Abstract
At least eight types of ubiquitin chain exist, and individual linkages affect distinct cellular processes. The only distinguishing feature of differently linked ubiquitin chains is their structure, as polymers of the same unit are chemically identical. Here, we have crystallized Lys 63-linked and linear ubiquitin dimers, revealing that both adopt equivalent open conformations, forming no contacts between ubiquitin molecules and thereby differing significantly from Lys 48-linked ubiquitin chains. We also examined the specificity of various deubiquitinases (DUBs) and ubiquitin-binding domains (UBDs). All analysed DUBs, except CYLD, cleave linear chains less efficiently compared with other chain types, or not at all. Likewise, UBDs can show chain specificity, and are able to select distinct linkages from a ubiquitin chain mixture. We found that the UBAN (ubiquitin binding in ABIN and NEMO) motif of NEMO (NF-κB essential modifier) binds to linear chains exclusively, whereas the NZF (Npl4 zinc finger) domain of TAB2 (TAK1 binding protein 2) is Lys 63 specific. Our results highlight remarkable specificity determinants within the ubiquitin system.
Keywords: ubiquitin linkage, deubiquitinase, ubiquitin binding domain, NF-κB signalling, TAK1/IKK/NEMO/NF-κB
Introduction
Protein ubiquitination is a post-translational modification involved in the regulation of diverse cellular processes. In addition to the well-studied roles of ubiquitin (Ub) in protein degradation, new roles of ubiquitin in cell signalling, DNA damage response and transport processes have emerged in recent years.
Ubiquitination involves the formation of an isopeptide linkage between the carboxy terminus of ubiquitin and the amino group of a substrate lysine side chain (Pickart, 2001). Importantly, ubiquitin itself has seven lysine residues, all of which can act as acceptors for further ubiquitination, generating polyubiquitin chains (Ikeda & Dikic, 2008). This ability of ubiquitin to form polymers is crucial to the versatility of the ubiquitin system. Lys 48-linked polyubiquitin targets substrate proteins for proteasomal degradation (Hershko & Ciechanover, 1998), whereas most non-proteolytic functions of ubiquitin chains are currently associated with Lys 63-linked ubiquitin polymers (Pickart & Fushman, 2004; Ikeda & Dikic, 2008). For example, Lys 63-linked polyubiquitin chains are assembled at cytokine receptor complexes upon stimulation, and are required to activate downstream kinase signalling through the TAK1 (TGF-β activated kinase 1)/IKK (inhibitor of κB kinase) pathway to nuclear factor-kappaB (NF-κB; Adhikari et al, 2007). TAK1 has been shown to bind to Lys 63-linked chains through its TAB2 (TAK1 binding protein 2) or TAB3 subunit, whereas the IKK complex contains a ubiquitin-binding domain (UBD) in its subunit NEMO. Ubiquitin binding to these kinases leads to the activation of IKK by TAK1 and subsequent NF-κB signalling (Adhikari et al, 2007).
In addition to the lysine-mediated polyubiquitin chain formation, the amino terminus of ubiquitin can be used to form polyubiquitin. In this head-to-tail linkage—hereafter referred to as a linear ubiquitin chain—a canonical peptide bond is formed between Ub1Gly 76 and Ub2Met 1. Several ubiquitin polygenes are encoded in eukaryotic cells and undergo post-translational processing to generate the cellular source of free ubiquitin monomers. However, linear linkages can also be assembled by an E3 ligase complex known as the linear ubiquitin chain assembly complex (LUBAC; Kirisako et al, 2006); however, the role of this modification is at present unclear.
Generating polymers of the same units poses the question of how proteins and enzymes recognize different linkages. Importantly, several specific UBDs and deubiquitinase (DUB) enzymes have been identified, indicating that such intrinsic specificity determinants are present. The key to understanding ubiquitin specificity lies in the structure of the ubiquitin chain itself. Crystal and nuclear magnetic resonance (NMR) structures of Lys 48-linked ubiquitin polymers have been determined (Eddins et al, 2007), whereas for Lys 63-linked chains only NMR data are available (Varadan et al, 2004); linear chains have not as yet been structurally characterized.
We crystallized linear and Lys 63-linked ubiquitin chains, and the structures reveal that both adopt highly similar open conformations. However, despite their similar structures, many DUBs and UBDs can discriminate between these two types of chains, emphasizing a remarkable specificity within the ubiquitin system.
Results And Discussion
Crystal structures of Lys 63 and linear diubiquitin
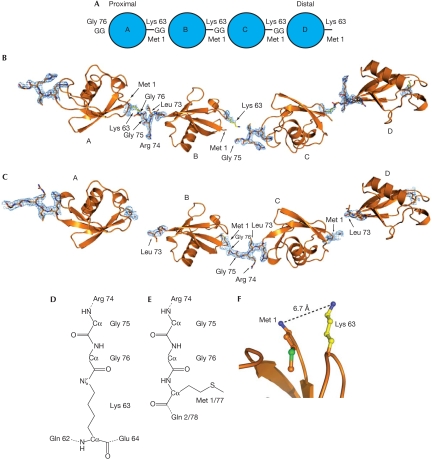
We crystallized a Lys 63-linked diubiquitin and a linear diubiquitin molecule. Both crystallized under similar conditions with the same space group and similar unit cell dimensions, and diffraction data were obtained at high resolution (1.90 and 2.25 Å; supplementary Table I online). The refined models comprise complete diubiquitin molecules, in which the distal molecule (mol) B is bound with its C terminus to Lys 63 or Met 1 of the proximal mol A (Fig 1). In the crystal lattice, neighbouring ubiquitin dimers extend this arrangement, positioning the C terminus of a symmetry-related diubiquitin (mol C in Fig 1 and in the remaining text) at the distal tip of the polymer, extending the chain virtually infinitely (Fig 1). This arrangement leads to an ambiguity as to which of the two molecules are linked—A–B or B–C. Discernible electron density linking mol A to mol B is visible for the Lys 63 linkage (Fig 1B; supplementary Fig 1A online), whereas slightly more defined density is present for the equivalent mol B to mol C in the linear chain (Fig 1C; supplementary Fig 1B online). Importantly, it is possible that the crystals represent mixtures of both arrangements. Furthermore, we could generate crystals of linear tetraubiquitin and Lys 63-linked triubiquitin that adopt the same lattice (data not shown, as these small crystals diffracted only to 4.5 Å). The linkages are partly disordered (Fig 1B,C) and show high temperature factors, consistent with high intrinsic flexibility or linkage ambiguity and therefore partial occupancy. Hence, the crystal structures represent models for extended, infinite Lys 63-linked and linear ubiquitin chains (Fig 1).
Figure 1.
Structure of Lys 63 and linear ubiquitin chains. (A) Nomenclature for polyubiquitin chains. The proximal molecule is linked through its carboxy terminus to a substrate lysine residue, or has a free carboxy-terminal diGly (GG) motif in unattached chains. (B,C) Four equivalent ubiquitin molecules, corresponding to two adjacent asymmetric units within the crystal lattice, are shown in cartoon representation. 2∣Fo∣−∣Fc∣ electron density at 1σ is drawn for the linkage residues between molecules A–B and C–D for Lys 63-linked diubiquitin, and for B–C in linear diubiquitin. (D) Chemical representation of the Lys 63 linkage. Other isopeptide linkages (for example, Lys 48 linkages) differ only in the type of neighbouring residues. (E) Representation of the peptide linkage in a linear ubiquitin chain between Gly 76 and Met 1 of the second molecule. (F) Close spatial location of Lys 63 and Met 1 (distance of 6.7 Å) allow similar conformation of linear and Lys 63-linked chains. Ub, ubiquitin.
The chemical environment of the isopeptide linkage in the Lys 63 chain differs significantly from the peptide bond between Gly 76 and Met 1 in the linear chain (Fig 1D,E). Despite these differences, the two polymers are able to adopt the same overall conformations (Fig 1B,C). Structural differences between linear and Lys 63-linked diubiquitin are confined to small shifts of the distal ubiquitin Gly 76 residue and to the immediate vicinity of the linkage in the proximal ubiquitin—that is, the Lys 63 side chain and N-terminal Met 1, respectively (Fig 1F). Only small changes in the relative disposition of the two ubiquitin monomers exist—that is, A to B and B to C—which are accountable for by the flexible linker (supplementary Fig 1C online). Importantly, the distance between ubiquitins is equivalent (supplementary Fig 1C online). Overall, linear and Lys 63 linkages can adopt equivalent conformations without restraints between monomers. This is an important feature, as it allows the most commonly used ubiquitin-binding surface, centred on Ile 44, to be aligned independently of the neighbouring ubiquitin moiety.
Comparison with other polyubiquitin structures
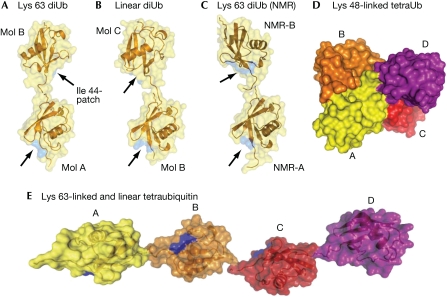
The diubiquitin crystal structures reveal that there are no contacts between individual molecules, apart from the linkage (Fig 2A,B). This is in agreement with NMR analysis of a Lys 63-linked diubiquitin, which shows no chemical shift perturbations in other than the linked residues (Varadan et al, 2004). Previous calculations of the average relative orientation from NMR data (Varadan et al, 2004) have indicated a different Lys 63-linked diubiquitin relative orientation in solution (Fig 2C) from those observed in our crystal structures. The three distinct orientations shown in Fig 2A–C indicate the lack of rotational restraints between individual ubiquitin moieties. Interestingly, monoubiquitin crystallizes in the same cubic setting as reported here (data not shown). This suggests that linear and Lys 63-linked ubiquitin chains have a high degree of flexibility and do not seem to be restrained in relative orientation between ubiquitin molecules.
Figure 2.
Similarities and differences between differentially linked ubiquitin polymers. (A–C) A semitransparent surface covers the ubiquitin molecules in cartoon representation, and the position of the hydrophobic surface patch formed by Ile 44-Val 70-Leu 8 is shown in blue on the surface, indicated by arrows. The diubiquitin molecules are aligned on the proximal ubiquitin moiety. (A) Structure of Lys 63-linked diubiquitin. (B) Diubiquitin orientations derived from the linear diubiquitin crystal structure (representing mol B/mol C in the Lys 63 structure). (C) Diubiquitin orientation derived from NMR analysis (Varadan et al, 2004; coordinates were kindly provided by D. Fushman). (D) Model of Lys 48-linked tetraubiquitin (pdb-id 2o6v; Eddins et al, 2007). (E) Model of linear or Lys 63-linked tetraubiquitin. NMR, nuclear magnetic resonance; Ub, ubiquitin.
The Lys 63-linked and linear ubiquitin structures are markedly different from Lys 48-linked ubiquitin dimers and ubiquitin tetramers, which adopt a compact conformation with extensive hydrophobic interactions at the interfaces (Eddins et al, 2007; Fig 2D). Further analysis and comparison can be found in the supplementary information online.
Deconjugation of Lys 48, Lys 63 and linear ubiquitin chains
Here, we examined various deubiquitinating enzymes from different families for their ability to hydrolyse Lys 48, Lys 63 and linear tetraubiquitin substrates. By definition, DUBs must recognize the linkage between ubiquitin molecules (or ubiquitin and substrates), and this chemical environment is markedly different between a lysine isopeptide and a peptide bond in linear chains (Fig 1D,E).
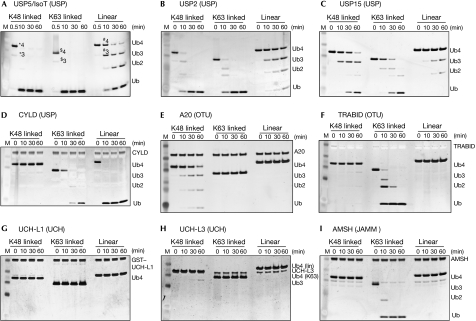
The ubiquitin specific protease (USP) family DUBs, IsoT (Fig 3A), USP2 (Fig 3B) and USP15 (Fig 3C), although cleaving both Lys 48- and Lys 63-linked chains equally well, showed significantly less activity towards linear ubiquitin chains. Another USP, the product of the cylindromatosis tumour suppressor gene CYLD, which acts as a negative regulator of NF-κB signalling and is Lys 63 specific (Komander et al, 2008), cleaved linear chains with similar if not higher activity compared with Lys 63 chains, whereas it did not hydrolyse Lys 48 chains (Fig 3D).
Figure 3.
Specificity of deubiquitinating enzymes. Time course analysis of degradation of tetraubiquitin by different DUBs was visualized by silver staining and performed as described by Komander et al (2008). (A) IsoT/USP5, (B) USP2, (C) USP15, (D) CYLD, (E) A20, (F) TRABID, (G) UCH-L1, (H) UCH-L3 and (I) AMSH. Polyubiquitin chains (n>2) run at different sizes on SDS–PAGE, labelled in (A): *4K48-Ub4, *3K48-Ub3, $4K63-Ub4, $3K63-Ub3, #4linear Ub4, #3linear Ub3. AMSH, associated molecule with the SH3 domain of STAM; DUB, deubiquitinase; GST, glutathione S-transferase; JAMM, JAB1/MPN/Mov34; M, marker; OTU, ovarian tumour; SDS–PAGE, SDS–polyacrylamide gel electrophoresis; Ub, ubiquitin; UCH, ubiquitin carboxy-terminal hydrolase; USP, ubiquitin specific protease.
The ovarian tumour (OTU) domains of A20 and TRABID showed marked specificity for particular chain types. In accordance with previous studies, we found that the A20 OTU domain hydrolysed only Lys 48 chains (Fig 3E), whereas the TRABID OTU domain showed Lys 63 specificity (Fig 3F; Komander & Barford, 2008; Tran et al, 2008). Neither OTU domain hydrolysed linear chains (Fig 3E,F).
Two members of the ubiquitin C-terminal hydrolase (UCH) family, UCH-L1 (Fig 3G) and UCH-L3 (Fig 3H), did not show discernible activity towards Lys 48, Lys 63 or linear ubiquitin chains. This is in agreement with previous structural data ( Johnston et al, 1999), as well as a recent systematic study, which showed that the active-site crossover loop in UCH-L1/L3 allows only binding to ubiquitins attached to an unfolded peptide, but not to folded substrates or polyubiquitin chains (Popp et al, 2009).
Finally, we tested the JAMM (JAB1/MPN/Mov34) metalloprotease DUB, AMSH (associated molecule with the SH3 domain of STAM; Clague & Urbe, 2006). AMSH is specific to Lys 63 chains, which it cleaved with high activity, and yet it was totally inactive towards both Lys 48 and linear ubiquitin polymers (Fig 3I). The recent structure of the JAMM domain DUB AMSH-LP in complex with Lys 63-linked diubiquitin showed that AMSH-LP exploits the open conformation of Lys 63-linked ubiquitin chains, and extends the flexible linker as much as possible. AMSH-LP forms a tight contact with the Lys 63 side chain and neighbouring residues, explaining the inability of AMSH to cleave linear and Lys 48-linked chains (Sato et al, 2008; supplementary information online).
The analysed members of the OTU, UCH and JAMM protease families failed to recognize peptide bonds within linear chains, and hence these proteins can be regarded as true isopeptide hydrolases; however, linear DUBs might exist within these families. USP DUBs show a more promiscuous activity towards differently linked chain types, but all the USP DUBs analysed here cleaved peptide bonds with lower efficiency—CYLD being the exception. To understand the specificity determinants of other DUB families, further DUB–diubiquitin complex structures are required.
The DUB specificity study also revealed another important feature of DUBs, which is that a particular DUB family is not endowed with intrinsic linkage selectivity. Within the OTU domain family, for example, highly specific members exist for Lys 48 and Lys 63 chains (Fig 3E,F). Similarly, USP domains, despite being generally more nonspecific in vitro, have evolved specific DUBs such as CYLD, which will not cleave Lys 48 chains (Fig 3D).
Binding of differentially linked ubiquitin chains to UBDs
Next, we analysed the ability of various UBDs to bind to Lys 48, Lys 63 and linear tetraubiquitin by using qualitative pull-down analysis (Fig 4). Differently linked ubiquitin tetramers have different electrophoretic mobility (Figs 3, 4), which allows easy identification of linkage types. Glutathione S-transferase–UBDs (GST–UBDs) were tested against individual chain types (Fig 4A), as well as against mixtures of tetraubiquitin chains (Fig 4B). Despite carefully adjusting the input levels by concentration measurements resulting in equivalent silver and Coomassie staining (Figs 3, 4C), none of the tested ubiquitin antibodies recognized different linkages equivalently, and Lys 48 chains consistently gave a weaker signal for the same amounts of input material (Fig 4A,B).
Figure 4.
Binding of ubiquitin tetramers to selected ubiquitin-binding domains. (A) Pull-down analysis with immobilized GST-tagged UBDs incubated with 1.5 μg tetraubiquitin of different linkages. Three lanes per linkage correspond to 5% of input tetraubiquitin, GST–UBD-bound tetraubiquitin and GST control-bound tetraubiquitin. Ponceau-stained membranes are shown in supplementary Fig 3 online. (B) Pull-down analysis as in (A), in which the different tetraubiquitins were mixed and used as inputs. The Ponceau-stained membrane shows the GST–UBDs. (C) The input samples from (B; first four lanes) were silver and Coomassie stained, which indicated the antibody does not recognize different linkages equivalently. (D) Potential mechanisms of UBD binding to differently linked polyubiquitin chains. (E) Summary of specificity for all proteins tested. Full-length TRABID is further analysed in supplementary Fig 4 online. ABIN2(FL), A2O binding inhibitor of NF-κB signalling 2 (full length); AMSH, associated molecule with the SH3 domain of STAM; CARD, caspase recruitment domain; cIAP1, cellular inhibitor of apoptosis 1; DUBs, deubiquitinases; GST, glutathione S-transferase; IB, immunoblotting; JAMM, JAB1/MPN/Mov34; MUD1, UBA domain-contining protein mud1; NZF, Npl4 zinc finger; OTU, ovarian tumour; TAB2, TAK1 binding protein 2; Ub, ubiquitin; UBA, ubiquitin associated; UBAN, ubiquitin binding in ABIN and NEMO; UBD, ubiquitin-binding domain; UCH, ubiquitin carboxy-terminal hydrolase; USP, ubiquitin specific protease.
In contrast to DUBs, UBDs rarely recognize the ubiquitin linkage, but generally interact with a hydrophobic ubiquitin surface centred on Ile 44 (Hurley et al, 2006). Owing to the similarity of Lys 63-linked and linear ubiquitin chain structures, we investigated whether UBDs that were known to bind to Lys 63 chains would also bind to linear polymers. The Lys 63-specific DUB TRABID (Fig 3F) contains three NZF (Npl4 zinc finger) zinc-finger UBDs preceding the OTU domain, which were shown to prefer Lys 63 over Lys 48 chains (Tran et al, 2008). The UBDs of TRABID recognized both Lys 63 and linear tetraubiquitin to a similar extent (Fig 4A, top panel). Furthermore, the TRABID UBDs could selectively bind to these chains from a mixture of Lys 48, Lys 63 and linear tetraubiquitin (Fig 4B). The spatial alignment of the NZF repeats might allow such binding to the conformationally similar chains (Fig 4D). Binding of linear and Lys 63 chains to the N terminus, as well as the marked specificity for Lys 63 chains of the C-terminal OTU domain of TRABID (Fig 3F), posed the question about DUB specificity of full-length TRABID. Therefore, we analysed the specificity of overexpressed full-length TRABID in human embryonic kidney 293 (HEK293) cells and found that it could hydrolyse Lys 63-linked isopeptide bonds but not the peptide bonds in linear chains (supplementary Fig 4 online). This suggests that the catalytic centre is unable to hydrolyse linear chains even if they are presented by the N terminus. Structural analysis of TRABID is likely to reveal the molecular basis for this specificity.
The C terminus of cIAP1 (cellular inhibitor of apoptosis 1) contains a UBA domain that interacts with both Lys 63-linked and linear tetraubiquitin (Fig 4A,B; Gyrd-Hansen et al, 2008). IAP molecules dimerize by virtue of their C-terminal RING domain; this could then allow linear or Lys 63-linked tetraubiquitin to interact with both UBA domains, in which the most distal and proximal ubiquitin interact with one UBA each, wrapping around the IAP dimer (Fig 4D). Such a binding mode has recently been suggested for the Cbl (casitas B-lineage lymphoma proto-oncogene)–UBA domain (Peschard et al, 2007). In support of this hypothesis, cIAP1 requires a minimum of four ubiquitin molecules for detectable binding (Gyrd-Hansen et al, 2008).
The recently described UBAN (ubiquitin binding in ABIN and NEMO) domains present in ABIN (A20-binding inhibitor of NF-κB signalling) proteins and NEMO (NF-κB essential modifier) were shown to interact with Lys 63 chains in vivo (Ea et al, 2006; Wagner et al, 2008). The pull-down analysis showed that NEMO UBAN bound exclusively to linear chains, and that full-length ABIN2 strongly preferred linear chains to Lys 63 chains (Fig 4A). In direct competition experiments, both proteins interacted only with linear tetraubiquitin, and no signal was detected for Lys 63 binding even after overexposure of the blots (Fig 4B and data not shown). While this manuscript was under revision, the linear E3 ligase LUBAC was reported to modify NEMO with linear chains, which was shown to be essential for the activation of IKK and NF-κB (Tokunaga et al, 2009).
The C-terminal NZF domain of TAB2 (Kanayama et al, 2004) showed the opposite specificity. TAB2 did not interact with linear polymers, but only with Lys 63-linked chains (Fig 4A,B). The data for TAB2, and also for NEMO and ABIN2, suggest that these UBDs recognize additional interfaces to the Ile 44 patch. We propose that these specific UBDs interact with the linkage region, in a manner similar to DUBs (Fig 4D). The co-crystal structure of NEMO bound to linear diubiquitin supports this hypothesis (Rahighi et al, 2009). Structural information will be required for the TAB2–ubiquitin interaction to understand these specificity determinants.
None of these described Lys 63-linked chain binders interacted detectably with Lys 48-linked tetraubiquitin. As a control for a Lys 48-binding UBD, the UBA domain of UBA domain-containing protein mud1 was tested and found to bind to Lys 48 chains, although not to Lys 63 or linear chains (Fig 4A,B), in accordance with published work (Trempe et al, 2005). This UBA domain inserts between the interacting Ile 44 patches within a Lys 48-linked dimer, and interacts with both ubiquitin moieties using two distinct surfaces on the UBA domain (Trempe et al, 2005; Fig 4D). It is noted that, in a manner similar to DUBs, UBA domains have evolved Lys 48- and Lys 63/linear-specific members, and several different ubiquitin-binding modes have been found for these domains (Trempe et al, 2005; Peschard et al, 2007).
In our qualitative pull-down analysis, fast off-rates might conceal binding of different types of chains; however, the binding data from individual chains are reproduced entirely when the UBDs are presented with a choice of polymers. Such direct competition emphasizes the fact that UBDs have intrinsic specificity for particular chain types, and suggests that these UBDs might be useful reagents.
Conclusions
Overall, our analysis shows that Lys 63 and linear ubiquitin chains are virtually equivalent in overall conformation, and form a structure in which individual ubiquitin moieties can be regarded as singular units that are rotationally unrestrained and highly flexible. Nevertheless, remarkable differences in specificity exist between chain types. Differential recognition and hydrolysis by DUBs can be explained by the chemically distinct isopeptide lysine linkage compared with the peptide linkage in linear chains. Differential recognition by UBDs is more difficult to rationalize, and further structural work will provide new insights into the principles of specific ubiquitin chain recognition by UBDs.
Both Lys 63 and linear chains have now been implicated in the activation of IKK and NF-κB (Adhikari et al, 2007; Tokunaga et al, 2009). Here, we have shown that the NF-κB regulator CYLD can hydrolyse both linkages efficiently and hence can inhibit both layers of pathway activation. Furthermore, the UBD analysis has shown that although the NEMO UBAN domain is highly specific for linear chains, the upstream kinase TAK1 relies on Lys 63 chains, as this is the only linkage its adaptor TAB2 can recognize.
Here, only three out of eight possible ubiquitin linkages have been analysed. New reagents—foremost differently linked ubiquitin chains—must be made available and their properties analysed to appreciate fully the intrinsic specificity of the ubiquitin system.
Methods
Ubiquitin chain synthesis and crystallization. Lys 63-linked and linear diubiquitin chains were produced and purified according to Komander et al (2008) and Reyes-Turcu et al (2008), respectively. Lys 63 diubiquitin crystals were obtained from protein concentrated to 5 mg/ml and grown after 7 days from 12% (w/v) PEG 3350, 5 mM NiCl2, 5 mM CoCl2, 5 mM CdCl2, 5 mM MgCl2 and 0.1 M HEPES (pH 7.5). Linear diubiquitin crystals were obtained from 22% PEG 3350 and 200 mM ZnAc. Before freezing in a nitrogen cryo-stream, the crystals were soaked in mother liquor containing 15% PEG 400.
In vitro deubiquitination assays. DUBs were diluted to 0.2 μg/μl in 150 mM NaCl, 25 mM Tris (pH 7.5) and 10 mM DTT, and preincubated at 23°C for 10 min. In a 30 μl reaction, 10 μl of the diluted enzyme was mixed with 3 μg of tetraubiquitin and 3 μl of 10 × DUB buffer (500 mM NaCl, 500 mM Tris (pH 7.5) and 50 mM DTT). Aliquots of 6 μl of the reaction were mixed with 6 μl 4 × LDS loading buffer (Invitrogen, Paisley, UK) at the time points indicated to stop the reaction. Samples (5 μl) were subjected to SDS–polyacrylamide gel electrophoresis analysis with subsequent silver staining using the Bio-Rad (Hemel Hempstead, UK) Silver Stain Plus kit according to the manufacturer's procedures.
Ubiquitin chain competition and pull-down assay. GST-tagged UBDs or GSTs (12.5 μg) were incubated with 20 μl glutathione sepharose 4B (GE Healthcare, Buckinghamshire, UK) for 1 h in 450 μl pull-down buffer (PDB; 150 mM NaCl, 50 mM Tris (pH 7.5), 5 mM DTT and 0.1% NP-40) and subsequently washed three times with PDB. The washed beads were incubated with 1.5 μg of individual chains, or for the competition assays with a mixture of 1 μg each of Lys 48, Lys 63 and linear tetraubiquitin in 450 μl PDB plus BSA (0.5 mg/ml), overnight at 4°C. The beads were washed five times with 500 μl PDB, mixed with 4 × LDS loading buffer and boiled for 2 min. SDS–polyacrylamide gel electrophoresis analysis using 4–12% gradient gels and a MES buffer system (Invitrogen) separated the differently linked tetraubiquitin chains. Western blotting was performed with rabbit ubiquitin antibody (Millipore, Livingston, UK; 07-375; 1:2000) and subsequent enhanced chemiluminescence (ECL) detection.
Further experimental details can be found in the supplementary information online.
Coordinates and structural factors have been submitted to the Protein Data Bank, accession numbers 2jf5 (K63-diUb) and 2w9n (linear diUb).
Supplementary information is available at EMBO reports online (http://www.emboreports.org)
Supplementary Material
Supplementary Information
Acknowledgments
We acknowledge Mark Roe for data collection, and Philip Cohen, Mariann Bienz, Sylvie Urbe, Michael Clague, Jane Endicott, Jean-Francois Trempe, Pascal Meier, Mads Gyrd-Hansen, Stefan Becker, Tom Nicholson and Paul Sheppard for constructs and reagents. D.K. was supported by a Beit Memorial Fellowship for Medical Research, and D.B. and P.O. acknowledge Cancer Research UK for financing the study. This study was supported in part by grants 5T32GM008367 and GM075426 to F.R-T., and GM30308 to K.D.W. from the National Institutes of Health.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adhikari A, Xu M, Chen ZJ (2007) Ubiquitin-mediated activation of TAK1 and IKK. Oncogene 26: 3214–3226 [DOI] [PubMed] [Google Scholar]
- Clague MJ, Urbe S (2006) Endocytosis: the DUB version. Trends Cell Biol 16: 551–559 [DOI] [PubMed] [Google Scholar]
- Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ (2006) Activation of IKK by TNFα requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell 22: 245–257 [DOI] [PubMed] [Google Scholar]
- Eddins MJ, Varadan R, Fushman D, Pickart CM, Wolberger C (2007) Crystal structure and solution NMR studies of Lys 48-linked tetraubiquitin at neutral pH. J Mol Biol 367: 204–211 [DOI] [PubMed] [Google Scholar]
- Gyrd-Hansen M et al. (2008) IAPs contain an evolutionarily conserved ubiquitin-binding domain that regulates NF-κB as well as cell survival and oncogenesis. Nat Cell Biol 10: 1309–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67: 425–479 [DOI] [PubMed] [Google Scholar]
- Hurley JH, Lee S, Prag G (2006) Ubiquitin-binding domains. Biochem J 399: 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda F, Dikic I (2008) Atypical ubiquitin chains: new molecular signals. ‘Protein Modifications: Beyond the Usual Suspects' review series. EMBO Rep 9: 536–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston SC, Riddle SM, Cohen RE, Hill CP (1999) Structural basis for the specificity of ubiquitin C-terminal hydrolases. EMBO J 18: 3877–3887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanayama A, Seth RB, Sun L, Ea CK, Hong M, Shaito A, Chiu YH, Deng L, Chen ZJ (2004) TAB2 and TAB3 activate the NF-κB pathway through binding to polyubiquitin chains. Mol Cell 15: 535–548 [DOI] [PubMed] [Google Scholar]
- Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, Sano S, Tokunaga F, Tanaka K, Iwai K (2006) A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J 25: 4877–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Barford D (2008) Structure of the A20 OTU domain and mechanistic insights into deubiquitination. Biochem J 409: 77–85 [DOI] [PubMed] [Google Scholar]
- Komander D, Lord CJ, Scheel H, Swift S, Hofmann K, Ashworth A, Barford D (2008) The structure of the CYLD USP domain explains its specificity for Lys 63-linked polyubiquitin and reveals a B box module. Mol Cell 29: 451–464 [DOI] [PubMed] [Google Scholar]
- Peschard P, Kozlov G, Lin T, Mirza IA, Berghuis AM, Lipkowitz S, Park M, Gehring K (2007) Structural basis for ubiquitin-mediated dimerization and activation of the ubiquitin protein ligase Cbl-b. Mol Cell 27: 474–485 [DOI] [PubMed] [Google Scholar]
- Pickart CM (2001) Mechanisms underlying ubiquitination. Annu Rev Biochem 70: 503–533 [DOI] [PubMed] [Google Scholar]
- Pickart CM, Fushman D (2004) Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol 8: 610–616 [DOI] [PubMed] [Google Scholar]
- Popp MW, Artavanis-Tsakonas K, Ploegh HL (2009) Substrate filtering by the active site crossover loop in UCHL3 revealed by sortagging and gain-of-function mutations. J Biol Chem 284: 3593–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahighi S et al. (2009) Specific recognition of linear ubiquitin chains by NEMO is important for NF-κB activation. Cell 136: 1098–1109 [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Shanks JR, Komander D, Wilkinson KD (2008) Recognition of polyubiquitin isoforms by the multiple ubiquitin binding modules of isopeptidase T. J Biol Chem 283: 19581–19592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Yoshikawa A, Yamagata A, Mimura H, Yamashita M, Ookata K, Nureki O, Iwai K, Komada M, Fukai S (2008) Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature 455: 358–362 [DOI] [PubMed] [Google Scholar]
- Tokunaga F et al. (2009) Involvement of linear polyubiquitylation of NEMO in NF-κB activation. Nat Cell Biol 11: 123–132 [DOI] [PubMed] [Google Scholar]
- Tran H, Hamada F, Schwarz-Romond T, Bienz M (2008) Trabid, a new positive regulator of Wnt-induced transcription with preference for binding and cleaving K63-linked ubiquitin chains. Genes Dev 22: 528–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempe JF, Brown NR, Lowe ED, Gordon C, Campbell ID, Noble ME, Endicott JA (2005) Mechanism of Lys 48-linked polyubiquitin chain recognition by the Mud1 UBA domain. EMBO J 24: 3178–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadan R, Assfalg M, Haririnia A, Raasi S, Pickart C, Fushman D (2004) Solution conformation of Lys 63-linked di-ubiquitin chain provides clues to functional diversity of polyubiquitin signaling. J Biol Chem 279: 7055–7063 [DOI] [PubMed] [Google Scholar]
- Wagner S et al. (2008) Ubiquitin binding mediates the NF-κB inhibitory potential of ABIN proteins. Oncogene 27: 3739–3745 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information