessed with a urine drug screen and ethnicity was determined by self-report. Subjects with a history of substance dependence or a history of major psychiatric illness were excluded from this study [8]. Patients and controls were not matched for sex and age in this genetic association study. This could be a potential confounding factor since epidemiologic data show that the incidence for cocaine dependence is higher among men; however, autosomal alleles seldom show sex differences in frequency and SNP frequencies are unlikely to change with age. Furthermore, no clear relationship has been established between age and genotype in a complex disease with variable age of onset [13, 25]. Genomic DNA was extracted from peripheral leukocytes within obtained blood samples by standard protocols. All protocols were approved by the Institutional Review Boards at Thomas Jefferson University and the University of Pennsylvania, and all subjects provided written informed consent before blood sample collection.
SNP selection and genotyping
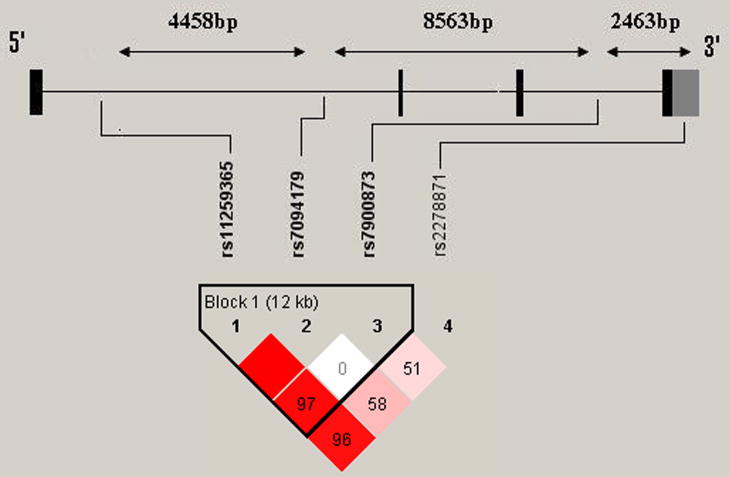
The CDNF gene is located on chromosome 10p13. CDNF contains 4 exons and spans 18,732bp (Ensembl Human Exon View accession ENST00000378441). SNPs for genotyping were selected using the tagging SNP algorithm based on available HapMap data with a minor allele frequency > 0.18 in the Yoruban population and a pairwise linkage disequilibrium (LD) r2 cutoff of > 0.8. Only 4 tagging SNPs fulfilled these criteria: SNP1: rs11259365 (intron 1); SNP2: rs7094179 (intron 1); SNP3: rs7900873 (intron 3); SNP4: rs2278871 (3′UTR). SNP genotyping was performed using Applied Biosystems Inc. (ABI) (Foster City, CA, USA) ‘Assays-on-demand’ as per manufacturer protocol. Quality control was maintained by genotyping 10% duplicates for cases and controls.
Statistical analyses
Genotype and allele frequencies were compared between groups using χ2 contingency analysis. A two-tailed type I error rate of 5% was chosen for the analysis. Linkage disequilibrium (LD) and haplotype frequencies were estimated using the Haploview software (version 4.1). Haplotype blocks were identified using the solid spine of LD method in Haploview [4]. Correction for multiple testing was performed using permutation correction by the Haploview program [4]. This approach corrects for multiple testing but takes into account the correlation between markers. Permutation correction is thus less conservative than the Bonferroni correction but is appropriate for independent tests with multiple markers [9]. For the single-marker analysis, 10,000 permutations were carried out to estimate the significance of the best results, correcting for the four loci tested. Haplotype analysis was performed using the Haploview software and P-values were corrected by permutation analysis as described above.
Our sample size had reasonable power to detect a disease association at a P-value less than or equal to 0.05, assuming an odds ratio of 1.5 and a minor allele frequency of 30% (99% for log-additive mode of inheritance, 92% for dominant, and 56% for recessive modes of inheritance). Power analysis was performed using the Quanto program [15].
RESULTS
None of the genotype distributions deviated significantly from those expected by Hardy-Weinberg equilibrium for cases or controls. LD measures and haplotype blocks across the CDNF gene are shown in Fig. 1. We observed moderate LD across the gene with one exception, specifically in the 3′ region, where weak LD was observed. Single marker analysis did not yield evidence for association on the genotypic level or allelic level for any of the SNPs (Table 1). After permutation correction, haplotype analysis did not produce any evidence for association with disease (Table 2). Genotyping success rates were between 98.1% and 99.2%. The mean concordance rate for all the markers was 97.4 ± 2.8% with respect to the 10% of samples that were genotyped twice for quality control. Allelic frequencies were consistent with those reported in the Hapmap database for Yoruba in Ibadan, Nigeria.
Figure 1.
Linkage disequilibrium measures (D′) across a schematic representation of the CDNF gene. LD patterns and haplotype blocks were defined by the “solid spine of LD” using the Haploview software. A standard color scheme is used to display LD pattern, with dark red for very strong LD, white for no LD, and shades of red for intermediate LD. Increasing intensity of red indicates increasing degrees of LD. The number inside each block indicates the calculated D′ value.
Table 1.
Genotype and Allele frequencies of variations in the CDNF gene.
| SNP | Sample | n | Genotype frequency | P* | Allele frequency | P** | ||
|---|---|---|---|---|---|---|---|---|
| rs11259365 | G/G | G/C | C/C | f(G) | ||||
| Cocaine | 350 | 0.623 | 0.320 | 0.057 | 0.910 | 0.783 | 0.768 | |
| Controls | 257 | 0.638 | 0.304 | 0.058 | 0.790 | |||
| rs7094179 | A/A | A/C | C/C | f(A) | ||||
| Cocaine | 351 | 0.453 | 0.430 | 0.117 | 0.995 | 0.668 | 0.920 | |
| Controls | 254 | 0.449 | 0.433 | 0.118 | 0.665 | |||
| rs7900873 | A/A | A/G | G/G | f(A) | ||||
| Cocaine | 349 | 0.464 | 0.458 | 0.077 | 0.527 | 0.693 | 0.749 | |
| Controls | 257 | 0.471 | 0.428 | 0.101 | 0.685 | |||
| rs2278871 | C/C | C/T | T/T | f(C) | ||||
| Cocaine | 338 | 0.453 | 0.456 | 0.092 | 0.645 | 0.680 | 0.888 | |
| Controls | 255 | 0.475 | 0.420 | 0.106 | 0.684 | |||
P values for comparison of genotype frequencies between cocaine dependent individuals and controls
P values for comparison of allele frequencies between cocaine dependent individuals and controls.
Table 2.
Analysis of common haplotype in the CDNF gene
| Haplotype block | Case frequencies | Control frequencies | Chi square | P Value | Permutation P value |
|---|---|---|---|---|---|
| Block 1 | |||||
| GAA | 0.457 | 0462 | 0.025 | 0.874 | 1.000 |
| CCA | 0.217 | 0.207 | 0.154 | 0.695 | 0.998 |
| GAG | 0.211 | 0.204 | 0.085 | 0.771 | 0.999 |
| GCG | 0.099 | 0.109 | 0.304 | 0.581 | 0.985 |
| GCA | 0.015 | 0.015 | 0.000 | 0.985 | 1.000 |
DISCUSSION
Dopaminergic brain systems have been implicated to play a major role in drug reward [20], thus making genes involved in these circuits plausible candidates for influencing susceptibility to substance use disorders. In fact, several genes coding for the dopaminergic system have been investigated in cocaine dependence including genes for the dopamine receptor D2 (DRD2) [16, 30, 33, 36] the dopamine receptor D3 (DRD3) [10, 14, 30], the dopamine receptor D4 (DRD4) [3], the dopamine transporter (DAT) [17, 18, 37] and the catechol-O-methy-transferase (COMT) gene [27]. The results of all these studies have been conflicting with some positive reports and some negative findings, possibly due to small sample sizes and the complex genetic nature of cocaine dependence. In this study we investigated whether polymorphisms in the CDNF gene, another gene involved in dopaminergic neurotransmission, confer risk to cocaine dependence. Neither single marker analysis nor haplotype analysis produced evidence in support of an association between the CDNF gene and cocaine dependence among individuals of African descent. While these results suggest that polymorphisms in CDNF do not play a major role in cocaine dependence, several limitations in our study must be considered carefully before excluding CDNF as a susceptibility gene for cocaine dependence.
One of the conceptual strengths, and at the same time a weakness of genetic association studies, lies in their design. The key concept of DNA population association studies is the use highly informative genetic markers as surrogates for the block structure of the human genome. Investigating markers across a gene that are in strong LD, and thus likely to be inherited together as a block, reduces the amount of tests needed for association analysis. While this approach has the advantage of detecting major susceptibility factors in LD using only a few selected markers, it fails to detect rare variants that may be causative and does not take into account the complex LD patterns that may exist in the tested population. We selected four Hapmap tagging SNPs in order to provide broad coverage of the CDNF gene. We observed moderate LD values between markers that spanned the gene with exception, however. The 3′ region of CDNF between SNP3 (rs7900873) and SNP4 (rs2278871) is approximately 2.5kb. The observed LD between these two markers was low (D′=0.51) (Figure 1), indicating that our coverage of this region was weak. Weak LD may be explained by a frequent occurrence of conversions [1] and homologous recombination in that region of the gene, a phenomenon termed as a “recombination hotspot”. Recombination hotspots are DNA sites at which recombination occurs at increased rates due to discrete recombination signaling sequences and interacting proteins [49]. They occur on average every 200 kilobases across the human genome [29, 31]. Given this weak LD, there may be polymorphisms between SNP3 and SNP4 that are associated with cocaine dependence which our study failed to detect. Additional studies are needed to genotype more SNPs in that region or conduct large scale sequencing of cases and controls to comprehensively cover the gene. Such gene coverage is necessary before conclusively excluding CDNF as a genetic risk factor of cocaine dependence.
Another limitation of this study was the sample size, which had limited statistical power to detect risk alleles that contribute small effects to the overall disorder. This limited power could explain our negative results. Additional studies with larger sample sizes should be conducted to test for polymorphisms in the CDNF that may contribute small effects to susceptibility to cocaine dependence. However, it must also be considered that larger sample sizes may increase genetic heterogeneity and contribute to undetected population stratification, which would negatively impact the interpretation of association analysis [28]. Unaccounted differences in population structure can create associations in and of themselves and lead to inaccurate interpretation of results [40]. The possibility of unaccounted differences in population structure is especially relevant to analyses involving individuals of African descent since there is substantial genetic heterogeneity among African Americans [35, 38, 46, 54]. Genomic controls and/or the utilization of family-based association studies may control for these stratification issues [2, 11, 44]. Future studies investigating associations between CDNF and cocaine dependence with larger samples sizes should utilize genomic controls or family-based association paradigms to control for stratification issues.
When conducting genetic association studies, the clinical phenotype might add additional heterogeneity which could obscure an association or lead to a false positive finding. Patients with co-morbid alcohol dependence/abuse and nicotine dependence were not excluded from our cocaine dependent population. While all patients were diagnosed with cocaine dependence in accordance with the DSM-IV criteria, co-morbid alcohol and nicotine use might have differed between patients. It has been shown that genetic factors play a role in nicotine dependence [53] and alcoholism [19], thus perhaps shared genetic factors contribute to all substance use disorders. Hence, unaccounted clinical heterogeneity within a population may have exacerbated genetic heterogeneity among the cocaine cases and may have led to false negative or positive findings. Although all patients were diagnosed according to DSM-IV criteria and the diagnosis of cocaine dependence was supported by urine drug screen data, the control subjects were assessed using semi-structured interviews but did not undergo urine drug testing. While drug testing is useful in establishing a diagnosis, it might not be useful for assessment of controls since it does not rule out past exposure or substance use. Unreported or minimized substance abuse in the control population is thus an important limitation that needs to be considered; however, even under the assumption that the control group had 1% of undetected cocaine dependence cases, assuming the general prevalence rate of cocaine dependence in the control population, this factor might have only minor impact when comparing cocaine cases to the group of controls.
In summary, our results do not support an association between polymorphism in the CDNF gene and cocaine dependence in individuals of African descent. However, additional studies using larger sample sizes, comprehensive SNP coverage, and independent, homogenous populations are necessary before excluding CDNF as a contributing genetic risk factor for cocaine dependence.
Acknowledgments
This work was supported by the Center for Neurobiology and Behavior, Department of Psychiatry, University of Pennsylvania. Financial support is gratefully acknowledged from National Institutes of Health grants K08MH080372 (F.W.L.), NIDA grants P60-051186 (C.P.O.) and P50-12756 (H.M.P), the VISN4 Mental Illness Research and Clinical Center grant from the Veterans Affairs Administration (D.W.O.), a grant from the Tzedakah Foundation (W.H.B.) and a grant from Philip and Marcia Cohen (W.H.B.). Most importantly, we thank the subjects who have participated in and contributed to these studies.
The NIMH control subjects were collected by the NIMH Schizophrenia Genetics Initiative ‘Molecular Genetics of Schizophrenia II’ (MGS-2) collaboration. The investigators and coinvestigators are: ENH/Northwestern University, Evanston, IL, MH059571 – Pablo V. Gejman, MD (Collaboration Coordinator; PI), Alan R. Sanders, MD; Emory University School of Medicine, Atlanta, GA, MH59587 – Farooq Amin, MD (PI); Louisiana State University Health Sciences Center; New Orleans, LA, MH067257 – Nancy Buccola APRN, BC, MSN (PI); University of California-Irvine, Irvine, CA, MH60870 – William Byerley, MD (PI); Washington University, St Louis, MO, U01, MH060879 – C. Robert Cloninger, MD (PI); University of Iowa, Iowa, IA, MH59566 – Raymond Crowe, MD (PI), Donald Black, MD; University of Colorado, Denver, CO, MH059565 – Robert Freedman, MD (PI); University of Pennsylvania, Philadelphia, PA, MH061675 – Douglas Levinson, MD (PI); University of Queensland, QLD, Australia, MH059588 – Bryan Mowry, MD (PI); Mt Sinai School of Medicine, New York, NY, MH59586– Jeremy Silverman, PhD (PI).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ardlie K, Liu-Cordero SN, Eberle MA, Daly M, Barrett J, Winchester E, Lander ES, Kruglyak L. Lower-than-expected linkage disequilibrium between tightly linked markers in humans suggests a role for gene conversion. Am J Hum Genet. 2001;69:582–589. doi: 10.1086/323251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bacanu SA, Devlin B, Roeder K. The power of genomic control. Am J Hum Genet. 2000;66:1933–1944. doi: 10.1086/302929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballon N, Leroy S, Roy C, Bourdel MC, Olie JP, Charles-Nicolas A, Krebs MO, Poirier MF. Polymorphisms TaqI A of the DRD2, BalI of the DRD3, exon III repeat of the DRD4, and 3′ UTR VNTR of the DAT: association with childhood ADHD in male African-Caribbean cocaine dependents? Am J Med Genet B Neuropsychiatr Genet. 2007;144:1034–1041. doi: 10.1002/ajmg.b.30540. [DOI] [PubMed] [Google Scholar]
- 4.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 5.Beitner-Johnson D, Nestler E. Morphine and cocaine exert common chronic actions on tyrosine hydroxylase in dopaminergic brain reward regions. J Neurochem. 1991;57:344–347. doi: 10.1111/j.1471-4159.1991.tb02133.x. [DOI] [PubMed] [Google Scholar]
- 6.Berhow M, Hiroi N, Kobierski L, Hyman S, Nestler E. Influence of cocaine on the JAK-STAT pathway in the mesolimbic dopamine system. J Neurosci. 1996;16:8019–8026. doi: 10.1523/JNEUROSCI.16-24-08019.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berhow M, Russell D, Terwilliger R, Beitner-Johnson D, Self D, Lindsay R, Nestler E. Influence of neurotrophic factors on morphine- and cocaine-induced biochemical changes in the mesolimbic dopamine system. Neuroscience. 1995;68:969–979. doi: 10.1016/0306-4522(95)00207-y. [DOI] [PubMed] [Google Scholar]
- 8.Berrettini WH, Persico AM. Dopamine D2 receptor gene polymorphisms and vulnerability to substance abuse in African Americans. Biological psychiatry. 1996;40:144–147. doi: 10.1016/0006-3223(96)00036-4. [DOI] [PubMed] [Google Scholar]
- 9.Camargo A, Azuaje F, Wang H, Zheng H. Permutation - based statistical tests for multiple hypotheses. Source Code Biol Med. 2008;3:15. doi: 10.1186/1751-0473-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Comings DE, Gonzalez N, Wu S, Saucier G, Johnson P, Verde R, MacMurray JP. Homozygosity at the dopamine DRD3 receptor gene in cocaine dependence. Molecular psychiatry. 1999;4:484–487. doi: 10.1038/sj.mp.4000542. [DOI] [PubMed] [Google Scholar]
- 11.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 12.Everitt BJ, Robbins TW. Neural systems of reinforcement for drug addiction: from actions to habits to compulsion. Nat Neurosci. 2005;8:1481–1489. doi: 10.1038/nn1579. [DOI] [PubMed] [Google Scholar]
- 13.Freedman ML, Reich D, Penney KL, McDonald GJ, Mignault AA, Patterson N, Gabriel SB, Topol EJ, Smoller JW, Pato CN, Pato MT, Petryshen TL, Kolonel LN, Lander ES, Sklar P, Henderson B, Hirschhorn JN, Altshuler D. Assessing the impact of population stratification on genetic association studies. Nature genetics. 2004;36:388–393. doi: 10.1038/ng1333. [DOI] [PubMed] [Google Scholar]
- 14.Freimer M, Kranzler H, Satel S, Lacobelle J, Skipsey K, Charney D, Gelernter J. No association between D3 dopamine receptor (DRD3) alleles and cocaine dependence. Addict Biol. 1996;1:281–287. doi: 10.1080/1355621961000124896. [DOI] [PubMed] [Google Scholar]
- 15.Gauderman WJ. Sample size requirements for association studies of gene-gene interaction. Am J Epidemiol. 2002;155:478–484. doi: 10.1093/aje/155.5.478. [DOI] [PubMed] [Google Scholar]
- 16.Gelernter J, Kranzler H, Satel SL. No association between D2 dopamine receptor (DRD2) alleles or haplotypes and cocaine dependence or severity of cocaine dependence in European- and African-Americans. Biological psychiatry. 1999;45:340–345. doi: 10.1016/s0006-3223(97)00537-4. [DOI] [PubMed] [Google Scholar]
- 17.Gelernter J, Kranzler HR, Satel SL, Rao PA. Genetic association between dopamine transporter protein alleles and cocaine-induced paranoia. Neuropsychopharmacology. 1994;11:195–200. doi: 10.1038/sj.npp.1380106. [DOI] [PubMed] [Google Scholar]
- 18.Guindalini C, Howard M, Haddley K, Laranjeira R, Collier D, Ammar N, Craig I, O’Gara C, Bubb VJ, Greenwood T, Kelsoe J, Asherson P, Murray RM, Castelo A, Quinn JP, Vallada H, Breen G. A dopamine transporter gene functional variant associated with cocaine abuse in a Brazilian sample. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:4552–4557. doi: 10.1073/pnas.0504789103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heath AC, Madden PA, Bucholz KK, Dinwiddie SH, Slutske WS, Bierut LJ, Rohrbaugh JW, Statham DJ, Dunne MP, Whitfield JB, Martin NG. Genetic differences in alcohol sensitivity and the inheritance of alcoholism risk. Psychological medicine. 1999;29:1069–1081. doi: 10.1017/s0033291799008909. [DOI] [PubMed] [Google Scholar]
- 20.Hyman SE, Malenka RC, Nestler EJ. Neural Mechanisms Of Addiction: The Role of Reward-Related Learning and Memory. Annual Review of Neuroscience. 2006;29:565–598. doi: 10.1146/annurev.neuro.29.051605.113009. [DOI] [PubMed] [Google Scholar]
- 21.Kalivas PW, McFarland K. Brain circuitry and the reinstatement of cocaine-seeking behavior. Psychopharmacology (Berl) 2003;168:44–56. doi: 10.1007/s00213-003-1393-2. [DOI] [PubMed] [Google Scholar]
- 22.Kalivas PW, O’Brien C. Drug addiction as a pathology of staged neuroplasticity. Neuropsychopharmacology. 2008;33:166–180. doi: 10.1038/sj.npp.1301564. [DOI] [PubMed] [Google Scholar]
- 23.Kendler KS, Karkowski LM, Neale MC, Prescott CA. Illicit psychoactive substance use, heavy use, abuse, and dependence in a US population-based sample of male twins. Archives of general psychiatry. 2000;57:261–269. doi: 10.1001/archpsyc.57.3.261. [DOI] [PubMed] [Google Scholar]
- 24.Kendler KS, Prescott CA. Cocaine use, abuse and dependence in a population-based sample of female twins. Br J Psychiatry. 1998;173:345–350. doi: 10.1192/bjp.173.4.345. [DOI] [PubMed] [Google Scholar]
- 25.Lewis SJ, Brunner EJ. Methodological problems in genetic association studies of longevity--the apolipoprotein E gene as an example. Int J Epidemiol. 2004;33:962–970. doi: 10.1093/ije/dyh214. [DOI] [PubMed] [Google Scholar]
- 26.Lindholm P, Voutilainen M, Laurén J, Peränen J, Leppänen V, Andressoo J, Lindahl M, Janhunen S, Kalkkinen N, Timmusk T, Tuominen R, Saarma M. Novel neurotrophic factor CDNF protects and rescues midbrain dopamine neurons in vivo. Nature. 2007;448:73–77. doi: 10.1038/nature05957. [DOI] [PubMed] [Google Scholar]
- 27.Lohoff FW, Weller AE, Bloch PJ, Nall AH, Ferraro TN, Kampman KM, Pettinati HM, Oslin DW, Dackis CA, O’Brien CP, Berrettini WH. Association between the catechol-O-methyltransferase Val158Met polymorphism and cocaine dependence. Neuropsychopharmacology. 2008;33:3078–3084. doi: 10.1038/npp.2008.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marchini J, Cardon LR, Phillips MS, Donnelly P. The effects of human population structure on large genetic association studies. Nature genetics. 2004;36:512–517. doi: 10.1038/ng1337. [DOI] [PubMed] [Google Scholar]
- 29.McVean GAT, Myers SR, Hunt S, Deloukas P, Bentley DR, Donnelly P. The Fine-Scale Structure of Recombination Rate Variation in the Human Genome. Science (New York, NY) 2004;304:581–584. doi: 10.1126/science.1092500. [DOI] [PubMed] [Google Scholar]
- 30.Messas G, Meira-Lima I, Turchi M, Franco O, Guindalini C, Castelo A, Laranjeira R, Vallada H. Association study of dopamine D2 and D3 receptor gene polymorphisms with cocaine dependence. Psychiatric genetics. 2005;15:171–174. doi: 10.1097/00041444-200509000-00006. [DOI] [PubMed] [Google Scholar]
- 31.Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A Fine-Scale Map of Recombination Rates and Hotspots Across the Human Genome. Science (New York, NY) 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- 32.Nestler EJ. Common molecular and cellular substrates of addiction and memory. Neurobiol Learn Mem. 2002;78:637–647. doi: 10.1006/nlme.2002.4084. [DOI] [PubMed] [Google Scholar]
- 33.Noble EP, Blum K, Khalsa ME, Ritchie T, Montgomery A, Wood RC, Fitch RJ, Ozkaragoz T, Sheridan PJ, Anglin MD, et al. Allelic association of the D2 dopamine receptor gene with cocaine dependence. Drug and alcohol dependence. 1993;33:271–285. doi: 10.1016/0376-8716(93)90113-5. [DOI] [PubMed] [Google Scholar]
- 34.Ortiz J, DeCaprio J, Kosten T, Nestler E. Strain-selective effects of corticosterone on locomotor sensitization to cocaine and on levels of tyrosine hydroxylase and glucocorticoid receptor in the ventral tegmental area. Neuroscience. 1995;67:383–397. doi: 10.1016/0306-4522(95)00018-e. [DOI] [PubMed] [Google Scholar]
- 35.Parra EJ, Kittles RA, Argyropoulos G, Pfaff CL, Hiester K, Bonilla C, Sylvester N, Parrish-Gause D, Garvey WT, Jin L, McKeigue PM, Kamboh MI, Ferrell RE, Pollitzer WS, Shriver MD. Ancestral proportions and admixture dynamics in geographically defined African Americans living in South Carolina. Am J Phys Anthropol. 2001;114:18–29. doi: 10.1002/1096-8644(200101)114:1<18::AID-AJPA1002>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 36.Persico AM, Bird G, Gabbay FH, Uhl GR. D2 dopamine receptor gene TaqI A1 and B1 restriction fragment length polymorphisms: enhanced frequencies in psychostimulant-preferring polysubstance abusers. Biological psychiatry. 1996;40:776–784. doi: 10.1016/0006-3223(95)00483-1. [DOI] [PubMed] [Google Scholar]
- 37.Persico AM, Vandenbergh DJ, Smith SS, Uhl GR. Dopamine transporter gene polymorphisms are not associated with polysubstance abuse. Biological psychiatry. 1993;34:265–267. doi: 10.1016/0006-3223(93)90081-n. [DOI] [PubMed] [Google Scholar]
- 38.Pfaff CL, Parra EJ, Bonilla C, Hiester K, McKeigue PM, Kamboh MI, Hutchinson RG, Ferrell RE, Boerwinkle E, Shriver MD. Population structure in admixed populations: effect of admixture dynamics on the pattern of linkage disequilibrium. Am J Hum Genet. 2001;68:198–207. doi: 10.1086/316935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pierce RC, Kumaresan V. The mesolimbic dopamine system: the final common pathway for the reinforcing effect of drugs of abuse? Neurosci Biobehav Rev. 2006;30:215–238. doi: 10.1016/j.neubiorev.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 40.Pritchard JK, Donnelly P. Case-control studies of association in structured or admixed populations. Theoretical Population Biology. 2001;60:227–237. doi: 10.1006/tpbi.2001.1543. [DOI] [PubMed] [Google Scholar]
- 41.Schoenbaum G, Stalnaker T, Shaham Y. A role for BDNF in cocaine reward and relapse. Nature Neuroscience. 2007;10:935–936. doi: 10.1038/nn0807-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shalev U, Grimm JW, Shaham Y. Neurobiology of relapse to heroin and cocaine seeking: a review. Pharmacol Rev. 2002;54:1–42. doi: 10.1124/pr.54.1.1. [DOI] [PubMed] [Google Scholar]
- 43.Sorg B, Chen S, Kalivas P. Time course of tyrosine hydroxylase expression after behavioral sensitization to cocaine. J Pharmacol Exp Ther. 1993;266:424–430. [PubMed] [Google Scholar]
- 44.Spielman RS, Ewens WJ. The TDT and other family-based tests for linkage disequilibrium and association. Am J Hum Genet. 1996;59:983–989. [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas M, Kalivas P, Shaham Y. Neuroplasticity in the mesolimbic dopamine system and cocaine addiction. British Journal of Pharmacology. 2008;154:327–342. doi: 10.1038/bjp.2008.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tian C, Hinds DA, Shigeta R, Kittles R, Ballinger DG, Seldin MF. A genomewide single-nucleotide-polymorphism panel with high ancestry information for African American admixture mapping. Am J Hum Genet. 2006;79:640–649. doi: 10.1086/507954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Volkow N, Wang G, Fowler J, Logan J, Gatley S, Hitzemann R, Chen A, Dewey S, Pappas N. Decreased striatal dopaminergic responsiveness in detoxified cocaine-dependent subjects. Nature. 1997;386:830–833. doi: 10.1038/386830a0. [DOI] [PubMed] [Google Scholar]
- 48.Vrana S, Vrana K, Koves T, Smith J, Dworkin S. Chronic cocaine administration increases CNS tyrosine hydroxylase enzyme activity and mRNA levels and tryptophan hydroxylase enzyme activity levels. Journal of Neurochemistry. 1993;61:2262–2268. doi: 10.1111/j.1471-4159.1993.tb07468.x. [DOI] [PubMed] [Google Scholar]
- 49.Wahls W, Smith G. A heteromeric protein that binds to a meiotic homologous recombination hot spot: correlation of binding and hot spot activity. Genes Dev. 1994;8:1693–1702. doi: 10.1101/gad.8.14.1693. [DOI] [PubMed] [Google Scholar]
- 50.Wise R. Neural mechanisms of the reinforcing action of cocaine. NIDA Res Monogr. 1984;50:15–33. [PubMed] [Google Scholar]
- 51.Wolf ME, Sun X, Mangiavacchi S, Chao SZ. Psychomotor stimulants and neuronal plasticity. Neuropharmacology. 2004;47(Suppl 1):61–79. doi: 10.1016/j.neuropharm.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 52.Wu J, Bell K, Najafi A, Widmark C, Keator D, Tang C, Klein E, Bunney B, Fallon J, Bunney W. Decreasing striatal 6-FDOPA uptake with increasing duration of cocaine withdrawal. Neuropsychopharmacology. 1997;17:402–409. doi: 10.1016/S0893-133X(97)00089-4. [DOI] [PubMed] [Google Scholar]
- 53.Xian H, Scherrer J, Grant J, Eisen S, True W, Jacob T, Bucholz K. Genetic and environmental contributions to nicotine, alcohol and cannabis dependence in male twins. Addiction. 2008;103:1391–1398. doi: 10.1111/j.1360-0443.2008.02243.x. [DOI] [PubMed] [Google Scholar]
- 54.Zhu X, Cooper RS, Elston RC. Linkage analysis of a complex disease through use of admixed populations. Am J Hum Genet. 2004;74:1136–1153. doi: 10.1086/421329. [DOI] [PMC free article] [PubMed] [Google Scholar]