Abstract
ARGONAUTE 1 (AGO1) slices endogenous messenger RNAs (mRNAs) during both microRNA (miRNA)- and short interfering RNA (siRNA)-guided post-transcriptional silencing. We have previously reported that AGO1 homeostasis is maintained through the repressive action of miR168 on AGO1 mRNA and the stabilizing effect of AGO1 protein on miR168, but siRNA-mediated AGO1 regulation has not been reported. Here, we show that AGO1-derived siRNAs trigger RNA DEPENDENT RNA POLYMERASE 6 (RDR6)-, SUPPRESSOR OF GENE SILENCING 3 (SGS3)- and SILENCING DEFECTIVE 5 (SDE5)-dependent AGO1 silencing, which also requires DICER-LIKE 2 (DCL2) and DCL4. By varying the efficacy of miR168-guided AGO1 mRNA cleavage, we show that siRNA-mediated AGO1 silencing depends on correct miRNA targeting, pointing to coordinated regulatory actions of the miRNA and siRNA pathways during the maintenance of AGO1 homeostasis. Finally, our results reveal that dcl2, dcl3 and dcl4 mutations similarly affect post-transcriptional gene silencing (PTGS) mediated by a sense transgene and PTGS mediated by inverted repeats, validating the branched pathway model proposed previously.
Keywords: ARGONAUTE, cosuppression, PTGS, miRNA, RNAi
Introduction
ARGONAUTE 1 (AGO1), one of 10 AGO proteins in Arabidopsis, is the main AGO protein mediating short interfering RNA (siRNA)-directed post-transcriptional gene silencing (PTGS) and microRNA (miRNA)-directed regulation (Vaucheret, 2008). miRNAs are processed from partially self-complementary miRNA transcripts whereas siRNA production from sense transgenes, viruses and endogenous trans-acting siRNA (TAS) loci entails stabilization of their non-self-complementary transcripts by SUPPRESSOR OF GENE SILENCING 3 (SGS3) followed by RNA DEPENDENT RNA POLYMERASE 6 (RDR6)-mediated conversion to double-stranded RNA (dsRNA; Chen, 2005; Jones-Rhoades et al, 2006; Mallory et al, 2008). SILENCING DEFECTIVE 5 (SDE5), a homologue of a human messenger RNA (mRNA) export factor, also influences the production of siRNA through an unidentified mechanism (Hernandez-Pinzon et al, 2007). Once mature, miRNAs and siRNAs load onto AGO1, which shows a preference for small RNAs beginning with 5′-uridine (Montgomery et al, 2008a; Mi et al, 2008; Takeda et al, 2008), and guide AGO1-mediated cleavage and translational repression of complementary RNAs (Baumberger & Baulcombe, 2005; Qi et al, 2005; Brodersen et al, 2008).
AGO1 mRNA is regulated by the miRNA miR168 in an AGO1-dependent manner (Rhoades et al, 2002; Vaucheret et al, 2004). Plants expressing a miR168-resistant version of AGO1 (4m-AGO1), which contains silent mutations that increase the number of mismatches between miR168 and AGO1 mRNA without altering AGO1 protein sequence, show developmental defects that can be rescued on expression of a compensatory miRNA (4m-miR168) that restores nucleotide pairing (Vaucheret et al, 2004). In addition to this regulatory loop, two additional mechanisms—transcriptional co-regulation of MIR168 and AGO1, and preferential stabilization of miR168 by AGO1—contribute to AGO1 homeostasis (Vaucheret et al, 2006).
Although AGO1 interacts with numerous siRNAs and mediates siRNA-guided PTGS, siRNA-mediated AGO1 regulation has not been reported. Several deep-sequencing analyses have shown that AGO1 transcripts give rise to both sense and antisense 21-nucleotide siRNAs that map downstream from, and generally in phase with, the miR168 cleavage site, a feature atypical of most miRNA targets (Lu et al, 2005; Axtell et al, 2006; Rajagopalan et al, 2006; Kasschau et al, 2007). This uncharacteristic production of siRNAs prompted us to test whether, in addition to miR168-directed regulation, AGO1 mRNA might be sensitive to siRNA-directed regulation. Here, we show that AGO1-derived siRNAs trigger AGO1 silencing in an RDR6-, SDE5- and SGS3-dependent manner, and that production of AGO1-derived siRNAs requires the action of DICER-LIKE 2 (DCL2) and DCL4, similar to viruses (Bouche et al, 2006; Deleris et al, 2006) and inverted repeat (IR) transgenes (Dunoyer et al, 2005, 2007; Fusaro et al, 2006). miR168-directed regulation of AGO1 is necessary for robust siRNA-directed AGO1 silencing, indicating that both the miRNA and siRNA pathways are involved in the maintenance of AGO1 homeostasis.
Results And Discussion
AGO1 is hyper-susceptible to cosuppression
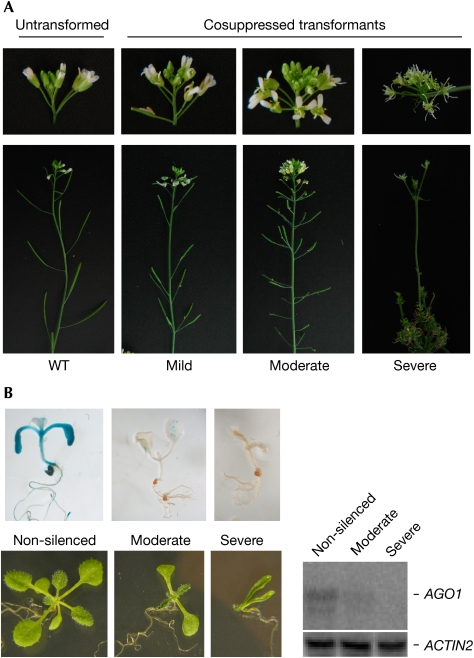
Plants showing typical ago1 mutant developmental defects, including serrated leaves, phyllotaxy defects, floral organ defects and reduced fertility (Fig 1), were obtained by the transformation of wild-type Arabidopsis with either an 8-kb AGO1 genomic fragment that contains all of the upstream and downstream regulatory elements required for the function of AGO1 (pAGO1:168-AGO1; Vaucheret et al, 2004), an AGO1 cDNA expressed under the control of the strong cauliflower mosaic virus 35S promoter (p35S:168-AGO1), a promoterless AGO1 cDNA (Δp:168-AGO1), or the 5′ 557 nt of the AGO1 mRNA translationally fused to Beta-glucuronidase (GUS) and expressed from the AGO1 promoter (pAGO1:168-AGO1-GUS). Developmental defects of ago1 mutants were not observed when these constructs were introduced in the PTGS-deficient rdr6 mutant (Fig 2), suggesting that these defects result from AGO1 cosuppression.
Figure 1.
Ectopic AGO1 copies trigger cosuppression. (A) Varying degrees of developmental defects resulting from AGO1 cosuppression, including spoon-shaped cotyledons, serrated leaves, altered floral phyllotaxis, floral organ defects and reduced fertility. Constructs that potentially encode a full-length AGO1 protein triggered mild and moderate phenotypes whereas constructs that encode a truncated AGO1 protein fused to Beta-glucuronidase (GUS) triggered moderate and severe phenotypes. (B) GUS staining and AGO1 messenger RNA (mRNA) analysis in non-silenced, moderate and severe cosuppressed pAGO1:168-AGO1-GUS transformants show that both the AGO1-GUS transgene and the endogenous AGO1 gene are cosuppressed. GUS intensity and the levels of AGO1 mRNA show that the severity of developmental defects correlates with increased cosuppression. The blot was hybridized for ACTIN2 as a control. AGO, ARGONAUTE; WT, wild type.
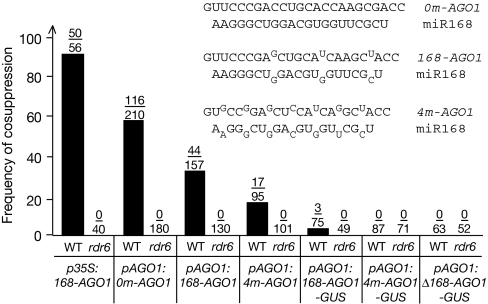
Figure 2.
miR168-directed AGO1 cleavage facilitates AGO1 cosuppression. Frequencies of AGO1 cosuppression observed after the transformation of wild-type (WT) plants and rna dependent rna polymerase 6 (rdr6) mutants with the indicated constructs. The fraction of plants showing the cosuppression phenotypes of AGO1 is indicated above each bar. The pairing between miR168 and WT and mutant AGO1 messenger RNAs is shown. AGO, ARGONAUTE.
The cosuppression of AGO1 follows the previously established traits of cosuppression. For example, AGO1 cosuppression efficiency was high using the strong 35S promoter (p35S:168-AGO1; Fig 2), which is consistent with reports showing that cosuppression efficiency depends on the strength of the transgene promoter (Van Blockland et al, 1994; Vaucheret et al, 1995; Que et al, 1997; Schubert et al, 2004). In addition, AGO1 cosuppression frequency using the pAGO1:168-AGO1-GUS construct, which shares 557 nt of homology with the endogenous AGO1 mRNA, was lower than that using the pAGO1:168-AGO1, p35S:168-AGO1 and Δp:168-AGO1 constructs, which all share more than 3 kb of homology with the endogenous AGO1 mRNA (Fig 2), which is consistent with reports showing that cosuppression efficiency decreases when shortening the length of homology between the transgene and the endogenous gene (Vaucheret et al, 1997; Crete & Vaucheret, 1999). Furthermore, the developmental defects of cosuppressed plants were more pronounced in pAGO1:168-AGO1-GUS plants than in pAGO1:168-AGO1, p35S:168-AGO1 and Δp:168-AGO1 plants (Fig 1), probably because the pAGO1:168-AGO1-GUS transgene, unlike the three other transgenes, does not encode a full-length functional AGO1 protein. Indeed, cosuppression degrades only a fraction of the target mRNA; therefore, it is easier to reach low levels of the target protein when only the endogenous mRNA encodes a functional protein than when both the endogenous gene and homologous transgene are contributing to the final pool of functional protein. Consistently, reduction of AGO1 mRNA accumulation below detectable levels was observed only in pAGO1:168-AGO1-GUS cosuppressed transformants (Fig 1; data not shown).
Cosuppression efficiency has been reported to vary greatly from gene to gene for reasons that are not fully understood. The frequency of AGO1 cosuppression triggered by the p35S:168-AGO1 construct (89%) was higher than the cosuppression frequencies generally reported for 35S-driven transgenes (usually 20–40%), although it was not as high as that triggered by the p35S-NIA2 construct (99%; Elmayan et al, 1998). Furthermore, the frequency of AGO1 cosuppression triggered by the pAGO1:168-AGO1 and Δp:168-AGO1 constructs was higher than that reported for other genomic or promoterless transgenes, which probably trigger cosuppression only when inserted into highly transcribed areas of the genome (Van Blockland et al, 1994; Vaucheret et al, 1995). The high frequency of AGO1 cosuppression suggests that, in addition to high transcription levels, determinants unique to AGO1 promote the triggering of AGO1 cosuppression.
miR168 complementarity modulates AGO1 cosuppression
AGO1 mRNA contains a miR168 complementarity site that is cleaved through the slicer activity of the AGO1 protein (Vaucheret et al, 2004; Baumberger & Baulcombe, 2005; Qi et al, 2005). As transgene-derived uncapped and unpolyadenylated RNAs have been shown to activate cosuppression (Gazzani et al, 2004; Luo & Chen, 2007), it is possible that miR168-guided cleavage of AGO1 mRNA, which produces uncapped and unpolyadenylated cleavage fragments, contributes to the hyper-susceptibility of AGO1 to cosuppression. To test this possibility, we engineered two constructs: pAGO1:4m-AGO1-GUS and pAGO1:Δ168-AGO1-GUS. The pAGO1:4m-AGO1-GUS construct contains the same sequence as pAGO1:168-AGO1-GUS but has silent mutations that create four additional mismatches between AGO1 mRNA and miR168, and render AGO1 resistant to miR168 regulation (Vaucheret et al, 2004). The pAGO1:Δ168-AGO1-GUS construct is expressed from the 1.5 kb AGO1 promoter and consists of the 5′ 387 nt of AGO1 mRNA translationally fused to GUS, but lacks the miR168 complementary site. Unlike the pAGO1:168-AGO1-GUS construct, the pAGO1:4m-AGO1-GUS and pAGO1: Δ168-AGO1-GUS constructs, both of which lack the miR168 complementary site, did not trigger AGO1 cosuppression (Fig 2), suggesting that miR168-directed AGO1 cleavage facilitates AGO1 cosuppression.
To assess whether the rate of miR168-guided AGO1 mRNA cleavage influences AGO1 cosuppression efficiency, constructs producing full-length AGO1 mRNAs that are either resistant or hyper-susceptible to miR168-guided cleavage were introduced into wild-type Arabidopsis plants and rdr6 mutants. The pAGO1:4m-AGO1 construct is similar to the pAGO1:168-AGO1 construct but contains silent mutations that create four additional mismatches between the AGO1 mRNA and miR168 and render AGO1 resistant to miR168 regulation (Vaucheret et al, 2004). By contrast, the pAGO1:0m-AGO1 construct repairs the three natural mismatches between AGO1 mRNA and miR168, producing an AGO1 mRNA that has an increased cleavage rate (Vaucheret et al, 2006). The frequency of AGO1 cosuppression was 50% lower in pAGO1:4m-AGO1 plants than in pAGO1:168-AGO1 transgenic plants (Fig 2). Although the frequency of cosuppression was reduced in pAGO1:4m-AGO1 plants, it was not abolished, suggesting that miR168-guided cleavage of AGO1 mRNAs arising from the native AGO1 gene is sufficient to trigger AGO1 cosuppression. Confirming the positive role of miR168-guided AGO1 mRNA cleavage during AGO1 cosuppression, the frequency of cosuppression was nearly 30% higher in pAGO1:0m-AGO1 plants than in pAGO1:168-AGO1 plants (Fig 2). In all cases, cosuppression was observed only in wild-type plants and never in rdr6 mutants.
AGO1 cosuppression correlates with AGO1 siRNA
miR168-guided AGO1 mRNA cleavage results in the production of uncapped and unpolyadenylated fragments, which could contribute to AGO1 cosuppression. However, other miRNA targets seem to be no more susceptible to cosuppression than non-targeted RNAs, suggesting that, in addition to miRNA-guided mRNA cleavage, other determinants promote the triggering of AGO1 cosuppression. In contrast to other protein-coding mRNAs targeted by miRNAs, AGO1 gives rise to 21-nt siRNAs that map downstream from, and generally in phase with, the AGO1 miR168 cleavage site (Lu et al, 2005; Axtell et al, 2006; Rajagopalan et al, 2006; Kasschau et al, 2007). More than 70% of the cloned siRNAs that are complementary to AGO1 mRNA begin with a 5′ uridine, a characteristic shared by small RNAs that associate preferentially with and function through AGO1. The production of siRNA following miR168-guided cleavage of AGO1 mRNA is reminiscent of the production of trans-acting short interfering RNA (tasiRNA) following miRNA-guided cleavage of TAS transcripts (Mallory & Bouche, 2008). The production of AGO1 siRNAs and the heightened susceptibility of AGO1 to cosuppression suggest that AGO1 is naturally subjected to siRNA-mediated self-regulation in addition to miR168 regulation, and that heightened AGO1 siRNA production in transgenic plants containing ectopic AGO1 copies could be responsible for AGO1 cosuppression. Consistent with their low cloning frequencies, we were unable to detect AGO1 siRNAs in untransformed wild-type plants by RNA gel blot analyses (Fig 3). However, both sense and antisense AGO1 siRNAs corresponding to sequences downstream from the miR168 complementary site were detected in p35S:168-AGO1 transgenic plants undergoing cosuppression (Fig 3; data not shown). AGO1 cosuppression was observed after the transformation of wild-type plants and ago7 mutants, but was never observed in the PTGS-deficient mutants rdr6, sgs3 or sde5 (Fig 3).
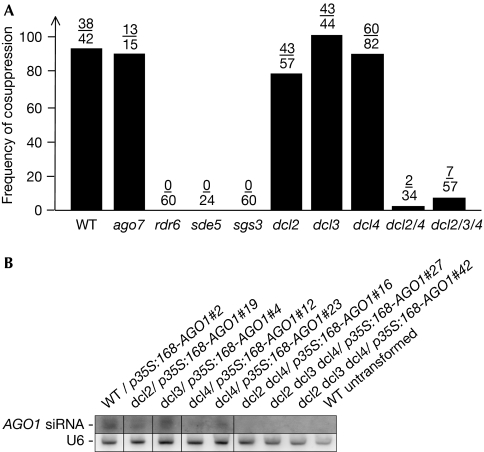
Figure 3.
AGO1 cosuppression requires SGS3, SDE5, RDR6, DCL2 and DCL4. (A) Frequencies of AGO1 cosuppression observed after the transformation of wild-type (WT) plants and the indicated mutants with p35S:168-AGO1. The fraction of plants showing the cosuppression phenotypes of AGO1 is indicated above each bar. (B) RNA gel blot analysis of AGO1 siRNAs in WT untransformed and p35S:168-AGO1-transformed WT and dcl mutant plants showing strong cosuppression of developmental defects. The accumulation of AGO1 siRNA depends on both DCL2 and DCL4. For both dcl4 and dcl2 dcl3 dcl4, two independent transformants are presented. The blot was hybridized for U6 snRNA as a control. AGO, ARGONAUTE; DCL, DICER-LIKE; RDR6, RNA DEPENDENT RNA POLYMERASE 6; SDE5, SILENCING DEFECTIVE 5; SGS3, SUPPRESSOR OF GENE SILENCING 3; siRNA, short interfering RNA; snRNA, small nuclear RNA.
DCL4 and DCL2 are required for AGO1 cosuppression
The mutants ago1, hua enhancer 1 (hen1), rdr6 and sgs3 have been isolated in genetic screens for mutants defective in sense transgene-mediated PTGS (Mourrain et al, 2000; Morel et al, 2002; Boutet et al, 2003) whereas nuclear rna polymerase d1a (nrpd1a), rdr2, rdr6, sde3, sde5 and sgs3 have been recovered in genetic screens for mutants defective in transgene/virus (amplicon)-based PTGS (Dalmay et al, 2000, 2001; Herr et al, 2005; Hernandez-Pinzon et al, 2007), indicating that neither of these two systems retrieved the entire set of PTGS components. Indeed, dcl mutants have not been recovered from these two screens. However, it has been shown that DCL2 and DCL4 act redundantly in the production of endogenous tasiRNAs, some viral siRNAs and IR transgene-derived siRNAs (Gasciolli et al, 2005; Xie et al, 2005; Blevins et al, 2006; Bouche et al, 2006; Deleris et al, 2006; Fusaro et al, 2006; Dunoyer et al, 2007). To determine the DCL that is responsible for AGO1 siRNA production, we introduced the p35S:168-AGO1 construct into dcl mutants and assayed AGO1 cosuppression frequencies and siRNA accumulation. AGO1 cosuppression was observed at high frequencies in wild-type plants and in dcl2, dcl3 and dcl4 single mutants, and AGO1 siRNAs were detectable in these mutants (Fig 3). By contrast, the frequency of AGO1 cosuppression and accumulation of AGO1 siRNAs were reduced in the dcl2 dcl4 double mutant and in the dcl2 dcl3 dcl4 triple mutant, indicating a redundant role for DCL2 and DCL4 during AGO1 siRNA production and AGO1 cosuppression, similar to that observed for tasiRNAs, viral siRNAs and IR transgene siRNAs (Gasciolli et al, 2005; Xie et al, 2005; Bouche et al, 2006; Deleris et al, 2006; Fusaro et al, 2006; Dunoyer et al, 2007). It is noted that the impact of AGO1 cosuppression was slightly more pronounced in dcl3 and dcl2 dcl3 dcl4 than in wild-type plants and dcl2 dcl4 mutants, respectively, suggesting an antagonistic role for DCL3 in cosuppression, similar to that reported for DCL3 in the pSUC-PDS IR-PTGS system (Smith et al, 2007).
Conclusions
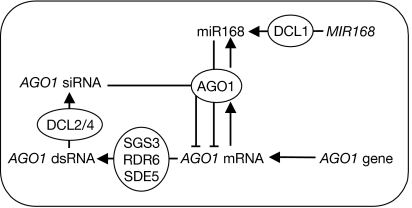
Our results show that siRNAs arising from AGO1 mRNA have the capacity to modulate the levels of AGO1 mRNA. This siRNA-mediated AGO1 mRNA degradation is miR168 cleavage dependent and requires SGS3, RDR6, SDE5 and DCL2/DCL4. Such regulation distinguishes AGO1 from other protein-coding miRNA targets, which undergo RNA degradation through the EXORIBONUCLEASE (XRN) and exosome pathways following miRNA-guided cleavage (Shen & Goodman, 2004; Souret et al, 2004; Gy et al, 2007). It remains to be tested whether the production of AGO1 siRNAs downstream from the miR168 complementary site is a special feature of miR168-directed AGO1 regulation, similar to the specific requirement for miR173- and miR390-directed regulation of TAS transcripts for the production of tasiRNAs (Montgomery et al, 2008a, 2008b). Together with AGO1-catalysed miR168-guided AGO1 mRNA cleavage (Vaucheret et al, 2004) and AGO1-mediated preferential stabilization of miR168 (Vaucheret et al, 2006), AGO1 siRNA-guided AGO1 mRNA cleavage helps AGO1 levels to be kept in check, indicating that the miRNA and siRNA pathways coordinately regulate the maintenance of AGO1 homeostasis (Fig 4).
Figure 4.
Model for the coordinated regulatory action of both the miRNA and siRNA pathways during the maintenance of AGO1 homeostasis. DCL1 processes MIR168 transcripts to produce the mature miR168, which associates with AGO1 protein and mediates cleavage of AGO1 messenger RNA (mRNA). AGO1 protein preferentially stabilizes miR168, which helps keep the levels of AGO1 mRNA, and thus the miRNA pathway, in check. In addition to miR168-directed regulation of AGO1, AGO1 mRNA also gives rise to transcripts that are substrates for SGS3, SDE5 and RDR6 and to the production of dsRNA, which is processed by DCL2 and DCL4 to produce siRNAs that regulate the level of AGO1 mRNA. AGO, ARGONAUTE; DCL, DICER-LIKE; dsRNA, double-stranded RNA; miRNA, microRNA; RDR6, RNA DEPENDENT RNA POLYMERASE 6; SDE5, SILENCING DEFECTIVE 5; SGS3, SUPPRESSOR OF GENE SILENCING 3; siRNA, short interfering RNA.
Methods
Plant material. All plants are in the Columbia ecotype. rdr6 (sgs2-1), sgs3-1, ago7 (zip-1), dcl2-1, dcl3-1 and dcl4-2 mutant alleles have been described previously (Elmayan et al, 1998; Mourrain et al, 2000; Hunter et al, 2003; Xie et al, 2004, 2005). sde5-3 corresponds to the T-DNA insertion line WiscDsLox429G09. Plants were transformed using the floral dip method (Clough & Bent, 1998). All plants were grown in standard long-day conditions (16 h light–8 h dark) at 22°C. Constructs were prepared as described in the supplementary information online.
Molecular analyses. High molecular and low molecular RNA gel blot analyses were performed as described previously (Gy et al, 2007). All RNAs were extracted from floral inflorescence at the same developmental stage. AGO1 mRNA probe was described previously (Vaucheret et al, 2006). GUS histochemical staining was performed as described previously (Elmayan et al, 2005).
Supplementary information is available at EMBO reports online (http://www.emboreports.org)
Acknowledgments
We thank David Bartel for the use of his lab facilities at the beginning of this project and for helpful discussions, the Nottingham Arabidopsis Stock Centre for the distribution of mutant seeds, and Bruno Letarnec and Herve Ferry for plant care. This study was supported by the Agence Nationale de la Recherche (ANR-06-BLAN-0203 and ANR-06-POGM-007 to H.V., and ANR-08-BLAN-0082-01 to A.C.M.).
Footnotes
The authors declare that they have no conflict of interest.
References
- Axtell MJ, Jan C, Rajagopalan R, Bartel DP (2006) A two-hit trigger for siRNA biogenesis in plants. Cell 127: 565–577 [DOI] [PubMed] [Google Scholar]
- Baumberger N, Baulcombe DC (2005) Arabidopsis ARGONAUTE1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA 102: 11928–11933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins T et al. (2006) Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res 34: 6233–6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouche N, Lauressergues D, Gasciolli V, Vaucheret H (2006) An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J 25: 3347–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutet S, Vazquez F, Liu J, Béclin C, Fagard M, Gratias A, Morel JB, Crété P, Chen X, Vaucheret H (2003) Arabidopsis HEN1: a genetic link between endogenous miRNA controlling development and siRNA controlling transgene silencing and virus resistance. Curr Biol 13: 843–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320: 1185–1190 [DOI] [PubMed] [Google Scholar]
- Chen X (2005) MicroRNA biogenesis and function in plants. FEBS Lett 579: 5923–5931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Crete P, Vaucheret H (1999) Expression and sequence requirements for nitrite reductase co-suppression. Plant Mol Biol 41: 105–114 [DOI] [PubMed] [Google Scholar]
- Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC (2000) An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101: 543–553 [DOI] [PubMed] [Google Scholar]
- Dalmay T, Horsefield R, Braunstein TH, Baulcombe DC (2001) SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J 20: 2069–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleris A, Gallego-Bartolome J, Bao J, Kasschau KD, Carrington JC, Voinnet O (2006) Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313: 68–71 [DOI] [PubMed] [Google Scholar]
- Dunoyer P, Himber C, Voinnet O (2005) DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat Genet 37: 1356–1360 [DOI] [PubMed] [Google Scholar]
- Dunoyer P, Himber C, Ruiz-Ferrer V, Alioua A, Voinnet O (2007) Intra- and intercellular RNA interference in Arabidopsis thaliana requires components of the microRNA and heterochromatic silencing pathways. Nat Genet 39: 848–856 [DOI] [PubMed] [Google Scholar]
- Elmayan T et al. (1998) Arabidopsis mutants impaired in cosuppression. Plant Cell 10: 1747–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmayan T, Proux F, Vaucheret H (2005) Arabidopsis RPA2: a genetic link among transcriptional gene silencing, DNA repair, and DNA replication. Curr Biol 15: 1919–1925 [DOI] [PubMed] [Google Scholar]
- Fusaro AF et al. (2006) RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep 7: 1168–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasciolli V, Mallory AC, Bartel DP, Vaucheret H (2005) Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr Biol 15: 1494–1500 [DOI] [PubMed] [Google Scholar]
- Gazzani S, Lawrenson T, Woodward C, Headon D, Sablowski R (2004) A link between mRNA turnover and RNA interference in Arabidopsis. Science 306: 1046–1048 [DOI] [PubMed] [Google Scholar]
- Gy I, Gasciolli V, Lauressergues D, Morel JB, Gombert J, Proux F, Proux C, Vaucheret H, Mallory AC (2007) Arabidopsis FIERY1, XRN2, and XRN3 are endogenous RNA silencing suppressors. Plant Cell 19: 3451–3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Pinzon I, Yelina NE, Schwach F, Studholme DJ, Baulcombe D, Dalmay T (2007) SDE5, the putative homologue of a human mRNA export factor, is required for transgene silencing and accumulation of trans-acting endogenous siRNA. Plant J 50: 140–148 [DOI] [PubMed] [Google Scholar]
- Herr AJ, Jensen MB, Dalmay T, Baulcombe DC (2005) RNA polymerase IV directs silencing of endogenous DNA. Science 308: 118–120 [DOI] [PubMed] [Google Scholar]
- Hunter C, Sun H, Poethig RS (2003) The Arabidopsis heterochronic gene ZIPPY is an ARGONAUTE family member. Curr Biol 13: 1734–1739 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B (2006) MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol 57: 19–53 [DOI] [PubMed] [Google Scholar]
- Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Carrington JC (2007) Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol 5: e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Tej SS, Luo S, Haudenschild CD, Meyers BC, Green PJ (2005) Elucidation of the small RNA component of the transcriptome. Science 309: 1567–1569 [DOI] [PubMed] [Google Scholar]
- Luo Z, Chen Z (2007) Improperly terminated, unpolyadenylated mRNA of sense transgenes is targeted by RDR6-mediated RNA silencing in Arabidopsis. Plant Cell 19: 943–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Bouche N (2008) MicroRNA-directed regulation: to cleave or not to cleave. Trends Plant Sci 13: 359–367 [DOI] [PubMed] [Google Scholar]
- Mallory AC, Elmayan T, Vaucheret H (2008) MicroRNA maturation and action––the expanding roles of ARGONAUTEs. Curr Opin Plant Biol 11: 560–566 [DOI] [PubMed] [Google Scholar]
- Mi S et al. (2008) Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133: 116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery TA, Howell MD, Cuperus JT, Li D, Hansen JE, Alexander AL, Chapman EJ, Fahlgren N, Allen E, Carrington JC (2008a) Specificity of ARGONAUTE7–miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133: 128–141 [DOI] [PubMed] [Google Scholar]
- Montgomery TA et al. (2008b) AGO1-miR173 complex initiates phased siRNA formation in plants. Proc Natl Acad Sci USA 105: 20055–20062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel JB, Godon C, Mourrain P, Béclin C, Boutet S, Feuerbach F, Proux F, Vaucheret H (2002) Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14: 629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain P et al. (2000) Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101: 533–542 [DOI] [PubMed] [Google Scholar]
- Qi Y, Denli AM, Hannon GJ (2005) Biochemical specialization within Arabidopsis RNA silencing pathways. Mol Cell 19: 421–428 [DOI] [PubMed] [Google Scholar]
- Que Q, Wang HY, English JJ, Jorgensen RA (1997) The frequency and degree of cosuppression by sense chalcone synthase transgenes are dependent on transgene promoter strength and are reduced by premature nonsense codons in the transgene coding sequence. Plant Cell 9: 1357–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan R, Vaucheret H, Trejo J, Bartel DP (2006) A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev 20: 3407–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP (2002) Prediction of plant microRNA targets. Cell 110: 513–520 [DOI] [PubMed] [Google Scholar]
- Schubert D, Lechtenberg B, Forsbach A, Gils M, Bahadur S, Schmidt R (2004) Silencing in Arabidopsis T-DNA transformants: the predominant role of a gene-specific RNA sensing mechanism versus position effects. Plant Cell 16: 2561–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Goodman HM (2004) Uridine addition after microRNA-directed cleavage. Science 306: 997. [DOI] [PubMed] [Google Scholar]
- Smith LM, Pontes O, Searle I, Yelina N, Yousafzai FK, Herr AJ, Pikaard CS, Baulcombe DC (2007) An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell 19: 1507–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souret FF, Kastenmayer JP, Green PJ (2004) AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol Cell 15: 173–183 [DOI] [PubMed] [Google Scholar]
- Takeda A, Iwasaki S, Watanabe T, Utsumi M, Watanabe Y (2008) The mechanism selecting the guide strand from small RNA duplexes is different among argonaute proteins. Plant Cell Physiol 49: 493–500 [DOI] [PubMed] [Google Scholar]
- Van Blokland R, Van der Geest N, Mol JNM, Kooter JM (1994) Transgene-mediated suppression of chalcone synthase expression in Petunia hybrida results from an increase in RNA turnover. Plant J 6: 861–877 [Google Scholar]
- Vaucheret H (2008) Plant ARGONAUTES. Trends Plant Sci 13: 350–358 [DOI] [PubMed] [Google Scholar]
- Vaucheret H, Palauqui JC, Elmayan T, Moffatt B (1995) Molecular and genetic analysis of nitrite reductase co-suppression in transgenic tobacco plants. Mol Gen Genet 248: 311–317 [DOI] [PubMed] [Google Scholar]
- Vaucheret H, Nussaume L, Palauqui JC, Quillere I, Elmayan T (1997) A transcriptionally active state is required for post-transcriptional silencing (cosuppression) of nitrate reductase host genes and transgenes. Plant Cell 9: 1495–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Vazquez F, Crete P, Bartel DP (2004) The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev 18: 1187–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Mallory AC, Bartel DP (2006) AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol Cell 22: 129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC (2004) Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2: E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Allen E, Wilken A, Carrington JC (2005) DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc Natl Acad Sci USA 102: 12984–12989 [DOI] [PMC free article] [PubMed] [Google Scholar]