In a recent study published in this issue of EMBO reports, Jobert and colleagues made the unanticipated discovery that a portion of the U1 small-nuclear (sn)RNA in human cells is associated with the transcription factor TAF15 (TATA box-binding protein (TBP)-associated factor 15; Jobert et al, 2009). The U1 snRNA is classically known as a component of the splicing machinery, which removes introns from precursor messenger RNAs (pre-mRNAs). U1 snRNA functions within a protein–RNA complex known as the U1 snRNP, which associates with the 5′ splice site to initiate the process of splicing. In addition to U1 snRNA, the U1 snRNP consists of seven Sm proteins and three U1-specific proteins, U1-A, U1-C and U1-70K (Patel & Bellini, 2008), and its structure has been recently solved (Pomeranz Krummel et al, 2009). The U1 snRNP ultimately functions—along with the U2, U4, U5 and U6 snRNPs, and many other factors—as part of a much larger spliceosome (Nilsen, 2003).
TAF15 is a protein known to associate with a distinct population of transcription factor (TF)IID (Bertolotti et al, 1996), which is a large multiprotein complex that functions in transcriptional regulation and core-promoter recognition at mRNA genes. TFIID is composed of TBP and a series of TAFs (Thomas & Chiang, 2006). The protein sequence of TAF15 contains an RNA-recognition motif (RRM), indicating that it might be an RNA-binding protein (Bertolotti et al, 1996).
The study by Jobert et al aimed to identify cellular RNAs that associate with TAF15; therefore, the RNAs that co-immunoprecipitated with TAF15 were 3′ end-labelled. The most abundant TAF15-associated RNA was confirmed to be U1 snRNA, whereas other snRNAs such as U2 and U4 did not co-immunoprecipitate with TAF15. Single point mutations within the RRM of TAF15 decreased the level of co-immunoprecipitated U1 snRNA, indicating that TAF15 and U1 snRNA might interact directly, although more complicated models for association are still possible. Surprisingly, other protein components of the U1 snRNP, including the three U1-specific proteins and two Sm proteins, were not detected in the anti-TAF15 immunoprecipitates. Reciprocally, TAF15 did not co-immunoprecipitate with the U1-70K protein. These data are consistent with U1 snRNA being part of two distinct complexes: the well-characterized U1 snRNP and a new complex containing TAF15. Previous studies aimed at identifying proteins that co-immunoprecipitate with U1 snRNA did not detect TAF15 (Hackl et al, 1994; Hochleitner et al, 2005), probably because the amount of TAF15 bound to the total cellular U1 snRNA is small relative to the amounts of the proteins that are known to be part of the U1 snRNP.
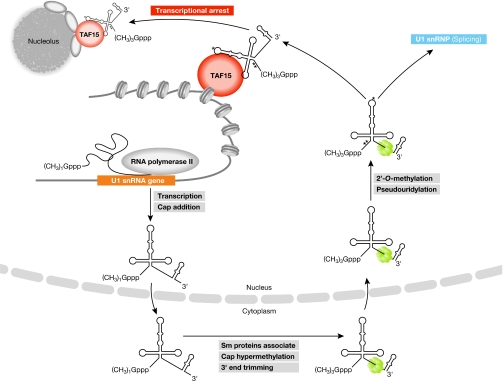
Jobert et al analysed whether the modifications that occur on U1 snRNA during the assembly of the U1 snRNP are also present on the U1 snRNA that co-purifies with TAF15. They found that the TAF15-associated U1 snRNA contained a 5′ tri-methyl guanosine cap, a 2′-O-methylated ribose on adenosine 70 and two pseudouridines, all of which are modifications found on the U1 snRNA contained in the classic U1 snRNP (Patel & Bellini, 2008). During the assembly of the U1 snRNP, cytoplasmic Sm proteins assemble around the Sm site in U1 snRNA before the complex is transported into the nucleus. When Jobert et al mutated the Sm site in U1 snRNA, the mutant was cytoplasmic, its 3′ end was not appropriately processed and it did not co-immunoprecipitate with Sm proteins. Intriguingly, the mutant U1 snRNA also did not co-immunoprecipitate with TAF15, indicating that the Sm site is important for biogenesis of the TAF15-containing complex. These data point to a model in which a portion of the U1 snRNA dissociates from snRNP proteins and associates with TAF15 after re-importation into the nucleus (Fig 1).
Figure 1.
Biogenesis of the U1 snRNA–TAF15 complex and its fate in the cell. U1 snRNA is transcribed in the nucleus and transported to the cytoplasm where the snRNA is modified, associates with Sm proteins (and other factors that are not depicted), and is re-imported into the nucleus for additional modification and assembly into the functional U1 snRNP. The mature processed and modified U1 snRNA is able to associate with TAF15 and incorporate into complexes that concentrate on chromatin through an unknown mechanism. When Pol II transcription is inhibited, the complex containing U1 snRNA and TAF15 localizes to perinucleolar cap structures. Pol II, RNA polymerase II; snRNA, small nuclear RNA; TAF15, TATA box binding protein-associated factor 15. Asterisks indicate snRNA modifications.
The subnuclear distributions of TAF15, U1 snRNA and the complex that contains these two factors were also investigated. TAF15 was found to associate predominantly with chromatin in an RNA-dependent manner. In addition, fourfold more U1 snRNA co-immunoprecipitated with TAF15 from chromatin-derived extracts than from nucleoplasmic extracts. When the nucleoplasmic and chromatin extracts were subjected to gel-filtration chromatography, TAF15 and U1-70K did not co-elute, again showing that these proteins are present in distinct complexes. U1 snRNA was present in both the TAF15-containing fraction and the U1-70K-containing fraction, although the amount of U1 snRNA was greater in the U1-70K fractions. The TAF15–U1 snRNA complex eluted at approximately 230 kDa, indicating that it probably contains other components, although this remains to be determined.
Given the enrichment of the complex on chromatin and the documented role of TAF15 in RNA polymerase II (Pol II) transcriptional regulation, the association between TAF15 and the U1 snRNA was analysed after transcriptional arrest. Under these conditions, the amount of U1 snRNA that co-immunoprecipitated with TAF15 increased, and both factors co-localized at perinucleolar caps, which are structures that form during the segregation of nucleolar components that occurs after transcriptional inhibition. Perinucleolar caps are known to accumulate many nuclear and nucleolar factors, although their function is not understood (Shav-Tal et al, 2005). Interestingly, the TAF15–U1 snRNA-containing complex localizes to the perinucleolar caps before the U1 snRNP, which is known to accumulate in caps 5–6 h after the induction of transcriptional arrest (Carmo-Fonseca et al, 1992).
Although a function for the TAF15–U1 snRNA-containing complex has yet to be elucidated, one can envisage that it would have a role in Pol II transcription. TAF15 is a component of a distinct population of TFIID and, in addition, has been shown to associate tightly with Pol II in cells and incorporate into preinitiation complexes in vitro (Bertolotti et al, 1996). U1 snRNA could be incorporated into preinitiation complexes along with TAF15, which would explain why both factors are found associated with chromatin. U1 snRNA has previously been implicated in Pol II transcriptional regulation, it interacts with the general transcription factor TFIIH and has been shown to stimulate transcription in vitro (Kwek et al, 2002); however, whether U1 snRNA influences TFIIH activity and/or transcription in cells is unknown. A stable interaction of U1 snRNA with the 5′ splice site has been shown to stimulate transcription and enhance the recruitment of general transcription factors to genes (Damgaard et al, 2008). Moreover, the DEAD (Asp–Glu–Ala–Asp) box polypeptide 17 (Ddx17) helicase, which is a known regulator of transcription, has been found associated with the U1 snRNP (Lee, 2002; Fuller-Pace & Ali, 2008). Whether the newly discovered association of U1 snRNA with TAF15 influences TFIIH activity or the functional connection between transcription and splicing awaits future studies. Apart from transcription, it is possible that association with TAF15 acts to sequester the U1 snRNA that is not contained in U1 snRNPs. The observation that U1 snRNA and TAF15 co-localize in perinucleolar caps after transcriptional inhibition is compelling; however, the function of these subnuclear structures is not understood.
The studies performed by the Tora laboratory document a potentially important new complex between a transcription factor and an RNA that functions in splicing. The discovery of the TAF15–U1 snRNA complex suggests the existence of yet undiscovered mechanisms to regulate gene expression at the level of transcription, splicing or possibly both.
References
- Bertolotti A, Lutz Y, Heard DJ, Chambon P, Tora L (1996) hTAF(II)68, a novel RNA/ssDNA-binding protein with homology to the pro-oncoproteins TLS/FUS and EWS is associated with both TFIID and RNA polymerase II. EMBO J 15: 5022–5031 [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Pepperkok R, Carvalho MT, Lamond AI (1992) Transcription-dependent colocalization of the U1, U2, U4/U6, and U5 snRNPs in coiled bodies. J Cell Biol 117: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard CK, Kahns S, Lykke-Andersen S, Nielsen AL, Jensen TH, Kjems J (2008) A 5′ splice site enhances the recruitment of basal transcription initiation factors in vivo. Mol Cell 29: 271–278 [DOI] [PubMed] [Google Scholar]
- Fuller-Pace FV, Ali S (2008) The DEAD box RNA helicases p68 (Ddx5) and p72 (Ddx17): novel transcriptional co-regulators. Biochem Soc Trans 36: 609–612 [DOI] [PubMed] [Google Scholar]
- Hackl W, Fischer U, Luhrmann R (1994) A 69-kD protein that associates reversibly with the Sm core domain of several spliceosomal snRNP species. J Cell Biol 124: 261–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochleitner EO, Kastner B, Frohlich T, Schmidt A, Luhrmann R, Arnold G, Lottspeich F (2005) Protein stoichiometry of a multiprotein complex, the human spliceosomal U1 small nuclear ribonucleoprotein: absolute quantification using isotope-coded tags and mass spectrometry. J Biol Chem 280: 2536–2542 [DOI] [PubMed] [Google Scholar]
- Jobert L, Pinzón N, Van Herreweghe E, Jády BE, Guialis A, Kiss T, Tora L (2009) Human U1 snRNA forms a new chromatin-associated snRNP with TAF15. EMBO Rep 10: 494–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwek KY, Murphy S, Furger A, Thomas B, O'Gorman W, Kimura H, Proudfoot NJ, Akoulitchev A (2002) U1 snRNA associates with TFIIH and regulates transcriptional initiation. Nat Struct Biol 9: 800–805 [DOI] [PubMed] [Google Scholar]
- Lee C-G (2002) RH70, a bidirectional RNA helicase, co-purifies with U1snRNP. J Biol Chem 277: 39679–39683 [DOI] [PubMed] [Google Scholar]
- Nilsen TW (2003) The spliceosome: the most complex macromolecular machine in the cell? Bioessays 25: 1147–1149 [DOI] [PubMed] [Google Scholar]
- Patel SB, Bellini M (2008) The assembly of a spliceosomal small nuclear ribonucleoprotein particle. Nucleic Acids Res 36: 6482–6493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeranz Krummel DA, Oubridge C, Leung AK, Li J, Nagai K (2009) Crystal structure of human spliceosomal U1 snRNP at 5.5 Å resolution. Nature 458: 475–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav-Tal Y, Blechman J, Darzacq X, Montagna C, Dye BT, Patton JG, Singer RH, Zipori D (2005) Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol Biol Cell 16: 2395–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MC, Chiang CM (2006) The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol 41: 105–178 [DOI] [PubMed] [Google Scholar]