Abstract
Two promoters from the human filarial parasite Brugia malayi have been mapped in detail. The essential domains of both promoters lacked canonical eukaryotic core promoter motifs. However, the largest contiguous essential domain in both promoters flanked and included the splice leader addition site. These findings suggested that the region flanking the trans-splicing addition site might represent a conserved core domain in B. malayi promoters. To test this hypothesis, the putative promoters of 12 trans-spliced genes encoding ribosomal protein homologues from B. malayi were isolated and tested for activity in a B. malayi transient transfection system. Of the 12 domains examined, 11 produced detectable reporter gene activity. Mutant constructs of the six most active promoters were prepared in which the spliced leader acceptor site and the10 nt upstream and downstream of the site were deleted. All deletion constructs exhibited >90% reduction in reporter gene activity relative to their respective wild type sequences. A conserved pyrimidine-rich tract was located directly upstream from the spliced leader splice acceptor site which contained a conserved T residue located at position −3. Mutation of the entire polypyrimidine tract or the conserved T individually resulted in the loss of over 90% of reporter gene activity. In contrast, mutation of the splice acceptor site did not significantly reduce promoter activity. These data suggest that the region surrounding the splice acceptor site in the ribosomal promoters represents a conserved essential domain which functions independently of splice leader addition.
Keywords: filariasis, transfection, promoter, trans splicing, promoter
Introduction
Approximately 140 million individuals worldwide are infected with the three major human filarial parasites, making these infections a significant cause of morbidity in the developing world [1, 2]. As a result of their public health importance, there has been a significant amount of scientific interest in these parasites. A landmark achievement in the research effort into the human filaria was the recent report of a draft genomic sequence for Brugia malayi [3]. The availability of a genomic sequence for this parasite has opened the way for studies into how these parasites regulate their gene expression. The study of the regulation of gene expression will potentially provide important insights into how this parasite has adapted to life in two very different host environments (vertebrate and insect) and how it survives in the face of an ongoing attack by the host’s immune system.
Brugia malayi can be transiently transfected by biolistic methods [4]. The biolistic transfection method has been used to explore both promoter structure and trans-splicing in B. malayi [4–8]. Recent studies have indicated that promoter structure in this parasite is relatively unique. Detailed mapping studies of the promoter of the B. malayi 70 kDa heat shock protein (BmHSP70) and the 12 kDa small subunit ribosomal protein (BmRPS12) have demonstrated that the essential domains of both of these promoters do not contain sequences normally emblematic of the core domain of a typical eucaryotic promoter, such as CAAT or TATAA boxes [7, 9]. Instead, both promoters contain essential domains which flank and include the splice leader (SL) splice acceptor site. The promoter activity of these domains is not related to SL addition, as transgenic transcripts produced from B. malayi transfected with constructs containing either promoter alone were not trans-spliced [5, 8]. These data together suggest the hypothesis that the region flanking the SL domain of B. malayi genes might form at least part of the core promoter domain in these organisms. In the current manuscript, we have tested this hypothesis, using the transient transfection system and targeted deletion analysis to evaluate the role that the SL splice addition site and its flanking domains play in the transcription of a number of B. malayi ribosomal protein (RP) promoters.
Materials and Methods
Amplification and cloning of the putative promoters from the RP Genes
The upstream domains of the RP genes were amplified from B. malayi genomic DNA, essentially as previously described [7]. In brief, specific primer pairs were developed for each upstream domain with assistance of the primer design program Oligo v4.0 [10]. In every case, the 3′ (non coding) primer was designed so that its 5′ end extended through position −1 relative to the open reading frame (ORF), ensuring that the complete upstream domain for each gene was included in the amplicon. The 5′ (coding orientation) primer was designed to be compatible with the 3′ primer, and was located roughly 1,000 nucleotides (nt) upstream from the start of the ORF for those upstream domains that were found not contain a predicted coding domain (CD) within 1,000 nt of the ORF. For genes where the ORF to CD distance was between 400 nt and 1000 nt, the entire intergenic sequence was amplified.
The PCR amplicons obtained from the PCR amplification reactions were cloned directly into the PCR2.1 cloning vector (Invitrogen, Carlsbad, CA), and clones containing the correctly sized inserts were isolated and their identity confirmed by DNA sequencing. The inserts in representative clones for each upstream domain were excised with Eco R1 and the inserts sub-cloned into the luciferase expression vector pGL3 Basic (Promega, Madison, WI) as previously described [9]. The identity and orientation of the insert in the sub-clones was confirmed by DNA sequence analysis of selected sub-clones.
Mutation of the SL splice acceptor sites and flanking domains of the RP promoters
Deletions were created using an inverse PCR strategy, essentially as previously described [7]. In brief, outward facing primers containing Spe1 sites at their 5′ ends were designed to be located upstream and downstream from the SL splice acceptor site in each promoter. In all cases, the 5′ outward facing primer was located at a position beginning 11 nt upstream of the SL splice acceptor site. For those promoters for which the SL splice acceptor site was located more than 10 nt from the start of the ORF, the downstream outward facing primer began 11 nt downstream of the SL splice acceptor site. For promoters in which the SL splice acceptor site was located less than 10 nt from the start of the ORF, the downstream primer was designed from the vector sequence located directly downstream from the 3′ end of the promoter. These primers were then used in inverse PCRs employing the pGL3 Basic clones containing the appropriate promoter as a template. The resulting amplicons were purified, digested with Spe1, and self-ligated, as previously described [7]. This resulted in the production of internal deletion mutant clones in which the SL splice acceptor site and its flanking domains were deleted and replaced with an Spe1 site. In promoters where the SL splice acceptor site was located ≥10 nt from the start of the ORF, this procedure produced clones in which the SL splice acceptor site and the 10 nt located 5′ and 3′ to the SL splice acceptor site were deleted. For those promoters in which the SL splice acceptor site was located <10 nt from the start of the ORF, the 10 nt upstream of the SL splice acceptor site and the entire sequence extending 3′ of the SL splice acceptor site to the beginning of the ORF were deleted.
For mutating the conserved polypyrimidine stretch, the −3 position conserved T and the AG trans-spliced addition site GeneTailor site directed mutagenesis system (Invitrogen) was used. The primers were designed according to the instructions provided in the kit manual and the native sequences were replaced by the most distantly related nucleotides (A was replaced by C and G with T, and vice versa).
Transient transfection and analysis of promoter activity
Isolated B. malayi embryos were transfected and promoter activity assayed by luciferase activity essentially as previously described [5]. In brief, embryos were isolated from gravid female parasites and transfected with the experimental DNA driving the expression of firefly luciferase mixed with a constant amount of an internal standard, consisting of the BmHSP70 promoter fragment driving the expression of renilla luciferase (construct BmHSP70(−659 to −1)/ren) [5]. Transfected embryos were maintained in culture for 48 hours before being assayed for transgene activity. Firefly luciferase activity was normalized to the amount of renilla luciferase activity in each sample to control for variations in transfection efficiency. Firefly/renilla activity ratios for each sample were further normalized to the activity ratio found in embryos transfected in parallel in each experiment with BmRPS12(−641 to −1)/luc). This permitted comparisons of data collected in experiments carried out on different days. Each construct was tested in at least two independent experiments, with each experiment containing triplicate transfections of each construct to be analyzed. The statistical significance of the differences in the activity of the native constructs and the paired SL deletion mutant constructs was assessed using a t-test. As part of the testing process, the assumption of equal variances was tested. In all cases, the variances tested unequal (p < 0.001) so an approximate test with degrees of freedom based on Satterthwaite’s approximation was used.
Hemi-nested PCR analysis of trans-splicing in transgenic mRNAs
RNA prepared from 48-hour post-transfection embryos was used as the template in hemi-nested RT-PCRs to detect evidence of trans-splicing, as previously described [5, 8]. In brief, total RNA was purified from each batch of transfected embryos using Trizol (Invitrogen) following the manufacturer’s instructions. A total of 1μg of this purified RNA was used as a template in a single tube RT-PCR amplification reaction, employing the Qiagen’s One Step reagent (Qiagen, Valencia, CA), following the manufacturer’s protocol. The one step reaction contained primers derived from the SL1 sequence (5′ GGTTTAATTACCCAAGTTTGAG 3′) and the luciferase gene encoded in the pGL3 basic vector (luc nc490; 5′ TTTGCAACCCCTTTTTGGAA 3′). A total of 5 μl of this amplification reaction was used as a template in a 50 μl hemi-nested PCR reaction, employing Pfu Turbo DNA polymerase (Invitrogen), the buffer and amplification conditions recommended by the manufacturer. The hemi-nested PCR reaction employed the SL primer and a nested primer derived from the luciferase gene (luc nc238; 5′ CGACGATTCTGTGATTTGTA 3′). The amplification reactions were analyzed by agarose gel electrophoresis.
Putative transcription factor and binding site and conserved sequence analysis
The P-match algorithm (http://www.gene-regulation.com/cgi-bin/pub/programs/pmatch/bin/p-match.cgi) was used to look for putative transcription factor binding sites in the SL domains and flanking regions in promoters showing activity in the transient transfection assay. P-match uses TRANSFAC 6.0 library as a source database and combines both pattern matching and nucleotide weight matrix approaches for predicting transcription factor binding sites [11]. A medium level of stringency for the prediction was used for the analysis, so that the sum of false-negatives and false-positives were kept to a minimum.
A total of 297 EST assemblies that were represented at a frequency of >1% in the Brugia malayi SL primed EST libraries JHU93SL-BmL4, JHU96SL-BmL2, JHU93SL-BmL4, JHU93SL-BmL3 and JHU93SL-BmL4 (http://www.nematodes.org/nembase3/libSpec.php) were downloaded from the Computational Biology and Functional Genomics Laboratory website (http://compbio.dfci.harvard.edu/tgi/gi/bmgi/searching/xpress_search.html). The genomic contigs of the assemblies were identified by using NCBI genomic BLAST tool (http://www.ncbi.nlm.nih.gov/sutils/blast_table.cgi?taxid=6231) and the sequences flanking the trans splice acceptor addition site were downloaded for those assemblies for which the SL addition site could be identified unambiguously. These sequences were then filtered to remove those encoding the ribosomal proteins. A total 118 sequences were finally analyzed for conserved residues flanking the splice acceptor.
Results
The overall goal of this study was to test the hypothesis that the SL addition domain and its flanking sequences were generally important in controlling transcription from B. malayi genes, perhaps serving as part of the core promoter domain. To accomplish this, it was necessary to identify genes that were transcribed at a level that was detectable using the biolistic transfection system. Furthermore, we felt that it was best to concentrate upon constitutively expressed genes, to prevent regulatory factors from confounding the level of expression from the core promoter domains to the greatest extent possible. Based upon these criteria, we decided to concentrate on the genes encoding homologues of the RPs in B. malayi. Previous studies had shown that the promoter for one member of this protein family, BmRPS12, was amenable to analysis using the transient transfection system [9]. As the genes for RPs are generally coordinately expressed [12], we hypothesized that the RPs represented a good candidate gene family for further evaluating the role of the SL domain in transcription in B. malayi.
To identify the putative RP promoter domains, the B. malayi EST NemBase2 EST database (www.nematodes.org/nematodeESTs/nembase.html) was searched with the terms “ribosome OR ribosomal” to identify ESTs encoding putative RPs. Of the approximately 49 RP genes expected to be present in the B. malayi genome [13], 44 EST clusters encoding putative RPs (including BmRPS12) were identified. The 43 novel EST clusters were then analyzed to identify those that included putative full length cDNA sequences, filtering for EST sequences that contained an SL sequence at their 5′ ends and encoded a putative full length ORF for a RP homologue. The putative ORFs were translated, and the derived amino acid sequences used as queries in BLAST searches. Clusters in which the ORF encoded a protein that exhibited the highest homology scores to RPs derived from other nematodes and in which the homology extended to the amino terminal end of both the derived amino acid sequence and the RP with the highest degree of homology in the database were examined further. A total of 20 EST clusters of the 43 putative novel homologues initially identified met these criteria. The 5′ 100 nt of each of these EST clusters (excluding the SL) were then extracted and used in BLAST searches querying the draft B. malayi genome sequence [3] to extract the 1000 nt extending upstream from the putative 5′ end of mature mRNA. Each of these upstream regions was then analyzed to determine if it contained an upstream predicted CD. Genes in which a CD was predicted to occur within the 400 nt upstream of the RP ORF were eliminated from the study. This was done because previous studies have suggested that some B. malayi genes are organized into operons [3], and it was expected that genes with a proximal 5′ CD might therefore be a downstream gene in such an operon. The filter was set at 400 nt, as previous studies have suggested that most B. malayi promoters extend no more than 400 nt upstream from the start of the ORF [4, 5, 9]. Of the 20 EST clusters determined to encode a putative full length mRNA, 16 met all of these criteria. The SL splice acceptor and addition sites in these genes were then mapped by comparison of the EST and genomic DNA sequences.
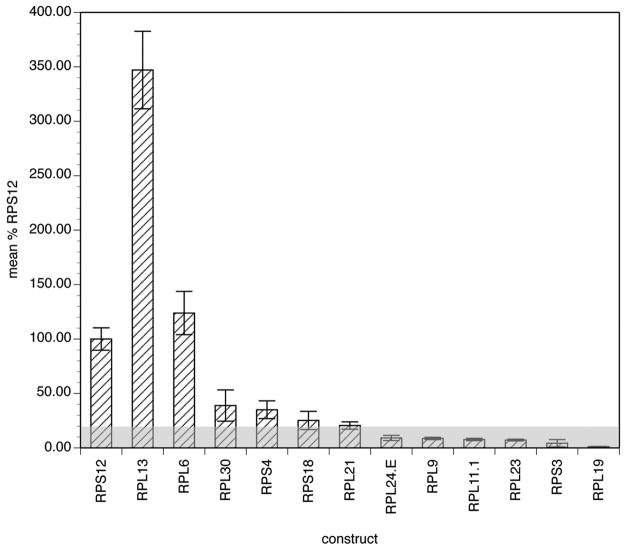
The 5′ untranslated regions (UTRs) from 12 of the 16 novel genes meeting the criteria were successfully amplified and cloned from B. malayi genomic DNA. These were sub-cloned into the luciferase reporter vector pGL3 Basic, and tested for promoter activity in the transient transfection system as described in Materials and Methods. Of the 12 novel 5′ UTRs examined, 11 produced detectable levels of luciferase activity in the transient transfection assay (defined as light units which were at least two fold greater than those obtained from untransfected embryos; Figure 1). The sole putative promoter that did not produce activity in the assay was derived from the B. malayi 19 kDa large subunit ribosomal protein (BmRPL19) gene. Mean activities in the active promoters ranged from 4% to 347% of the activity seen in embryos transfected in parallel with the native BmRPS12 promoter (Figure 1).
Figure 1.
Promoter activity of RP 5′ UTRs in transfected B. malayi embryos
Transfections and luciferase reporter assays were conducted as described in Materials and Methods. Activities are expressed as a percentage of the mean activity of three independent transfections normalized to embryos transfected with the BmRPS12 promoter carried out in parallel, as described in Materials and Methods. Bars represent the mean activity and error bars the standard deviation of six independent transfections. The gray rectangle highlights the activity cutoff for further consideration, as discussed in the text.
Of the 11 promoter sequences found to produce activity in the transient transfection assay, six produced mean levels of activity that were ≥20% of that seen with the BmRPS12 native promoter (Figure 1). Based upon prior experience with this assay, this level of activity was felt to be necessary to reliably detect a significant difference in activities of native and mutant promoters. The six promoters exhibiting activities ≥20% of that exhibited by the BmRPS12 promoter were therefore chosen for further study.
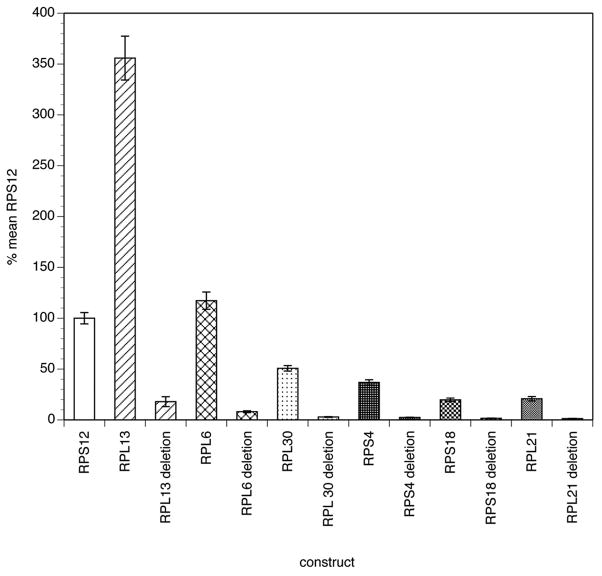
Internal deletions removing the SL splice acceptor sites and the 10 nt surrounding the SL splice acceptor sites from these six RP promoters were then prepared and tested for luciferase activity in the transient transfection system. In every case, deletion of the SL splice acceptor site and its flanking sequences resulted in an approximately 95% reduction in promoter activity relative to that obtained from the native promoter sequence, suggesting that in all cases the SL domain and its flanking sequences were essential for promoter function (Figure 2).
Figure 2.
Activity of native and SL domain-deleted RP promoters in transfected B. malayi embryos
Transfections and luciferase reporter assays were conducted as described in Materials and Methods. Activities are expressed as a percentage of the mean activity of three independent transfections normalized to embryos transfected with the BmRPS12 promoter carried out in parallel, as described in Materials and Methods. Bars represent the mean activity and error bars the standard deviation of six independent transfections. Patterns highlight the paired promoter constructs analyzed, with each pattern representing the native promoter sequence and the sequence in which the SL domain and its flanking nucleotides were deleted. The activity in all of the deletion constructs was significantly reduced relative to that seen in embryos transfected with the corresponding native promoter (p < 0.0001; t-test).
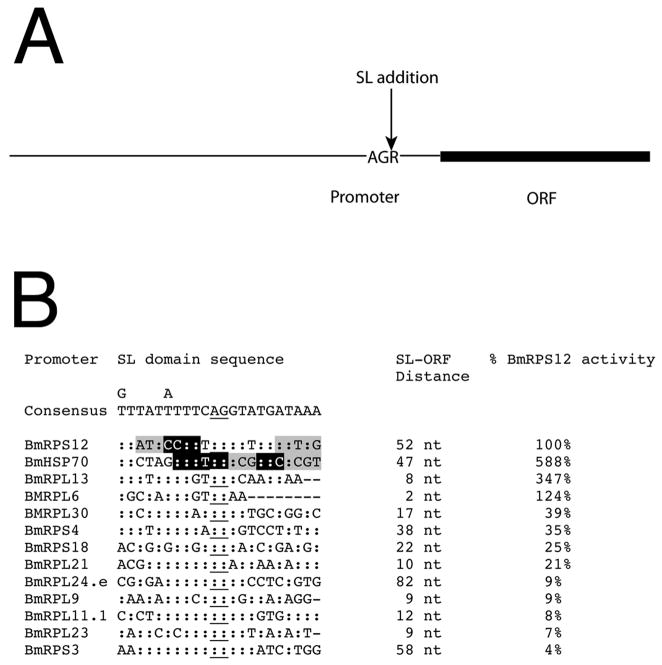
The distance between the SL addition sites and the start of the ORF varied from 2 to 82 nt (Figure 3). The SL-ORF distance did not clearly correlate with level of activity exhibited by the promoters in the transient transfection assay (Figure 3). An alignment of the SL splice acceptor sites and the corresponding flanking domains revealed that, apart from the SL splice acceptor site itself, there were few strictly conserved residues in these domains. The most conserved feature was the presence of a pyrimidine-rich motif located immediately upstream of the SL splice acceptor site (Figure 3). The only strictly conserved residue in this motif was a T located at position −3 relative to the SL splice acceptor site. Portions of this motif, including the conserved T residue at position −3, were previously shown to be necessary for promoter activity in detailed promoter mapping studies of the BmHSP70 and BmRPS12 promoters (Figure 3).
Figure 3.
Alignment of the SL domains of RP promoters with activity in transiently transfected embryos
Panel A: A schematic of a typical B. malayi promoter. The promoter domain extends roughly 400 nt upstream of the start of the ORF and contains SL splice acceptor and SL addition sites. The splice leader addition site is located a variable distance away from the ORF. The schematic is not drawn to scale. Panel B: The SL splice acceptor sites and 10 nt flanking the addition site for RP upstream domains shown to exhibit promoter activity in transiently transfected embryos are presented. The SL splice acceptor site is indicated by underlining. The sequence labeled “consensus” indicates the most common nucleotide present at each position. “:” indicates identity to the most common nucleotide. “-“ indicate nucleotides that do not exist in the sequence (i.e., the SL splice acceptor site fell <10nt of the start codon of the ORF, and therefore the promoter sequence tested did not include any nucleotides in this position). In the case of the BmHSP70 and BmRPS12 promoters, residues highlighted in grey indicate nucleotides which, when mutated, resulted in a loss of 50 – 90% of promoter activity, while residues highlighted in black indicate those nucleotides which, when mutated, resulted in a loss of >90% of promoter activity. The distance from the SL addition site to the start of the ORF for each promoter and the promoter activity relative to that exhibited by the BmRPS12 promoter are also presented. Data for the promoter activity of BmHSP70 and the mapping of the essential residues of the BmHSP70 and BmRPS12 promoters were extracted from previous publications [7, 9].
The SL addition domains were also examined to identify putative transcription factor binding sites using the P-match algorithm, as described in Materials and Methods. P-match predicted no putative transcription factor binding sites in the majority of SL domains. The software predicted binding sites for transcription factor c-Rel in BmRPS12, BmRPS4 and BmRPL23 in the non-coding strand at positions 12 (GGTTT), 13 (GGTCC) and 12 (GGTTT) respectively. The putative binding sites were located within or just downstream from the SL splice acceptor site itself, and did not include the upstream pyrimidine-rich motif discussed above. Furthermore, this region was not essential for activity in the BmRPS12 promoter (Figure 3). This region did contain residues that were essential for activity in the BmHSP70 promoter (Figure 3); however this region of the BmHSP70 promoter did not encode a predicted binding factor for c-Rel.
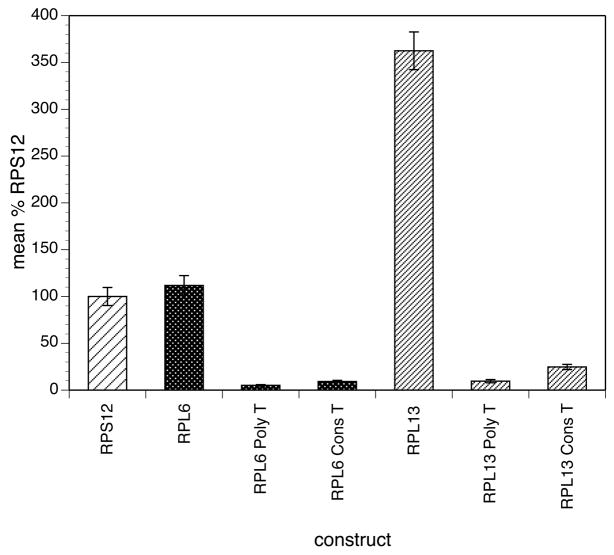
The polypyrimidine tract and the conserved T residue at position −3 relative to the splice acceptor site was present in all of the RPs that produced detectable reporter gene activity in the transient transfection system, as well as in the BmHSP70 promoter. Furthermore, parts of this motif were found to be essential for activity in the detailed mapping studies conducted on both the BmRPS12 and BmHSP70 promoters [7, 9]. It was therefore of interest to determine if these conserved features were essential for reporter gene activity in other promoters as well. To test this hypothesis, replacement mutants were constructed in the BmRPL13 and BmRPL6 promoters. In these mutants, either the entire polypyrimidine tract or the single conserved T residue at position −3 were mutated as described in Materials and Methods. The mutated constructs were then tested for promoter activity in the transient transfection assay. Mutation of the polypyrimidine tract or both RPL13 and RPL6 resulted in a loss of greater than 95% of the reporter gene activity seen in the native constructs (Figure 4). Similarly, mutation of the conserved T residue alone resulted in the loss of greater than 90% of the reporter activity produced by the native constructs (Figure 4).
Figure 4.
Effect of mutations in the upstream polypyrimidine tract on activity of the BmRPL6 and BmRPL 13 promoters
Transfections and luciferase reporter assays were conducted as described in Materials and Methods. Activities are expressed as a percentage of the mean activity of three independent transfections normalized to embryos transfected with the BmRPS12 promoter carried out in parallel, as described in Materials and Methods. Bars represent the mean activity and error bars the standard deviation of six independent transfections. Patterns highlight the promoter constructs analyzed, with each pattern representing the native promoter sequence and its associated mutations.
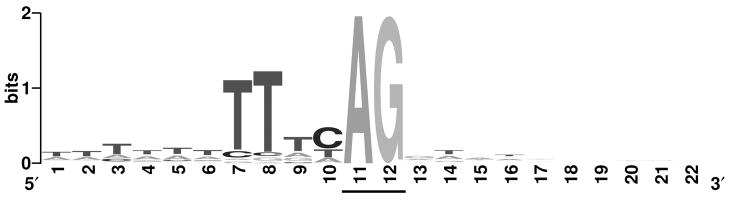
As the polypyrimidine tract appeared to be conserved and essential for reporter gene activity in all four of the promoters examined in detail to date, it was of interest to determine if this motif was conserved in other B. malayi genes. To accomplish this, 118 genes with clearly identifiable trans-splicing acceptor sites (excluding the RP genes) were analyzed for conserved domains in the 40nt flanking the splice acceptor site. Similar to what was seen in the RP promoters, a conserved stretch of residues consisting primarily of pyrimidines was found to be located at positions −1 to −4 relative to the splice acceptor (Figure 5) The most conserved residues were Ts located at positions −3 and −4, with the most conserved residue a T at position −3 (Figure 5). The T residue at −3 was present in 87.3% of the genes analyzed, while T was present 84% of the time at position −4.
Figure 5.
Conserved residues in flanking the trans-splice acceptor site in genes known to produce trans-spliced RNAs
Genes known to be trans-spliced were identified as described in Materials and Methods. The figure was produced using WebLogo (version 2.8.2) software (http://weblogo.berkeley.edu/) [16, 17]. The overall height of each stack of nucleotides represents the extent of sequence conservation at the position indicated. The height of each nucleotide represents its relative frequency at that position. The conserved splice acceptor sequence is indicated by underlining.
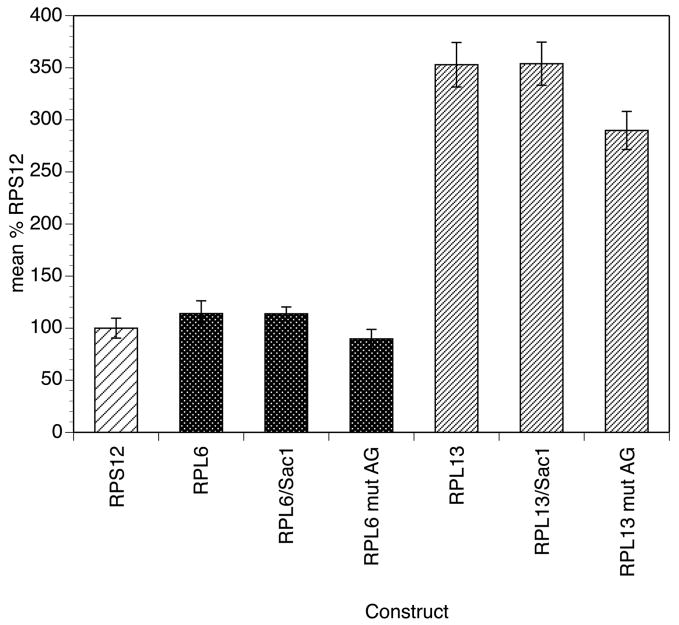
Previous studies have demonstrated that the reporter gene activity generated from both the BmHSP70 and the BmRPS12 promoters was independent of the trans-splicing process, as constructs containing these promoters alone in the absence of necessary downstream sequences produced transgenic mRNAs that were not trans-spliced [5, 6, 8]. Hemi-nested SL-mediated PCR assays targeting the transgenic mRNAs were carried out to determine if this was also the case for the six most active RP promoters. No amplification products were obtained from any of the RNA preparations extracted from embryos transfected with these promoters, demonstrating that, as is the case for the BmHSP70 and BmRPS12 promoters, the transgenic mRNAs produced from these promoters were not trans-spliced (data not shown). Although these studies demonstrated that the transgenic mRNAs from the ribosomal protein promoters did not have an SL at their 5′ end, they did not rule out the possibility that the mRNAs were processed through some other sort of cis-splicing pathway, perhaps involving upstream sequences present in the vector. If this was the case and if this processing was necessary to generate a functional mRNA, the regions surrounding the splice acceptor might still be primarily playing a role in an mRNA processing process which was essential for reporter gene activity. To test this hypothesis, the plasmids containing the RPL6 and RPL13 promoters were linearized with the restriction enzyme Sac1, which cut the plasmids in the polylinker just upstream from the 5′ end of the promoters. If cis-splicing to an upstream vector sequence was necessary for the activity seen, we predicted that linearizing the plasmid would eliminate this cis-splicing activity, resulting in a loss of reporter gene activity. However linearization of the plasmid DNA had no apparent effect upon reporter gene activity (Figure 6), suggesting that a cryptic cis-splicing activity utilizing upstream vector sequences was not necessary for promoter activity.
Figure 6.
Effect of Sac1 digestion and mutation of the trans splice acceptor on activity of the BmRPL6 and BmRPL 13 promoters
Transfections and luciferase reporter assays were conducted as described in Materials and Methods. Activities are expressed as a percentage of the mean activity of three independent transfections normalized to embryos transfected with the BmRPS12 promoter carried out in parallel, as described in Materials and Methods. Bars represent the mean activity and error bars the standard deviation of six independent transfections. Patterns highlight the groups of promoter constructs analyzed, as in previous figures.
If the role of the region surrounding the SL splice acceptor site was indeed to mediate some sort of splicing resulting in mRNA maturation, we further hypothesized that the splice acceptor site itself would be essential in this process. To test this hypothesis, the splice acceptor sites in the RPL6 and RPL 13 promoters were mutated and the mutant constructs tested for activity in the transient transfection system. The two constructs in which the splice acceptor sites were mutated exhibited levels of reporter gene activity which were roughly 80% of those seen in the wild type constructs (Figure 6), suggesting that any mRNA processing mediated by the splice acceptor site was not essential for reporter gene activity.
Discussion
In previous detailed mapping studies of the BmHSP70 and BmRPS12 promoters, motifs corresponding to the canonical CAAT and TATAA boxes normally found to represent the core domain of a typical eucaryotic promoter were not found to be essential for promoter activity [7, 9]. However, the region surrounding the SL splice acceptor site was found to be the largest domain necessary for activity in both promoters. In both cases, mutation of residues in the region located roughly 10nt upstream and downstream from the SL splice acceptor site resulted in a loss of over 90% of reporter gene activity [7, 9]. These data suggested that this region might represent a conserved essential domain of B. malayi promoters. The data presented here support this hypothesis. Deletion of the SL splice acceptor site and the 10 nucleotides located upstream and downstream of this site resulted in a greater than 95% reduction in reporter gene activity in all six RP promoters examined. Together, these data suggest that the region flanking the SL splice acceptor site represents a conserved essential domain in B. malayi promoters.
The data presented here, in conjunction with other studies suggest that the role of the residues encoded within the SL addition domain is not directly associated with the trans-splicing process. As shown above and in previous studies [5, 6, 8], transgenic mRNAs produced from constructs containing B. malayi promoters alone are not trans-spliced. Trans-splicing of such transgenic mRNAs can occur, but this process requires sequences encoded downstream from the start codon [5, 6, 8]. These data suggested the nucleotides flanking the SL splice acceptor site were playing some role in transcription that is at least partly independent of trans-slicing. This conclusion was further strengthened by the studies reported here, in which mutation of the splice acceptor sites in the RPL6 and RPL13 promoters resulted in levels of reporter gene activity that were decreased only 20% from the activities seen in the wild type constructs. It is thus possible that some sort of processing mediated by the splice acceptor site might be occurring in the transgenic mRNAs derived from these promoters, but this process does not greatly affect the amount of protein translated from these transgenic mRNAs.
The 12 RP promoters analyzed in this study produced widely varying levels of luciferase reporter gene activity in the transient transfection assay, ranging from essentially no activity to levels that were roughly four times those exhibited from the BmRPS12 promoter. It is possible that some of this variation in activity is a result of differences in the SL domains among these promoters. However, it is clear from the previous mapping studies of both the BmHSP70 and BmRPS12 promoters that regions located upstream from the SL addition domain are also necessary for full promoter activity. For example, the BmHSP70 promoter contains a canonical heat shock response element encoded in the region extending from 172 nt to 185 nt upstream of the SL splice acceptor, deletion of which results in a loss of roughly 80% of the activity of the native promoter [7]. Similarly, the BmRPS12 promoter contains a structure consisting of 5 3/4 44 nt direct repeat units extending from positions −11 to −264 relative to the SL addition site. Deletion of this repeat element results in a loss of roughly 80% of the native promoter activity [9]. Because the promoter fragments utilized in these studies were rather short, it is possible that important upstream domains were excluded from some of the RP promoters examined here. Thus, it is difficult to use the results presented above as a way to determine how polymorphisms in the SL addition domain might affect promoter activity. Such studies will require placing the various SL addition domains in the context of identical upstream domains.
A comparison of a large number of trans-spliced genes suggests that a semi-conserved pyrimidine-rich motif is located just upstream from the SL splice acceptor site. Interestingly, mutations in this polypyrimidine tract result in a loss of reporter gene activity in all four of the B. malayi promoters examined to date (BmHSP70, BmRPS12, BmRPL13 and BmRPL6). Taken together, these data suggest that the polypyrimidine motif might be the region responsible for the role that the SL addition domain plays in promoter activity. However, such semi-conserved pyrimidine rich domains also form part of the splice acceptor site in conventionally cis-spliced eukaryotic type 2 introns [13]. Thus, there are several potential roles that the polypyrimidine tract may play in the process of mRNA production, maturation and translation. First, it is possible that this motif plays an important role in initiating transcription of the mRNA. Second, it is possible that the polypyrimidine tract plays a role in transcript maturation, although the studies reported here suggest that any such processing event must be independent of the SL splice acceptor site. Finally, as the SL addition domain does not encode any motifs that are recognized by any of the factors known to be involved in the formation of the pre-initiation complex of a typical eukaryotic RNA polymerase [7, 9], it is possible that some of the factors involved in recognizing the SL addition site may also play a role in initiating or promoting transcription in B. malayi trans-spliced genes.
Identification of the exact start of transcription would be useful in determining if the SL addition domain is involved directly in initiating transcription. Attempts to map the start site of transcription in the transgenic transcripts produced by the RP promoters were unsuccessful (data not shown). However, studies of trans-splicing in the free living nematode Caenorhabditis elegans have demonstrated that trans-splicing requires the presence of an “outron” located upstream of the SL addition site, which is an intron-like sequence lacking a splice donor site [14]. Recent studies have shown that outrons in C. elegans are characterized by the presence of a U rich “OU element” located 50–75 nt upstream from the SL addition site [15]. Presumably, this element is involved in the trans-splicing process, and if so, one would predict that the start of transcription of trans-spliced genes in C. elegans would be located upstream of this element as well. However, a search of B. malayi genes with defined SL addition sites did not identify a homologue of the C. elegans Ou element in the region extending 50–75nt upstream of the SL addition site (data not shown).
EST analyses have suggested that a majority of mRNAs in B. malayi are trans-spliced. However, not all B. malayi genes produce trans-spliced mRNAs. The data presented above suggests that the SL addition domain encodes sequences that are necessary for transcriptional activity in genes producing trans-spliced mRNAs. However, it is not known if those genes producing non-trans-spliced mRNAs use a similar mechanism to initiate transcription as those producing trans-spliced messages, or if the mechanism used to initiate and promote transcription in non trans-spliced genes is distinct from that used in trans-spliced genes. In this regard, it is interesting to note that putative homologues of several proteins instrumental in forming the pre-initiation complex of RNA polymerase, such as the TATA box binding protein and TATA box binding protein associated factor are listed in the annotation of predicted CDs the B. malayi genome (data not shown). It will be of interest to determine if these represent true homologues of these transcription factors and if so whether they are specifically involved in initiating and promoting transcription of those genes encoding non-trans spliced mRNAs. Studies investigating this hypothesis are currently underway.
Acknowledgments
We would like to thank Drs. John Adams and Naomi Lang-Unnasch for critical reading of the manuscript. We also acknowledge the use of the B. malayi genomic sequence produced and maintained by TIGR. TIGR’s sequencing effort is part of the International Brugia Genome Sequencing Project and was supported by an award from the National Institute of Allergy and Infectious Diseases, National Institutes of Health. Preliminary sequence data from this project are deposited regularly into the GSS division of GenBank. This work was supported by a grant from National Institute of Allergy and Infectious Diseases to TRU (Project# R01AI048562).
Abbreviations
- BmHSP70
Brugia malayi 70 kDa heat shock protein
- BmRPL
Brugia malayi large subunit ribosomal protein
- BmRPS
Brugia malayi small subunit ribosomal protein
- CD
Coding domain
- nt
nucleotides
- ORF
Open reading frame
- PCR
polymerase chain reaction
- RT-PCR
reverse transcriptase polymerase chain reaction
- SL
Spliced leader
- UTR
Untranslated region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.World Health Organization. Lymphatic filariasis. World Health Organization; Geneva: 2000. [Google Scholar]
- 2.World Health Organization Expert Committee. Onchocerciasis and its control. World Health Organization; Geneva: 1995. [PubMed] [Google Scholar]
- 3.Ghedin E, Wang S, Spiro D, et al. Draft genome of the filarial nematode parasite Brugia malayi. Science. 2007;317:1756–60. doi: 10.1126/science.1145406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Higazi TB, Merriweather A, Shu L, et al. Brugia malayi: Transient transfection by microinjection and particle bombardment. Exp Parasitol. 2002;100:95–102. doi: 10.1016/S0014-4894(02)00004-8. [DOI] [PubMed] [Google Scholar]
- 5.Shu L, Katholi C, Higazi T, et al. Analysis of the Brugia malayi HSP70 promoter using a homologous transient transfection system. Mol Biochem Parasitol. 2003;128:67–75. doi: 10.1016/s0166-6851(03)00052-5. [DOI] [PubMed] [Google Scholar]
- 6.Higazi TB, Unnasch TR. Intron encoded sequences necessary for trans splicing in transiently transfected Brugia malayi. Mol Biochem Parasitol. 2004;137:181–84. doi: 10.1016/j.molbiopara.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 7.Higazi TB, DeOliveira A, Katholi CR, et al. Identification of elements essential for transcription in Brugia malayi promoters. J Mol Biol. 2005;353:1–13. doi: 10.1016/j.jmb.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 8.Liu C, de Oliveira A, Higazi TB, et al. Sequences necessary for trans-splicing in transiently transfected Brugia malayi. Mol Biochem Parasitol. 2007;156:62–73. doi: 10.1016/j.molbiopara.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oliveira AD, Katholi CR, Unnasch TR. Characterization of the promoter of the Brugia malayi 12kda small subunit ribosomal protein (rps12) gene. Int J Parasitol. 2008;38:1111–19. doi: 10.1016/j.ijpara.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rychlik W. Oligo. National Biosciences; Plymouth, MN: 1992. [Google Scholar]
- 11.Chekmenev DS, Haid C, Kel AE. P-match: Transcription factor binding site search by combining patterns and weight matrices. Nucleic Acids Res. 2005;33:W432–7. doi: 10.1093/nar/gki441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li B, Nierras CR, Warner JR. Transcriptional elements involved in the repression of ribosomal protein synthesis. Mol Cell Biol. 1999;19:5393–404. doi: 10.1128/mcb.19.8.5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alberts B, Johnson A, Lewis J, et al. Molecular biology of the cell. New York: Garland Science; 2002. [Google Scholar]
- 14.Conrad R, Thomas J, Spieth J, et al. Insertion of part of an intron into the 5′ untranslated region of a Caenorhabditis elegans gene converts it into a trans-spliced gene. Mol Cell Biol. 1991;11:1921–6. doi: 10.1128/mcb.11.4.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graber JH, Salisbury J, Hutchins LN, et al. C. elegans sequences that control trans-splicing and operon pre-mRNA processing. RNA. 2007;13:1409–26. doi: 10.1261/rna.596707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneider TD, Stephens RM. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crooks GE, Hon G, Chandonia JM, et al. Weblogo: A sequence logo generator. Genome Res. 2004;14:1188–90. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]