Abstract
The sequence of small-subunit rRNA varies in an orderly manner across phylogenetic lines and contains segments that are conserved at the species, genus, or kingdom level. By directing oligonucleotide primers at sequences conserved throughout the eubacterial kingdom, we amplified bacterial 16S ribosomal DNA sequences with the polymerase chain reaction. Priming sites were located at the extreme 5' end, the extreme 3' end, and the center of 16S ribosomal DNA. The isolates tested with these primers included members of the genera Staphylococcus, Coxiella, Rickettsia, Clostridium, Neisseria, Mycobacterium, Bilophila, Eubacterium, Fusobacterium, and Lactobacillus and the family Enterobacteriaceae. Initially, the yields from the reactions were erratic because the primers were self-complementary at the 3' ends. Revised primers that were not self-complementary gave more reproducible results. With the latter primers, 0.4 pg of Escherichia coli DNA consistently gave a visible band after amplification. This method should be useful for increasing the amounts of bacterial 16S ribosomal DNA sequences for the purposes of sequencing and probing. It should have a broad range of applications, including the detection and identification of known pathogens that are difficult to culture. This approach may make it possible to identify new, nonculturable bacterial pathogens.
Full text
PDF

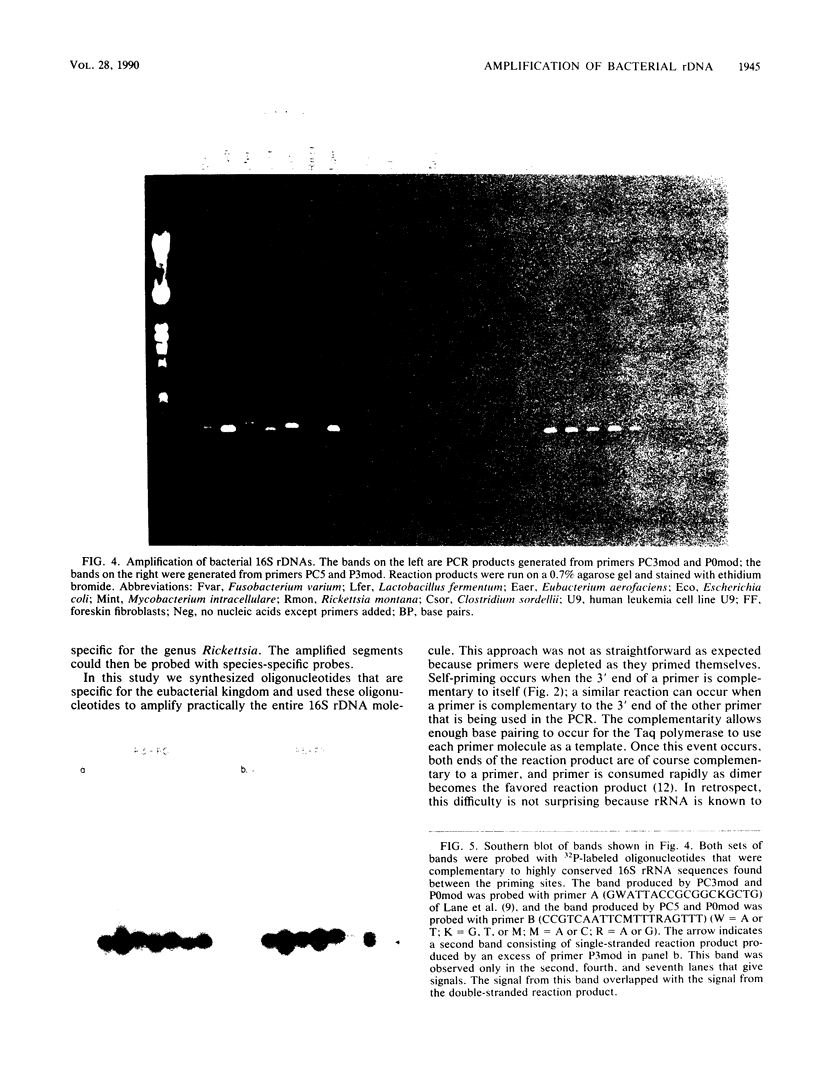

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chen K., Neimark H., Rumore P., Steinman C. R. Broad range DNA probes for detecting and amplifying eubacterial nucleic acids. FEMS Microbiol Lett. 1989 Jan 1;48(1):19–24. doi: 10.1016/0378-1097(89)90139-0. [DOI] [PubMed] [Google Scholar]
- Drake T. A., Hindler J. A., Berlin O. G., Bruckner D. A. Rapid identification of Mycobacterium avium complex in culture using DNA probes. J Clin Microbiol. 1987 Aug;25(8):1442–1445. doi: 10.1128/jcm.25.8.1442-1445.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelke D. R., Hoener P. A., Collins F. S. Direct sequencing of enzymatically amplified human genomic DNA. Proc Natl Acad Sci U S A. 1988 Jan;85(2):544–548. doi: 10.1073/pnas.85.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni S. J., DeLong E. F., Olsen G. J., Pace N. R. Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. J Bacteriol. 1988 Feb;170(2):720–726. doi: 10.1128/jb.170.2.720-726.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell R. R., Weiser B., Woese C. R., Noller H. F. Comparative anatomy of 16-S-like ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1985;32:155–216. doi: 10.1016/s0079-6603(08)60348-7. [DOI] [PubMed] [Google Scholar]
- Göbel U., Maas R., Haun G., Vinga-Martins C., Stanbridge E. J. Synthetic oligonucleotide probes complementary to rRNA for group- and species-specific detection of mycoplasmas. Isr J Med Sci. 1987 Jun;23(6):742–746. [PubMed] [Google Scholar]
- Huysmans E., De Wachter R. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 1986;14 (Suppl):r73–118. doi: 10.1093/nar/14.suppl.r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innis M. A., Myambo K. B., Gelfand D. H., Brow M. A. DNA sequencing with Thermus aquaticus DNA polymerase and direct sequencing of polymerase chain reaction-amplified DNA. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9436–9440. doi: 10.1073/pnas.85.24.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D. J., Pace B., Olsen G. J., Stahl D. A., Sogin M. L., Pace N. R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci U S A. 1985 Oct;82(20):6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell L. G., Merril C. R. Affinity generation of single-stranded DNA for dideoxy sequencing following the polymerase chain reaction. Anal Biochem. 1989 May 1;178(2):239–242. doi: 10.1016/0003-2697(89)90631-3. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Scharf S., Faloona F., Mullis K. B., Horn G. T., Erlich H. A., Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985 Dec 20;230(4732):1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Wilson K. H., Blitchington R., Hindenach B., Greene R. C. Species-specific oligonucleotide probes for rRNA of Clostridium difficile and related species. J Clin Microbiol. 1988 Dec;26(12):2484–2488. doi: 10.1128/jcm.26.12.2484-2488.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K. H., Blitchington R., Shah P., McDonald G., Gilmore R. D., Mallavia L. P. Probe directed at a segment of Rickettsia rickettsii rRNA amplified with polymerase chain reaction. J Clin Microbiol. 1989 Dec;27(12):2692–2696. doi: 10.1128/jcm.27.12.2692-2696.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]