Abstract
G-substrate, an endogenous substrate for cGMP-dependent protein kinase, exists almost exclusively in cerebellar Purkinje cells, where it is possibly involved in the induction of long-term depression. A G-substrate cDNA was identified by screening expressed sequence tag databases from a human brain library. The deduced amino acid sequence of human G-substrate contained two putative phosphorylation sites (Thr-68 and Thr-119) with amino acid sequences [KPRRKDT(p)PALH] that were identical to those reported for rabbit G-substrate. G-substrate mRNA was expressed almost exclusively in the cerebellum as a single transcript. The human G-substrate gene was mapped to human chromosome 7p15 by radiation hybrid panel analysis. In vitro translation products of the cDNA showed an apparent molecular mass of 24 kDa on SDS/PAGE which was close to that of purified rabbit G-substrate (23 kDa). Bacterially expressed human G-substrate is a heat-stable and acid-soluble protein that cross-reacts with antibodies raised against rabbit G-substrate. Recombinant human G-substrate was phosphorylated efficiently by cGMP-dependent protein kinase exclusively at Thr residues, and it was recognized by antibodies specific for rabbit phospho-G-substrate. The amino acid sequences surrounding the sites of phosphorylation in G-substrate are related to those around Thr-34 and Thr-35 of the dopamine- and cAMP-regulated phosphoprotein DARPP-32 and inhibitor-1, respectively, two potent inhibitors of protein phosphatase 1. However, purified G-substrate phosphorylated by cGMP-dependent protein kinase inhibited protein phosphatase 2A more effectively than protein phosphatase 1, suggesting a distinct role as a protein phosphatase inhibitor.
Keywords: cerebellum, long-term depression, protein phosphatase inhibitor, gene mapping
In the central nervous system, the cerebellum plays an important role in motor coordination and motor learning (1, 2), and because of its simple neuronal circuit, it has allowed extensive studies of the mechanisms underlying these functions. One established mechanism underlying cerebellar motor learning is long-term depression (LTD), which is an activity-dependent and long-lasting depression of synaptic transmission from parallel fibers onto Purkinje cells (3–5). As Purkinje cells are the sole origin of output from the cerebellum, modification of this output signal by LTD plays a significant role in motor learning (4, 6, 7).
A specific role for cGMP in the induction of LTD has been suggested (6). Moreover, nitric oxide (NO) is known to increase cGMP synthesis in cerebellar neurons (8). In turn, accumulating evidence has shown that NO plays an essential role in the induction of cerebellar LTD (9–12) and LTD-dependent motor learning such as vestibulo-ocular reflex adaptation (13) and interlimb adaptation in perturbed locomotion (14). Even though Purkinje cells lack NO synthase, most other cerebellar cells, such as the granule, basket, and Bergman glia cells, do synthesize NO. The target(s) for NO is reported to be inside the Purkinje cell (7, 15), where diffusing NO activates guanylyl cyclase (16) and therefore would be expected to increase cGMP-dependent protein kinase (PKG) activity. While each component involved in the cascade activating PKG has been shown to be essential, less is known about downstream components under the influence of PKG.
G-substrate purified from rabbit cerebellum was shown to be one of a few preferred substrates for PKG (17–22). Immunohistochemical studies reveal that G-substrate is concentrated in cerebellar Purkinje cells with PKG (18, 23, 24) in rat, mouse, and rabbit. These lines of evidence led us to consider that G-substrate is a downstream component of PKG and may also take part in the induction of cerebellar LTD. In the present study, we have cloned and characterized human G-substrate. The results obtained indicated that G-substrate mRNA is expressed almost exclusively in cerebellum. Moreover, recombinant human G-substrate expressed in Escherichia coli had characteristics similar to those of the protein purified from rabbit cerebellum. Notably, purified G-substrate phosphorylated by cGMP-dependent protein kinase was an effective inhibitor of protein phosphatase 2A (PP2A), suggesting a distinct role for the protein as a protein phosphatase inhibitor. The molecular cloning and expression of recombinant G-substrate should facilitate the investigation of the physiological role of the protein in cerebellar function.
MATERIALS AND METHODS
Molecular Cloning of Human G-Substrate and Sequencing.
The blast program (25–27) was utilized to search the human expressed sequence tag (EST) database (National Center for Biotechnology Information dbest; ref. 28) using the partial amino acid sequence obtained for rabbit G-substrate (21). Three potential clones were obtained from the IMAGE Consortium (Genome Systems, St. Louis) and sequenced. The clone 46041 (GenBank accession no. H09006) from normalized human infant brain cDNA library (29) was found to contain a full-length human G-substrate and was characterized further in this study. The cDNA clone was characterized by double-strand DNA sequencing using GeneRapid Seq 4X4 (Amersham–Pharmacia) with the thermo sequenase Cy5.5 dye terminator cycle sequencing kit (Amersham–Pharmacia). All regions of the clones were sequenced at least twice in both strands.
Tissue Distribution of Human G-Substrate mRNA.
A 275-bp PCR fragment (nucleotides 47–321) of human G-substrate cDNA was labeled by using the Ready-To-Go DNA labeling system (Amersham–Pharmacia) with [α-33P]dCTP and purified by spin column (Boehringer Mannheim). A human multiple tissue Northern blot (CLONTECH) and a human RNA master blot (CLONTECH) were probed with a 33P-labeled probe (1.5 × 106 cpm/ng of DNA) according to the manufacturer’s protocol.
Radiation Hybrid Panel Mapping.
The radiation hybrid panel mapping was performed with Stanford Human Genome Center G3 Radiation Hybrid Panel (Research Genetics, Huntsville, AL), using primers 5′-TTTCTGCTCTCATTTTTGTCATCGT-3′ and 5′-CAGAAGATGCAGAAACCGAACC-3′ for the 229-bp fragment of 3′ untranslated region (nucleotides 592–820) of human G-substrate cDNA. The result was analyzed on the RH server at the Stanford Human Genome Center’s web site (http://www-shgc.stanford.edu) and the relative map position was estimated.
In Vitro Translation of Human G-Substrate.
The cDNA coding full-length human G-substrate was inserted into pBluescript (Stratagene), and the T7 promoter of the vector was utilized for transcription. In vitro translation was carried out by using a Single Tube Protein System 3 (Novagen) in the presence of [35S]methionine (NEN) according to the manufacturer’s instructions. The resulting products were analyzed by SDS/PAGE and autoradiography.
Bacterial Expression of Human G-Substrate.
Human G-substrate cDNA was cloned into pTrcHis (Invitrogen) for bacterial expression of the protein as a hexahistidine-tagged fusion protein (6×His-hG-substrate). The 6×His-hG-substrate was expressed in E. coli BL21 (DE3) (Novagen) as described previously (30). The crude bacterial extracts were heated for 5 min at 95°C, the supernatants were applied to a metal-chelating column saturated with Ni2+ (chelating-Sepharose FF, Amersham–Pharmacia), and the bound recombinant proteins were eluted by stepwise gradients of imidazole. Purification was analyzed by SDS/PAGE (31).
Immunoblot Analysis.
SDS/PAGE was performed by the method of Laemmli (31). Proteins were transferred to poly(vinylidene difluoride) membrane (PVDF; Bio-Rad) in 10 mM 3-(cyclohexylamino)-1-propanesulfonic acid (Caps)–NaOH (pH 10.5) containing 10% (vol/vol) methanol. Membranes were blocked with nonfat dry milk in 20 mM Tris⋅HCl (pH 7.5) containing 0.1% Tween 20 and 150 mM NaCl (TTBS), then incubated with antibodies raised against rabbit G-substrate (32). Immunodetection used protein G conjugated with horseradish peroxidase (Bio-Rad) and the ECL (enhanced chemiluminescence) system (Amersham–Pharmacia). Protein concentration was determined by the method of Bradford (33), using BSA as standard (E2801% = 6.54).
Phosphorylation of Recombinant Human G-Substrate.
Recombinant human G-substrate was phosphorylated with the catalytic subunit of purified bovine heart cAMP-dependent protein kinase (PKA, 0.3 μg/ml; Sigma) in 50 mM [bis(2-hydroxyethyl)amino]tris(hydroxymethyl)methane (Bistris)⋅ HCl (pH 6.5) containing 50 μM ATP, 12.5 mM MgCl2, 0.25 mM EGTA, and 0.3 mg/ml BSA, or recombinant bovine PKG Iα (0.25 μg/ml; Calbiochem) in 50 mM Hepes–NaOH (pH 7.5), containing 50 μM ATP, 2 mM MgCl2, 10 μM cGMP, and 100 μM 3-isobutyl-1-methylxanthine at 30°C. The extent of phosphorylation was determined by including trace amounts of [γ-33P]ATP (NEN) in the reaction mixtures. Incorporation of [33P]phosphate was analyzed by SDS/PAGE and autoradiography. Phosphoamino acid analysis of the [33P]phosphorylated human G-substrate was carried out according to Hildebrandt and Fried (34) followed by thin-layer chromatography (35).
Protein Phosphatase Assay.
Rabbit G-substrate was purified according to Aswad and Greengard (19) and phosphorylated by PKG purified from bovine lung (36) to 2 mol of phosphate per mol of G-substrate (19). Inhibitor 1 (I-1) was purified from skeletal muscle as described (37). The catalytic subunits of protein phosphatase 1 (PP1) and PP2A were purified from rabbit skeletal muscle (38). The activities of PP1 and PP2A were assayed by using 32P-phosphorylase phosphorylated by phosphorylase kinase (38).
RESULTS
Molecular Cloning of Human G-Substrate.
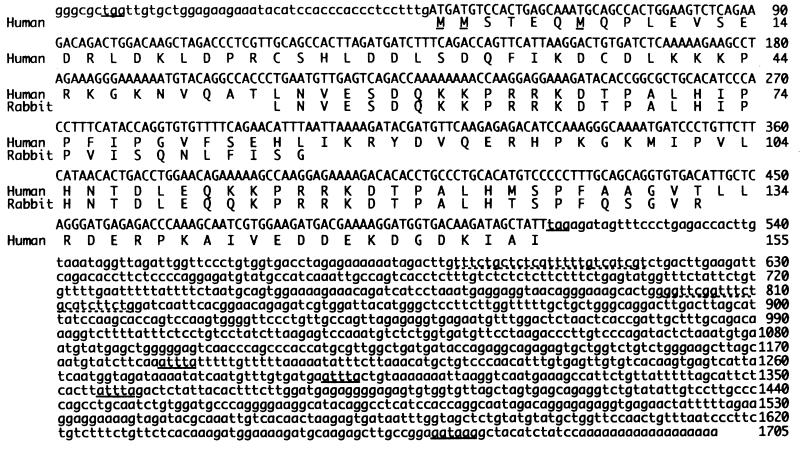
EST clones that encoded sequences similar to partial amino acid sequences of rabbit G-substrate were obtained, sequenced, and further characterized. Clone 46041 (GenBank accession no. H09006) from a normalized human brain cDNA library (29) contained full-length cDNA for human G-substrate and was used for further characterization. The cDNA clone contained an insert of 1.7 kb, a poly(A)+ tail at the 3′ end, and a short 5′-untranslated region. The cDNA has three mRNA destabilization signals (ATTTA; ref. 39) and one poly(A)+ signal (AATAAA) after the stop codon in the open reading frame (Fig. 1). The clone also contained an in-frame stop codon in the 5′-untranslated region (Fig. 1). The observed mRNA size of 1.85 kb (Fig. 2) and the insert size of 1.7 kb in clone 46041 suggest an additional 0.15 kb of 3′- and/or 5′-untranslated region for the human mRNA.
Figure 1.
Nucleotide and deduced amino acid sequences of human G-substrate. Human G-substrate sequence was aligned with the partial sequences of rabbit G-substrate (21). The cDNA contains three closely grouped potential initiation methionines (underlined). The deduced amino acid sequence has two phosphorylation sites (Thr-68 and Thr-119), as previously identified in rabbit G-substrate. The poly(A)+ signal (AATAAA), mRNA destabilization signals (ATTTA), and in-frame stop codon in the 5′-untranslated region are underlined. The primers used for radiation hybrid analysis are designated with broken underlines. The nucleotide sequence of human G-substrate has been submitted to GenBank (accession no. AF097730).
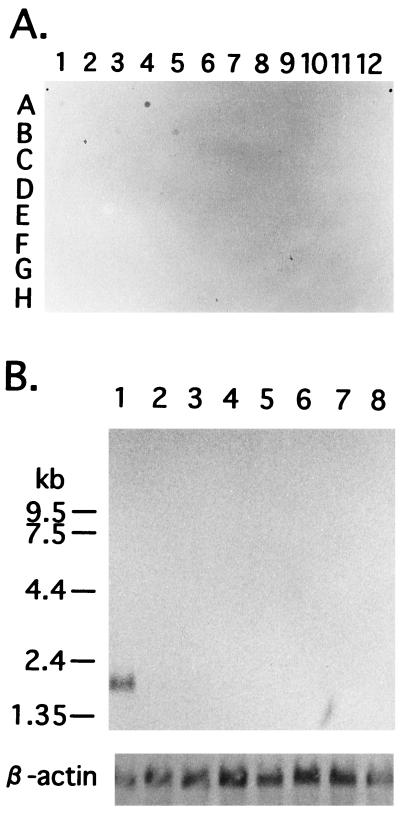
Figure 2.
Human G-substrate mRNA distribution in various human tissues. A human RNA master blot (CLONTECH) and a human multiple tissue Northern blot (CLONTECH) were probed with a 33P-labeled 275-bp PCR fragment of human G-substrate cDNA (nucleotides 47–321). (A) Human RNA master blot analysis. Each dot contains approximately 2 μg of poly(A)+ RNA. The sources of mRNA were as follows: A1, whole brain; A2, amygdala; A3, caudate nucleus; A4, cerebellum; A5, cerebral cortex; A6, frontal lobe; A7, hippocampus; A8, medulla oblongata; B1, occipital lobe; B2, putamen; B3, substantia nigra; B4, temporal lobe; B5, thalamus; B6, thalamic nucleus; B7, spinal cord; B8, no sample; C1, heart; C2, aorta; C3, skeletal muscle; C4, colon; C5, bladder; C6, uterus; C7, prostate; C8, stomach; D1, testis; D2, ovary; D3, pancreas; D4, pituitary gland; D5, adrenal gland; D6, thyroid gland; D7, salivary gland; D8, mammary gland; E1, kidney; E2, liver; E3, small intestine; D4, spleen; E5, thymus; E6, peripheral leukocyte; E7, lymph node; E8, bone marrow; F1, appendix; F2, lung; F3, trachea; F4, placenta; F5-F8, no sample; G1, fetal brain; G2, fetal heart; G3, fetal kidney; G4 fetal liver; G5, fetal spleen; G6, fetal thymus; and G7, fetal lung. (B) Human multiple tissue Northern blot analysis. Each lane contains approximately 2 μg of poly(A)+ RNA from each tissue. The sources of mRNA were as follows: lane 1, cerebellum; lane 2; cerebral cortex; lane 3, medulla; lane 4, spinal cord; lane 5, occipital pole; lane 6, frontal lobe; lane 7, temporal lobe; and lane 8, putamen. The blot was stripped and reprobed with 33P-labeled 2-kb fragment of human β-actin to assess the amount of poly(A)+ RNA loading in each lane.
Deduced Amino Acid Sequence of Human G-Substrate.
The full-length cDNA of 1,705 bp contained an open reading frame of 465 bp encoding 155 amino acids with a predicted molecular mass of 17,849.84 Da (Fig. 1). The predicted human G-substrate polypeptide is hydrophilic, with 64 charged amino acid residues and 36 basic amino acid residues (11 Arg, 19 Lys, and 6 His), resulting in a calculated pI of 8.6. Three closely positioned potential initiation methionines were observed in the open reading frame (Fig. 1). None of the cDNA sequences around the three methionines matched the Kozak sequences exactly (40), and further characterization is required to identify the translation initiation site. A remarkable similarity was found between the amino acid sequences surrounding the two phosphorylation sites (Thr-68 and Thr-119). There were 17 identities over a sequence of 21 residues in the amino acid sequences (Gln-60 to Val-80 and Gln-111 to Val-131). These results support the idea that an internal gene duplication occurred in G-substrate as suggested previously for rabbit G-substrate (21).
Distribution of Human G-Substrate mRNA in the Brain.
Nucleic acid hybridization analysis was used to examine the tissue distribution of human G-substrate mRNA (Fig. 2). By using an RNA master blot (CLONTECH), mRNA was detected almost exclusively in cerebellum. Extremely long exposures gave faint spots in whole brain, substantia nigra, thalamus, and pituitary gland, but human G-substrate mRNA was not observed in any other region of the central nervous system or in nonneuronal tissues. This observation was confirmed by Northern blotting analysis using a multiple tissue Northern Blot (CLONTECH). The human G-substrate transcript was detected as a single band at 1.85 kb and was found readily in cerebellum, but with extremely long exposures also gave faint bands on all tissues (not shown). The expression of mRNA almost exclusively in the cerebellum is in good agreement with the distribution of the protein in rat, mouse, and rabbit previously observed by using rabbit G-substrate antibodies (17, 18, 23, 24).
Chromosomal Localization of the Human G-Substrate Gene.
Radiation hybrid panel mapping was performed to map the human G-substrate gene. The result showed that this gene was tightly linked to the marker sWSS341 (D7S1414) located on chromosome 7p15 with a distance of 17.02 cR (centiray) and lod (logarithm of odds) score of 10.48. On the average, 1 cR equals 20 kb on chromosome 7. This result supported the idea that the human G-substrate gene is located on chromosome 7p15.
Characterization of Human G-Substrate.
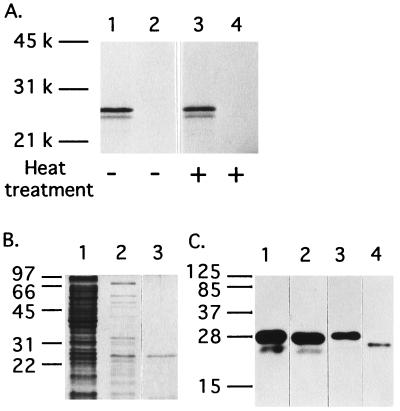
The full-length cDNA of human G-substrate was transcribed and translated in vitro with a rabbit reticulocyte lysate system. Two translation products were observed at approximately 24 kDa and 23 kDa on SDS/PAGE, with the 24-kDa band being the major product (Fig. 3A). This observation suggests that, as an initiation site, one of the first two methionines was used more frequently than the third methionine (Fig. 1). The apparent molecular mass of in vitro translated human G-substrate (24 kDa) was very similar to that of purified rabbit G-substrate (23 kDa) estimated on SDS/PAGE (19). Further, the in vitro translated human G-substrate was a heat-stable protein, and more than 90% of the protein remained in the supernatant after heat treatment (Fig. 3A). These physical properties are also found for purified rabbit G-substrate protein (19).
Figure 3.
Analysis of the in vitro translation products of human G-substrate cDNA and bacterially expressed recombinant human G-substrate. (A) The in vitro transcription–translation products were subjected to SDS/PAGE followed by autoradiography. Lane 1, pBluescript containing full-length human cDNA; lane 2, no template DNA; lane 3, pBluescript containing full-length human cDNA with heat treatment; lane 4, no template DNA with heat treatment. (B) Human G-substrate was expressed as a 6×His-tagged fusion protein in E. coli (BL21), purified, and analyzed by SDS/PAGE. Lane 1, crude extract; lane 2, heated crude extract; lane 3, after purification by chelating-Sepharose. The gel was stained with Coomassie brilliant blue. (C) Immunoblot of 6×His-tagged human G-substrate proteins with antibodies against rabbit G-substrate; lane 1, crude extract; lane 2, heated crude extract; lane 3, after purification by chelating-Sepharose; lane 4, purified rabbit G-substrate (10 ng).
Human G-substrate was expressed as a hexahistidine-tagged fusion protein (6×His-hG-substrate) in E. coli BL21 (DE3) and purified by metal-chelating chromatography (Fig. 3B). The 6×His-hG-substrate readily cross-reacted with antibodies generated against rabbit G-substrate (Fig. 3C). As with rabbit G-substrate (19) and in vitro translated human G-substrate, bacterially expressed 6×His-hG-substrate was also a heat-stable (Fig. 3B) and acid-soluble (0.5 M H2SO4) protein (data not shown). There was a discrepancy in molecular mass calculated from the cDNA sequence (17,849 Da) and that obtained by SDS/PAGE of the in vitro translated products of human G-substrate (24 kDa) (Fig. 3A). This difference could be attributed to an anomalous electrophoretic mobility of rabbit or human G-substrate on SDS/PAGE because of low detergent binding, as reported for I-1 (41) and dopamine- and cAMP-regulated phosphoprotein of 32,000 Da (DARPP-32) (42).
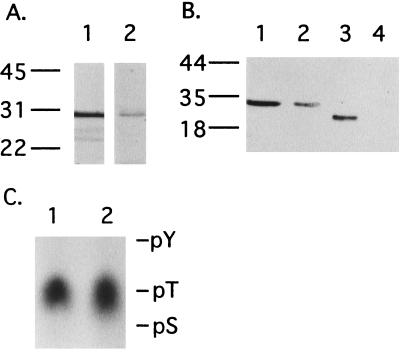
The recombinant 6×His-hG-substrate was phosphorylated by PKG with a stoichiometry of 2 mol of phosphate per mol of protein, suggesting that both phosphorylation sites were good substrates for the kinase (Fig. 4A). PKA also phosphorylated the 6×His-hG-substrate but at a much slower rate similar to the case of rabbit G-substrate (20). Phosphoamino acid analyses of 6×His-hG-substrate phosphorylated by PKG or PKA showed that the modification occurred exclusively on threonine residues of G-substrate (Fig. 4C). Immunoblot analysis showed that the phosphorylated human G-substrate was recognized by antibodies specific for the phosphorylated form of rabbit G-substrate (Fig. 4B), which confirmed that the sites of phosphorylation were the same as the ones in rabbit G-substrate (21, 32).
Figure 4.
Phosphorylation of bacterially expressed recombinant human G-substrate (6×His-hG-substrate). The recombinant proteins (0.5 μg) were phosphorylated by PKG or PKA as described in Materials and Methods. (A) Autoradiogram showing recombinant proteins phosphorylated by PKG or PKA in the presence of [γ-33P]ATP Mg salt for 15 min. Lane 1, 6×His-hG-substrate phosphorylated by PKG; lane 2, 6×His-hG-substrate phosphorylated by PKA. (B) Immunoblot of the recombinant proteins with antibodies specific for phosphorylated rabbit G-substrate. The phosphorylation reactions were carried out for 15 min, then the reaction mixtures were subjected to immunoblot analysis. Lane 1, 6×His-hG-substrate phosphorylated by PKG; lane 2, 6×His-hG-substrate phosphorylated by PKA; lane 3, rabbit G-substrate (10 ng) phosphorylated by PKG; lane 4, rabbit dephospho-G-substrate (10 ng). (C) Phosphoamino acid analysis of the recombinant human G-substrate phosphorylated by PKG or PKA. Each lane contained approximately 5 × 103 cpm of [33P]phosphate. Lane 1, 6×His-hG-substrate phosphorylated by PKG; lane 2, 6×His-hG-substrate phosphorylated by PKA. The positions of standard phosphoamino acids are indicated: pY, phosphotyrosine; pT, phosphothreonine; pS, phosphoserine.
Inhibition of Protein Phosphatases by G-Substrate.
The amino acid sequences surrounding the sites of phosphorylation in G-substrate are related to those surrounding Thr-34 and Thr-35 of DARPP-32 and I-1, respectively, two potent inhibitors of PP1 (Fig. 5). In addition, the physicochemical properties of G-substrate, DARPP-32, and I-1 are similar in that all three proteins are heat stable and acid soluble. We therefore assessed the ability of purified G-substrate to act as a protein phosphatase inhibitor. Dephospho-G-substrate, at concentrations of up to 20 μM, did not significantly inhibit the phosphorylase phosphatase activities of PP1 or PP2A (not shown). However, phospho-G-substrate inhibited both PP1 and PP2A effectively, with Ki values of 1.51 ± 0.76 and 0.27 ± 0.11 μM, respectively (Table 1). For both PP1 and PP2A, phospho-G-substrate inhibited phosphatase activity in a noncompetitive manner (not shown). As a control, phospho-I-1 was shown to inhibit PP1 potently in a noncompetitive manner, and to inhibit PP2A weakly in a competitive manner (Table 1, and data not shown).
Figure 5.
Comparison of amino acid sequence of human G-substrate, DARPP-32, and I-1. The regions around the phosphorylation sites of DARPP-32 (Thr-34), I-1 (Thr-35), and G-substrate (Thr-68 and Thr-119) are boxed. The regions in DARPP-32 and I-1 that contain the R/KKIQF motif necessary for inhibition of PP1 are also boxed.
Table 1.
Effect of phospho-G-substrate and phospho-I-1 on the phosphorylase phosphatase activities of PP1 and PP2A catalytic subunits
| Inhibitor |
Ki, μM
|
|
|---|---|---|
| PP1 | PP2A | |
| G-substrate | 1.51 ± 0.76 | 0.27 ± 0.11 |
| I-1 | 0.005 | 4.0 |
Ki values were obtained from secondary plots of data obtained from Lineweaver–Burk analysis (0.5–5.0 μM [32P]phosphorylase). For G-substrate, n = 3.
DISCUSSION
A human G-substrate cDNA was identified by screening an EST database, using the partial amino acid sequence of rabbit G-substrate. The major products of in vitro translated human G-substrate had an apparent molecular mass of 24 kDa, similar to that of rabbit G-substrate. Further, in vitro translated and bacterially expressed recombinant proteins were similar to the rabbit counterpart in their physical characteristics such as heat stability and molecular mass, and the recombinant protein was readily recognized by antibodies raised against rabbit G-substrate. In addition to the remarkable conservation found in the vicinity of phosphorylation sites (Thr-68 and Thr-119), 50 of the 60 amino acid residues obtained from rabbit G-substrate phosphorylated by PKG were identical to the corresponding regions of human G-substrate (Fig. 1). These results together with the amino acid sequence deduced from the EST cDNA strongly support the idea that the isolated clone is the human homolog of rabbit G-substrate. No other related protein was found in protein databases, leading us to conclude that this is the first G-substrate cDNA to be isolated from any species. The human G-substrate gene was mapped to chromosome 7p15, and it is interesting to note that there are several neuronal genetic diseases that have also been mapped to this location.
As discussed above, the amino acid sequences surrounding the sites of phosphorylation in G-substrate are related to those surrounding Thr-34 and Thr-35 of DARPP-32 and I-1 (Fig. 5), respectively, two potent inhibitors of PP1. However, in our study phospho-G-substrate inhibited the catalytic subunit of PP2A more effectively than it did that of PP1, suggesting a distinct role for the protein as a PP2A inhibitor. In cells, the catalytic subunit of PP2A interacts with A and B subunits to form heterotrimeric complexes (43). The substrate specificity and intracellular localization of PP2A may be regulated by the B subunit within this heterotrimeric complex (44). Cerebellar Purkinje cells contain both Bα and Bβ subunits (44), and it will be necessary to assess the interaction between phospho-G-substrate and heterotrimeric PP2A complexes prepared from cerebellum. Preliminary studies indicate that phospho-G-substrate does indeed inhibit PP2A holoenzymes prepared from cerebellum (P. Simonelli, H.-C. Li, A.C.N., and P.G., unpublished results). Two other inhibitors of PP2A, I1PP2A and I2PP2A, have been previously described (45). Though G-substrate is less potent than I1PP2A and I2PP2A (45), it is not known if these endogenous PP2A inhibitors are present in cerebellum. We have also observed that G-substrate inhibits the PP2A catalytic subunit with an IC50 of 70 nM when histone H1 is used as a substrate (data not shown). These observations suggest that G-substrate may work as a specific PP2A inhibitor in cerebellum.
Even though G-substrate is similar to DARPP-32 and I-1 in the region surrounding the phosphorylation sites of the proteins, it was a poor PP1 inhibitor, being much less potent than I-1 (this study and ref. 41) or DARPP-32 (46). The reduced PP1 inhibitory activity may reflect the absence of the R/KKIQF motif, which is present in DARPP-32 (47) and I-1 (48) (Fig. 5) and other PP1-binding proteins and is essential for inhibition of PP1. The importance of the R/KKIQF motif in I-1 was shown by mutating Ile of RKIQF to Asn, which resulted in a 30- to 50-fold increase in the IC50 for PP1 inhibition (48). In addition, synthetic peptides of DARPP-32 lacking the KKIQF motif failed to inhibit PP1 (49). G-substrate has a KKKPR sequence instead of R/KKIQF, N-terminal to the first phosphorylation site (Thr-68) (Fig. 5). This KKKPR sequence is presumably not functional in terms of binding to PP1. However, it is possible that the KKKPR sequence might play a role in binding to the catalytic subunit of PP2A. Analysis of the effect of mutations in G-substrate will make it possible to better understand the function of the KKKPR sequence and the influence that each of the phosphorylation sites has with respect to inhibition of PP2A and PP1.
In the present study, the mRNA for human G-substrate was found to be almost exclusively localized in the cerebellum, in good agreement with previous results obtained from immunohistological studies using anti-rabbit G-substrate antibodies (17, 18, 23, 24). Although our experiments were not extended to specific cerebellar cell types, it is likely that human G-substrate is also present in Purkinje cells. Protein phosphatase inhibitors have been implicated in models of neuronal plasticity, in LTD in the hippocampus (50), and in sensitization in Aplysia (51). Inhibition of PP2A by phospho-G-substrate may therefore also play a role in the regulation of LTD in cerebellar Purkinje cells. G-substrate may also interact with signaling proteins in addition to PP2A. Identification of G-substrate-binding proteins and generation of G-substrate knockout mice should provide further insight into the physiological role of the protein in the induction of cerebellar LTD and neuronal plasticity.
Acknowledgments
We thank Dr. R. Kado for critical reading of the original manuscript. This work was supported by U.S. Public Health Service Grants MH-40899 and DA-10044 (to P.G. and A.C.N.).
ABBREVIATIONS
- DARPP-32
dopamine- and cAMP-regulated phosphoprotein of 32,000 Da
- EST
expressed sequence tag
- I-1
protein phosphatase inhibitor 1
- LTD
long-term depression
- PKA
cAMP-dependent protein kinase
- PKG
cGMP-dependent protein kinase
- PP1 and PP2A
protein phosphatase 1 and 2A
Note Added in Proof
The amino acid sequence of mouse G-substrate has recently been reported (52).
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF097730).
References
- 1.Ito M. The Cerebellum and Neural Control. New York: Raven; 1984. [Google Scholar]
- 2.Thach W T. Trends Cognit Sci. 1998;2:331–337. doi: 10.1016/s1364-6613(98)01223-6. [DOI] [PubMed] [Google Scholar]
- 3.Ito M, Sakurai M, Tongroach P. J Physiol (London) 1982;324:113–134. doi: 10.1113/jphysiol.1982.sp014103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ito M. Trends Cognit Sci. 1998;2:313–321. doi: 10.1016/s1364-6613(98)01222-4. [DOI] [PubMed] [Google Scholar]
- 5.Daniel H, Levenes C, Crepel F. Trends Neurosci. 1998;21:401–407. doi: 10.1016/s0166-2236(98)01304-6. [DOI] [PubMed] [Google Scholar]
- 6.Ito M. Annu Rev Neurosci. 1989;12:85–102. doi: 10.1146/annurev.ne.12.030189.000505. [DOI] [PubMed] [Google Scholar]
- 7.Lisberger S G. Cell. 1998;92:701–704. doi: 10.1016/s0092-8674(00)81397-5. [DOI] [PubMed] [Google Scholar]
- 8.Garthwaite J, Charles S L, Chess-Williams R. Nature (London) 1988;336:385–388. doi: 10.1038/336385a0. [DOI] [PubMed] [Google Scholar]
- 9.Ito M, Karachot L. NeuroReport. 1990;1:129–132. doi: 10.1097/00001756-199010000-00012. [DOI] [PubMed] [Google Scholar]
- 10.Shibuki K, Okada D. Nature (London) 1991;349:326–328. doi: 10.1038/349326a0. [DOI] [PubMed] [Google Scholar]
- 11.Hartell N. NeuroReport. 1994;5:833–836. doi: 10.1097/00001756-199403000-00024. [DOI] [PubMed] [Google Scholar]
- 12.Lev-Ram V, Jiang T, Wood J, Lawrence D S, Tsien R Y. Neuron. 1997;18:1025–1038. doi: 10.1016/s0896-6273(00)80340-2. [DOI] [PubMed] [Google Scholar]
- 13.Nagao S, Ito M. NeuroReport. 1991;2:193–196. doi: 10.1097/00001756-199104000-00008. [DOI] [PubMed] [Google Scholar]
- 14.Yanagihara D, Kondo I. Proc Natl Acad Sci USA. 1996;93:13292–13297. doi: 10.1073/pnas.93.23.13292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lev-Ram V, Makings L R, Keitz P F, Kao J P Y, Tsien R Y. Neuron. 1995;15:407–415. doi: 10.1016/0896-6273(95)90044-6. [DOI] [PubMed] [Google Scholar]
- 16.Stone J R, Marletta M A. Biochemistry. 1996;35:1094–1099. doi: 10.1021/bi9519718. [DOI] [PubMed] [Google Scholar]
- 17.Schlichter D J, Casnellie J E, Greengard P. Nature (London) 1978;273:61–62. doi: 10.1038/273061a0. [DOI] [PubMed] [Google Scholar]
- 18.Schlichter D J, Detre J A, Aswad D W, Chehrazi B, Greengard P. Proc Natl Acad Sci USA. 1980;77:5537–5541. doi: 10.1073/pnas.77.9.5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aswad D W, Greengard P. J Biol Chem. 1981;256:3487–3493. [PubMed] [Google Scholar]
- 20.Aswad D W, Greengard P. J Biol Chem. 1981;256:3494–3500. [PubMed] [Google Scholar]
- 21.Aitken A, Bilham T, Cohen P, Aswad D, Greengard P. J Biol Chem. 1981;256:3501–3506. [PubMed] [Google Scholar]
- 22.Nairn A C, Hemmings H C, Jr, Greengard P. Annu Rev Biochem. 1985;54:931–976. doi: 10.1146/annurev.bi.54.070185.004435. [DOI] [PubMed] [Google Scholar]
- 23.Detre J A, Nairn A C, Aswad D W, Greengard P. J Neurosci. 1984;4:2843–2849. doi: 10.1523/JNEUROSCI.04-11-02843.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qian Y, Chao D S, Santillano D R, Cornwell T L, Nairn A C, Greengard P, Lincoln T M, Bredt D S. J Neurosci. 1996;16:3130–3138. doi: 10.1523/JNEUROSCI.16-10-03130.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karlin S, Altschul S F. Proc Natl Acad Sci USA. 1990;87:2264–2268. doi: 10.1073/pnas.87.6.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karlin S, Altschul S F. Proc Natl Acad Sci USA. 1993;90:5873–5877. doi: 10.1073/pnas.90.12.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boguski M S, Lowe T M, Tolstoshev C M. Nat Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- 29.Soares M B, Bonaldo M F, Jelene P, Su L, Lawton L, Efstratiadis A. Proc Natl Acad Sci USA. 1994;91:9228–9232. doi: 10.1073/pnas.91.20.9228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Endo S, Zhou X, Connor J H, Wang B, Shenolikar S. Biochemistry. 1996;35:5220–5228. doi: 10.1021/bi952940f. [DOI] [PubMed] [Google Scholar]
- 31.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 32.Nairn A C, Detre J A, Casnellie J E, Greengard P. Nature (London) 1982;299:734–736. doi: 10.1038/299734a0. [DOI] [PubMed] [Google Scholar]
- 33.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 34.Hildebrandt E, Fried V A. Anal Biochem. 1989;177:407–412. doi: 10.1016/0003-2697(89)90075-4. [DOI] [PubMed] [Google Scholar]
- 35.Neufeld E, Goren H J, Boland D. Anal Biochem. 1989;177:138–143. doi: 10.1016/0003-2697(89)90028-6. [DOI] [PubMed] [Google Scholar]
- 36.Walter U, Miller P, Wilson F, Menkes D, Greengard P. J Biol Chem. 1980;255:3757–3762. [PubMed] [Google Scholar]
- 37.Cohen P, Foulkes J G, Holmes C F B, Nimmo G A, Tonks N K. Methods Enzymol. 1988;159:427–437. doi: 10.1016/0076-6879(88)59042-0. [DOI] [PubMed] [Google Scholar]
- 38.Cohen P, Alemany S, Hemmings B A, Resink T J, Stralfors P, Tung H Y. Methods Enzymol. 1988;159:390–408. doi: 10.1016/0076-6879(88)59039-0. [DOI] [PubMed] [Google Scholar]
- 39.Shaw G, Kamen R. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 40.Kozak M. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nimmo G A, Cohen P. Eur J Biochem. 1978;87:341–351. doi: 10.1111/j.1432-1033.1978.tb12383.x. [DOI] [PubMed] [Google Scholar]
- 42.Girault J-A, Hemmings H C, Jr, Williams K R, Nairn A C, Greengard P. J Biol Chem. 1989;264:21748–21759. [PubMed] [Google Scholar]
- 43.Shenolikar S, Nairn A C. Adv Second Messenger Phosphoprotein Res. 1991;23:1–121. [PubMed] [Google Scholar]
- 44.Strack S, Zaucha J A, Ebner F F, Colbran R J, Wadzinski B E. J Comp Neurol. 1998;392:515–527. [PubMed] [Google Scholar]
- 45.Li M, Guo H, Damuni Z. Biochemistry. 1995;34:1988–1996. doi: 10.1021/bi00006a020. [DOI] [PubMed] [Google Scholar]
- 46.Hemmings H C, Jr, Greengard P, Tung H Y, Cohen P. Nature (London) 1984;310:503–505. doi: 10.1038/310503a0. [DOI] [PubMed] [Google Scholar]
- 47.Brene S, Lindefors N, Ehrlich M, Taubes T, Horiuchi A, Kopp J, Hall H, Sedvall G, Greengard P, Persson H. J Neurosci. 1994;14:985–998. doi: 10.1523/JNEUROSCI.14-03-00985.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Endo S, Connor J H, Forney B, Zhang L, Ingebritsen T S, Lee E Y C, Shenolikar S. Biochemistry. 1997;36:6986–6992. doi: 10.1021/bi970418i. [DOI] [PubMed] [Google Scholar]
- 49.Hemmings H C, Jr, Nairn A C, Elliott J I, Greengard P. J Biol Chem. 1990;265:20369–20376. [PubMed] [Google Scholar]
- 50.Mulkey R M, Endo S, Shenolikar S, Malenka R C. Nature (London) 1994;369:486–488. doi: 10.1038/369486a0. [DOI] [PubMed] [Google Scholar]
- 51.Endo S, Critz S D, Byrne J H, Shenolikar S. J Neurochem. 1995;64:1833–1840. doi: 10.1046/j.1471-4159.1995.64041833.x. [DOI] [PubMed] [Google Scholar]
- 52.Hall K U, Collins S P, Gamm D M, Massa E, DePaoli-Roach A A, Uhler M D. J Biol Chem. 1991;274:3485–3496. doi: 10.1074/jbc.274.6.3485. [DOI] [PubMed] [Google Scholar]