Abstract
Eukaryotic ribosome biogenesis and translation are linked processes that limit the rate of cell growth. Although ribosome biogenesis and translation are mainly controlled by distinct factors, eukaryotic initiation factor 6 (eIF6) has been found to regulate both processes. eIF6 is a necessary protein with a unique anti-association activity, which prevents the interaction of 40S ribosomal subunits with 60S subunits through its binding to 60S ribosomes. In the nucleolus, eIF6 is a component of the pre-ribosomal particles and is required for the biogenesis of 60S subunits, whereas in the cytoplasm it mediates translation downstream from growth factors. The translational activity of eIF6 could be due to its anti-association properties, which are regulated by post-translational modifications; whether this anti-association activity is required for the biogenesis and nuclear export of ribosomes is unknown. eIF6 is necessary for tissue-specific growth and oncogene-driven transformation, and could be a new rate-limiting step for the initiation of translation.
Keywords: RACK1, ribosomal biogenesis, translation
Glossary
Abce1 ABC family protein E1
EF-2 elongation factor-2
Efl1 elongation factor-like 1 protein
eIF6 eukaryotic initiation factor 6
GABP GA-binding protein
Hcr1 high copy suppressor of RPG1
Met-tRNAi initiator methionyl-transfer RNA
miRNA microRNA
Mnk mitogen-activated protein kinase (MAPK)-interacting kinase
mRNA messenger RNA
mTORC1 mammalian target of rapamycin complex 1
PIC preinitiation complex
PKC protein kinase C
pre-40S 40S precursor particle
pre-60S 60S precursor particle
pre-rRNA precursor ribosomal RNA
RACK1 receptor for active C kinase 1
RISC RNA-induced silencing complex
Rli1 RNase L inhibitor 1
rRNA ribosomal RNA
S6K1 S6 kinase 1
SBDS Shwachman–Bodian–Diamond syndrome
Sdo1 yeast orthologue of Shwachman–Bodian–Diamond syndrome protein
siRNA small-interfering RNA
snoRNA small nucleolar RNA
Tif6 translation initiation factor 6
tRNA transfer RNA
Introduction
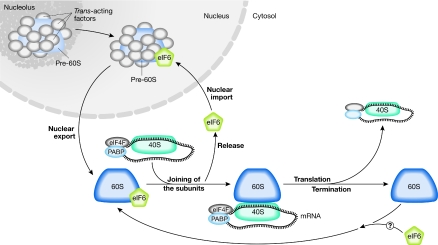
Cell growth and cell-cycle progression require the duplication of the ribosomal apparatus and the coordinated translation of specific mRNAs. In cycling cells, up to 50% of the cellular transcription machinery is estimated to be used for the synthesis of rRNA, ribosomal proteins and trans-acting factors of ribosome biogenesis (Warner, 1999). More than 600 gene products are involved in ribosome biogenesis, and at least 67 of them are necessary for the biogenesis of the large subunit alone (http://www.yeastgenome.org/cgi-bin/GO/goTerm.pl?goid=42254). After the ribosomes have been synthesized, both the rate of translation and which mRNAs are translated are tightly regulated in the cytoplasm, thereby shaping the gene-expression profile of a given cell (Sonenberg & Hinnebusch, 2009). Most of the genes that have been identified in the context of ribosome biogenesis do not have a role in translation and vice versa. However, a few yeast genes—such as Hcr1/eIF3j (Valasek et al, 2001) and Rli1/Abce1 (Dong et al, 2004; Kispal et al, 2005)—seem to have a dual role, acting both as initiation factors (IFs) and in the maturation of ribosomal subunits. Among the genes necessary for both ribosome biogenesis and translation is eIF6, which could mediate a continuum between the maturation of the large 60S subunit in the nucleus and translation in the cytoplasm (Fig 1). Furthermore, eIF6 is rate limiting for cell growth and could mediate a new regulatory step in the initiation of translation downstream from growth factors.
Figure 1.
eIF6 in ribosome biogenesis and translational control. In the nucleolus, eIF6 associates with immature large ribosomal subunits (pre-60S) and other regulatory proteins (trans-acting factors). eIF6 is found during pre-60S subunit maturation in the nucleoplasm and is exported to the cytosol. Here, eIF6 release allows the 60S to join the 40S subunit, and the active, translating 80S complex is formed. eIF6 shuttling between the nucleus and cytoplasm allows the proper formation of pre-60S. Whether eIF6 joins the 60S and prevents its premature association with 40S after the termination of translation is unknown. eIF6, eukaryotic initiation factor 6; PABP, polyA-binding protein.
eIF6: a unique and tightly regulated protein
The primary sequence of eIF6 was elucidated by expression cloning (Si et al, 1997), and was found to encode a protein of 245 amino acids that is 77% identical between yeast and humans. Eubacteria, in which transcription and translation are coupled, have no eIF6 homologues. Genomic sequencing projects have subsequently shown that eIF6 is a single gene in all species studied, with the exception of Arabidopsis. The presence of eIF6 pseudogenes has not been reported and although truncated isoforms of eIF6 are present in some nucleotide databases, they have not been observed at the protein level. As such, a strong pressure seems to exist against eIF6 duplication and/or evolution.
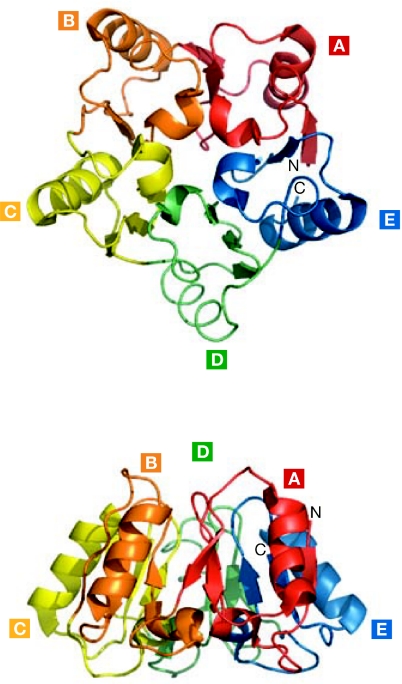
Intriguingly, the primary sequence of eIF6 is evolutionarily unique and has no conserved motifs, which is consistent with the fact that it folds into a unique structure in archaebacteria and Saccharomyces cerevisiae (Fig 2). According to the X-ray data (Groft et al, 2000), eIF6 has a cyclic fold that was named pentein because it is formed by five stretches of quasi-identical α/β-subdomains arrayed around a fivefold axis of pseudosymmetry. A structure-based sequence alignment of all five subdomains of aIF6—the archaebacteria eIF6 orthologue—shows no intramolecular similarity, suggesting that each is derived from the evolution of five independent genes. The structure has a cavity that contains 16 well-ordered water molecules with a limited degree of motility (Groft et al, 2000). Additionally, the semiconserved carboxy-terminal tail was proposed as a candidate region for eIF6 regulation due to its flexibility. In summary, eIF6 is a rigid, uniquely folded protein, the structure of which still retains mysteries regarding its mechanism of action.
Figure 2.
Unique structure of eIF6. The eIF6 protein is highly conserved and has a unique star-like structure known as pentein, which is formed by five quasi-identical α/β-subdomains (A–E) that enclose a cavity filled by 16 ordered water molecules (Groft et al, 2000). Here we show top and side views of the Saccharomyces cerevisiae structure, in which the cavity is closed. The structure of eIF6 is relatively rigid and has no structural homologue. The S. cerevisiae eIF6 Protein Data Bank access code is 1G62. eIF6, eukaryotic initiation factor 6.
Several studies have characterized the steady-state levels of eIF6 in metazoan tissues and shown that the protein is expressed at varying degrees; high levels of eIF6 can be detected in the brain and epithelia, and low levels in muscle. There is variability even within the same organ; eIF6 is strongly expressed in stem cells and cycling cells, but undetectable in some postmitotic cells (Donadini et al, 2001). The expression of eIF6 can be induced in response to different stimuli, such as a rupture of the epithelial barrier (Wood et al, 1999), mast-cell activation (Cho et al, 1998) and T-cell activation (Biffo et al, 1997), implying that tissues with a high growth demand have a rapid and sustained increase in eIF6 expression. The regulation of eIF6 expression at a molecular level is incompletely understood. The eIF6 gene is at least partly regulated by GABP, which is a global regulator of ribosomal protein transcription (Donadini et al, 2006), and could also be regulated by dynamic chromatin changes (Vreugde et al, 2006). In addition, the mammalian TORC1 complex integrates signals elicited by nutrients and growth factors to regulate many processes, including ribosome biogenesis and translation (Mayer & Grummt, 2006), and in mammals the myc oncogene also seems to regulate the transcriptional activation of genes associated with ribosomal homeostasis (White, 2008). However, neither rapamycin—an effective inhibitor of mTORC1 activity—nor Myc overexpression leads to a rapid change in eIF6 levels (A.M. & A.B., unpublished data). Therefore, a more complete understanding of the factors that regulate eIF6 gene expression is necessary, especially considering the important role of eIF6 in cell growth. The regulation of eIF6 levels has not been linked to post-transcriptional events, such as translational efficiency or protein stability. We speculate that eIF6 gene expression is under the control of both global changes that induce rapid cell proliferation and additional tissue-specific elements. However, with the exception of the involvement of GABP, the events that lead to the induction of eIF6 gene expression in response to different stimuli are still unclear.
eIF6 is necessary for ribosome biogenesis
The cloning of the eIF6 gene allowed the first series of genetic studies, which showed that eIF6 is required for ribosome biogenesis. Ribosome biogenesis is a complex process that involves several trans-acting factors and ultimately leads to the production of mature ribosomal subunits—40S and 60S in eukaryotic cells. Most of the ribosomal assembly occurs in the nucleolus, and leads to the rapid export of the 40S small subunit and the delayed export of the large 60S subunit. However, the final steps of ribosomal maturation are cytoplasmic (Henras et al, 2008). Briefly, the same rRNA precursor—which is known as 35S in S. cerevisiae and 47S in mammals—is transcribed by RNA polymerase I and processed into smaller fragments that will generate both the 18S rRNA that is incorporated into 40S subunits, and the 25S and 5.8S rRNAs that are incorporated into 60S subunits. A third rRNA, 5S, is transcribed by RNA polymerase III, and is incorporated into 60S subunits. The pre-rRNAs are subject to uridine isomerization, which forms pseudouridine, and to methylation inside a common pre-ribosomal particle known as 90S. The processing of rRNA requires snoRNAs, which act as guides for the modifying enzymes, and essential exonucleases and endonucleases that cleave the pre-rRNA, leading to the separation of pre-40S particles from pre-60S particles. The subsequent maturation of the pre-ribosomal particles takes place in the cytoplasm and requires trans-acting factors, which include nucleases, RNA helicases and several proteins of unknown function (Fatica & Tollervey, 2002).
In this context, three independent studies have shown that the deletion of the yeast eIF6 homologue, Tif6, leads to a loss of 60S ribosomal subunits that can be rescued by the ectopic expression of human eIF6 (Sanvito et al, 1999; Si & Maitra, 1999; Wood et al, 1999). These genetic observations were accompanied by biochemical and morphological evidence, which identified eIF6 in molecular complexes ranging from the 60S pre-ribosome to the almost mature 60S subunit (Fatica & Tollervey, 2002; Volta et al, 2005). In agreement with these findings, a pool of eIF6 was found to localize to the nucleolus of both mammalian and yeast cells (Lebreton et al, 2006; Sanvito et al, 1999). How does eIF6 regulate 60S biogenesis at a molecular level? rRNA pulse-chase experiments have shown that yeast cells depleted of Tif6 have defective pre-rRNA processing, which reduces the formation of mature 25S and 5.8S rRNAs relative to 18S rRNA (Basu et al, 2001). Therefore, eIF6 might act in the biogenesis of the 60S subunit, rather than in its stabilization.
Surprisingly, eIF6 has also been shown to be active in ribosome biogenesis. Shwachman–Bodian–Diamond syndrome—which is characterized by bone-marrow failure and a predisposition to leukaemia—is caused by a deficiency in the conserved SBDS protein. Mutations of the yeast orthologue of SBDS, Sdo1, result in an impaired biogenesis of the 60S subunit that is rescued by gain-of-function Tif6 mutants (Menne et al, 2007). It therefore appears that Tif6 can act as a controller of growth in the pathway of ribosome biogenesis and export, raising interesting questions about the relationship between gain-of-function mutations and their mechanistic role.
Intriguingly, a role for eIF6 in mammalian ribosome biogenesis has not been formally shown. In mammalian cells, eIF6 can be downregulated by using siRNA until only 25% of wild-type levels are expressed. In these conditions, ribosomal biogenesis is still normal, but only the cytoplasmic pool of eIF6 is reduced using this technology, whereas nucleolar eIF6 is totally retained (Gandin et al, 2008). We consider these findings to provide evidence that, in mammalian cells, a minor pool of eIF6 is sufficient for ribosome biogenesis. Mammalian eIF6 is probably needed for ribosome biogenesis given that its total depletion results in lethality before embryo implantation (Gandin et al, 2008), human eIF6 rescues yeast Tif6 mutations (Sanvito et al, 1999; Si & Maitra, 1999; Wood et al, 1999) and mammalian eIF6 is nucleolar (Lam et al, 2005).
eIF6 as an anti-association factor
Gene expression is shaped at many levels, including translation of mRNAs into proteins. Translation can be divided into four phases: initiation, elongation, termination and recycling. Most translational control is regulated during initiation, which requires the interplay of at least 30 gene products that encode IFs and miRNAs, and cis sequences in the mRNA (Sonenberg & Hinnebusch, 2009). During initiation, the small 40S ribosomal subunit associates with the ternary complex formed by eIF2–GTP–Met-tRNAi, leading to the formation of the 43S PIC, which then associates with the eIF4F complex and mRNA, thereby constituting the 48S initiation complex. The 48S complex scans to find the first initiation codon (ATG), and triggers the recruitment of the large 60S subunit, leading to the formation of an active 80S complex, which translates mRNA and elongates the peptide (Sonenberg & Dever, 2003). At a stop codon, the 80S ribosomes terminate translation, fall off from the mRNA and are recycled to start a new round of translation. The 40S ribosomal subunits have been long known to bind free 60S subunits in the absence of an mRNA, leading to an inactive 80S particle that needs to be dissociated into free subunits to allow the binding of mRNA to the 40S. This process is energy consuming and unproductive and, therefore, factors that inhibit the association of the 40S and 60S ribosomal subunits in the absence of mRNA were thought to exist. In this regard, eIF6 was initially identified on the basis of its anti-association activity in wheat germ (Russell & Spremulli, 1979) and calf liver (Valenzuela et al, 1982), where it binds to the 60S subunit and prevents its association with the 40S subunit. Notably, eIF6 cannot dissociate preformed 80S complexes. These early data indicated that eIF6 might be important in keeping ribosomes dissociated—either after their biogenesis or during their recycling from translated mRNA—rather than in stimulating their dissociation. The site of aIF6 binding to 60S has been recently mapped to the 40S–60S interface, which is consistent with this interpretation (Benelli et al, 2009). Furthermore, although eIF6 is dispensable for translation in vitro, Russell & Spremulli showed that low concentrations of eIF6 have a slight stimulatory effect on translation, whereas higher concentrations inhibit it (Russell & Spremulli, 1979). This finding indicates that the anti-association activity of eIF6 must be regulated.
The role of eIF6 in translation
The yeast experiments ruled out a role of eIF6 in translation. However, yeast and mammalian eIF6 have several properties (Table 1). Among them is the mainly cytoplasmic localization of mammalian eIF6, which points to a potential role in the control of translation. Two studies have underscored a role for eIF6 in mammalian translation: one suggests that eIF6 can be recruited by the miRNA machinery (Chendrimada et al, 2007), and the other that eIF6 expression is controlled by growth factors and/or mitogens, and positively regulates translation (Gandin et al, 2008).
Table 1.
Comparison of eIF6 features in yeast and mammals
| Properties | Yeast | Mammalian |
|---|---|---|
| Essential | Yes | Yes |
| Nucleolar localization | Yes | Yes |
| Cytoplasmic localization | A minor pool | Up to 70% |
| Function in ribosome biogenesis | Yes, predominant | Likely, not demonstrated |
| Anti-association activity | Yes | Yes |
| Role in translation | No | Yes |
| Expression | Constitutive | Broad variation among cells and tissues |
| Phosphorylation | Yes, on Ser 174 | Yes, on at least Ser 174 and Ser 235 |
| Interaction with Asc1/RACK1 | Possible, it canpurify with Asc1 | Yes, direct |
Asc1, Cyp1 absence of growth suppressor; eIF6, eukaryotic initiation factor 6; RACK1, receptor for active C kinase 1.
miRNA-dependent translational repression is a powerful way to regulate gene expression. Most studies show that a crucial step in miRNA-dependent repression is cap-complex formation, which occurs before 80S-complex formation (Filipowicz et al, 2008). However, it has also been reported that miRNA can act downstream from and repress the formation of the 80S complex (Wang et al, 2008). As eIF6 can repress 80S formation, it is tempting to speculate that its recruitment by the miRNAs could mediate miRNA-regulated translational repression. Indeed, Chendrimada and colleagues showed that eIF6 regulates miRNA-mediated translational repression in mammalian HeLa cells and during the development of Caenorhabditis elegans. Furthermore, eIF6 was reported to associate with the RISC complex (Chendrimada et al, 2007). However, this clear-cut mechanism has been challenged by independent experiments that failed to detect eIF6 in the RISC complex (Hock et al, 2007) and observed that the downregulation of eIF6 in Drosophila does not affect miRNA-based repression (Eulalio et al, 2008). From these data we can arrive at two alternative conclusions: either eIF6 mediates miRNA-translational repression only in specific cellular contexts; or the effects of eIF6 on translational repression by miRNA in C. elegans and HeLa cells were due to indirect effects on translation and growth. It will be crucial to perform in vitro reconstituted assays with active eIF6 to further address this issue.
A positive role for eIF6 in global translation is supported by results obtained in knockout mice (Gandin et al, 2008). eIF6-null mice are embryonic lethal; however, heterozygous mice are viable and have 50% of the eIF6 protein. Intriguingly, this reduction in eIF6 occurs only in the cytoplasmic pool, whereas nuclear levels are normal, which leads to proper biogenesis of the 60S ribosomal subunit. However, the liver of eIF6-heterozygous mice presents an accumulation of inactive 80S complexes, and hepatocytes derived from such livers have a normal basal level of translation but cannot upregulate it in response to insulin. Impaired translation in response to insulin stimulation is also observed in primary fibroblasts and adipocytes. The expression of eIF6 is rate limiting for tissue growth, as mice haploinsufficient for eIF6 have smaller livers than wild-type animals and reduced white fat mass. The deficit in insulin-stimulated translation that occurs in eIF6+/− cells correlates with a high insulin sensitivity in tissues. Hepatocytes, adipocytes and fibroblasts from heterozygous eIF6 cells show a delayed G1-to-S phase progression but are normal in size, and have normal senescence and apoptosis (Gandin et al, 2008). Therefore, it seems that eIF6 haploinsufficiency could regulate the translation of specific mRNAs involved in cell-cycle progression, which is unexpected considering that the regulation of 60S-subunit availability should not interfere with mRNA selection.
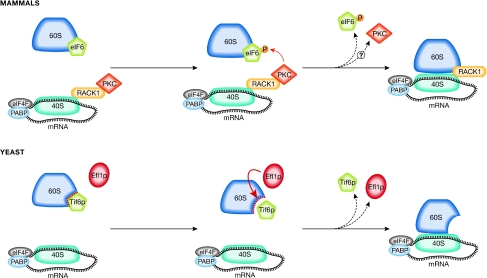
The molecular mechanism by which the depletion of eIF6 reduces translation and generates an increase of vacant 80S complexes (inactive ribosomes) is unknown; however, the mechanism whereby it is released from the 60S subunit might be important in this context. Two models have been independently proposed for the release of eIF6 from the 60S subunit (Fig 3). In one model, release occurs through interaction with a RACK1–PKC complex, which phosphorylates eIF6 and allows its release from 60S (Ceci et al, 2003). RACK1 also acts as a scaffold receptor for PKC and simultaneously binds to the small ribosomal subunit (Nilsson et al, 2004). In this model, activated PKC would translocate from endomembranes to 40S subunits containing RACK1, and activate 60S subunits through the phosphorylation and release of eIF6. This speculative model lacks genetic validation, although it is supported by the facts that PKC-induced eIF6 phosphorylation can also be observed during Xenopus development (Carotenuto et al, 2005) and the RACK1 platform can regulate translation through recruitment of PKCβII (Grosso et al, 2008). However, as PKC stimulation by agonists can lead to the activation of other kinases—including S6K1 and Mnk—that have been reported to act on the translational machinery directly (Waskiewicz et al, 1997), further work is required to establish this possibility. In yeast, an alternative model for the release of eIF6 from 60S has been proposed; an allosteric change in 60S subunits, mediated by the Efl1 protein (Senger et al, 2001), would lead to the release of eIF6. This release mechanism seems to regulate the maturation of the 60S subunit rather than mRNA translation (Fig 3). The most notable aspects of this elegant mechanism are that Efl1—which bears similarities to EF-2—is loosely associated with ribosomes and has a GTPase activity, and that point mutants of eIF6 can suppress the severe growth defect phenotype of Efl1 deletion. Therefore, we speculate that an Efl1-mediated allosteric change of 60S subunits might also be important during the process of translation initiation and not only at maturation. As for the RACK1–PKC-mediated release of eIF6, further work, particularly genetic, is needed to confirm this model.
Figure 3.
eIF6 release regulates the interaction of the two ribosomal subunits. Two models for eIF6 release have been described for mammals and yeast. In mammalian cells, this event is modulated by the RACK1–PKC complex. PKC is recruited to the 40S subunit by RACK1 and phosphorylates eIF6 on Ser 235, leading to its release from 60S. In yeast, a structural rearrangement of the large subunit is mediated by the Efl1 protein, which facilitates the release of Tif6 from 60S and its subsequent recycling to the nucleolus. Efl1, elongation factor-like 1 protein; eIF6, eukaryotic initiation factor 6; PABP, polyA-binding protein; PKC, protein kinase C; RACK1, receptor for active C kinase 1; Tif6, translation initiation factor 6.
Conclusions
The eIF6 protein is remarkably conserved, and is almost identical in species ranging from archaebacteria to humans, and yet its sequence and structure are unique. Genetic and biochemical studies have converged to show that eIF6 is necessary for ribosome biogenesis, and can control the association of the small and the large ribosomal subunits. In addition, its anti-association activity can be controlled by extracellular signalling pathways, and seems to be rate limiting for translation downstream from growth-factor and oncogene signalling. However, a number of unresolved and exciting issues remain to be elucidated (Sidebar A).
Sidebar A | In need of answers.
How is the anti-association activity of eukaryotic initiation factor 6 (eIF6) achieved mechanistically?
Does eIF6 reassociate to the 60S ribosomal subunit after the termination of translation?
How is the translational activity of eIF6 regulated?
Does eIF6 regulate global translation or that of specific classes of messenger RNA?
Does eIF6 affect tumorigenesis in mouse models and humans?
Where does mammalian eIF6 interact with the 60S subunit?
Why do Tif6 point mutants rescue 60S biogenesis in Shwachman–Bodian–Diamond syndrome mutants?
The initiation factor eIF6 seems to have a role in tumorigenesis, although its involvement is insufficiently characterized and remains puzzling. eIF6 is abundant in colon cancer (Sanvito et al, 2000) and aggressive leukaemias (Harris et al, 2004), and it was long thought that the upregulation of eIF6 was a by-product of the increased growth rate that is a characteristic of these cancers. However, recent data have indicated that a 50% reduction of eIF6 leads to a 90% reduction in oncogene-mediated transformation by activated Ras and Myc (Gandin et al, 2008). These novel data raise two unexpected questions: whether eIF6 is regulating the translation of specific mRNAs involved in tumorigenesis and whether its activity is downstream of oncogenic signalling. The fact that eIF6 haploinsufficient cells are resistant to oncogenic transformation implies that the translation of some mRNAs, specifically those controlled by growth factors, is more sensitive to eIF6 depletion than the translation of other mRNAs. This observation is in line with a new, emerging role of 60S subunits in directly (or perhaps indirectly) regulating the translation of selected mRNAs (Barna et al, 2008). The observation that eIF6 activity affects transformation raises the hope that targeting its biochemical activity could be therapeutically beneficial. The design of a high-throughput anti-association assay will make screening for compounds that increase or decrease the affinity of eIF6 binding to the 60S subunit possible.
Another important issue is understanding which signalling pathways control eIF6 activity and modulate its role in tumorigenesis, especially because eIF6 is the second clear case—in addition to eIF4F—of a rate-limiting IF that is induced downstream from growth-factor activity. Initiation proceeds through the sequential steps of the formation of the 43S PIC, the 48S PIC and the 80S complex. The formation of the 43S PIC is rate limiting and is controlled by eIF2 activity, which is regulated by eIF2α kinases (Wek et al, 2006). The formation of the 48S PIC is regulated by the cap complex, eIF4F assembly (which is made possible by mTORC1), Mnk1 and ribosomal S6 kinases (Sonenberg & Hinnebusch, 2009). In this logical flow of events, we surmise that eIF6 is the third rate-limiting controller of translation downstream from insulin and growth factors, at the level of 80S formation. The identification of the signalling pathways that activate eIF6 in vivo is a major undertaking that will shed light on how extracellular signals regulate the translational machinery.
Whether the essential role of eIF6 in ribosome biogenesis requires its anti-association activity is also unclear. One can speculate that a nucleolar anti-association factor such as eIF6 could prevent the improper association of pre-40S and pre-60S, allowing their export to the cytoplasm. The development of techniques to trace the transport of single molecules, in this case pre-ribosomes (in the presence of eIF6 blockers), will help us to determine whether this is the case. If the export of pre-40S is not affected by eIF6, then the function of eIF6 in ribosome biogenesis is probably independent of its anti-association activity. In such a scenario, the observation that the gain-of-function mutants of Tif6 are able to recover quasi-lethal mutations of two proteins in the pathway of 60S biogenesis will assume a new meaning, indicating that eIF6 is a controller of the efficiency of 60S ribosomal subunit biogenesis. It will also be important to define whether the gain-of-function mutants of Tif6 (eIF6) that suppress quasi-lethal mutations of Sdo1 and Efl1 in yeast also affect translation, ribosome biogenesis, and transformation and tumorigenesis in mammals. In this regard, if point mutants of eIF6 can also rescue the phenotype of mutations of the SBDS protein in mouse models, and if we are able to design in vitro-screening methods to search for modulators of eIF6 activity, this ancient and evolutionarily conserved gene will become a therapeutic target.
Thirty years after its biochemical isolation, we know that the anti-association activity of eIF6 is important for translational control downstream from growth factors and insulin, that eIF6 acts in the ribosome-biogenesis pathway and that gain-of-function mutations can suppress quasi-lethal mutations. Understanding whether eIF6 acts in a coordinated pathway or stands on its own, as does its unique sequence, will be a major challenge.

Annarita Miluzio

Anne Beugnet

Viviana Volta

Stefano Biffo
Acknowledgments
S.B. aknowledges P.C. Marchisio for his continuous support. We thank all the enthusiastic fellows who worked on eIF6, S. Vavassori for Fig 1 and the Italian Association for Cancer Research (AIRC), the Association for International Cancer Research (AICR), TELETHON, Fondazione Cariplo for funding.
References
- Barna M, Pusic A, Zollo O, Costa M, Kondrashov N, Rego E, Rao PH, Ruggero D (2008) Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature 456: 971–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu U, Si K, Warner JR, Maitra U (2001) The Saccharomyces cerevisiae TIF6 gene encoding translation initiation factor 6 is required for 60S ribosomal subunit biogenesis. Mol Cell Biol 21: 1453–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benelli D, Marzi S, Mancone C, Alonzi T, la Teana A, Londei P (2009) Function and ribosomal localization of aIF6, a translational regulator shared by archaea and eukarya. Nucleic Acids Res 37: 256–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffo S, Sanvito F, Costa S, Preve L, Pignatelli R, Spinardi L, Marchisio PC (1997) Isolation of a novel beta4 integrin-binding protein (p27(BBP)) highly expressed in epithelial cells. J Biol Chem 272: 30314–30321 [DOI] [PubMed] [Google Scholar]
- Carotenuto R, De Marco N, Biffo S, Wilding M, Vaccaro MC, Marchisio PC, Capriglione T, Russo GL, Campanella C (2005) Phosphorylation of p27(BBP)/eIF6 and its association with the cytoskeleton are developmentally regulated in Xenopus oogenesis. Cell Mol Life Sci 62: 1641–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceci M, Gaviraghi C, Gorrini C, Sala LA, Offenhauser N, Marchisio PC, Biffo S (2003) Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature 426: 579–584 [DOI] [PubMed] [Google Scholar]
- Chendrimada TP, Finn KJ, Ji X, Baillat D, Gregory RI, Liebhaber SA, Pasquinelli AE, Shiekhattar R (2007) MicroRNA silencing through RISC recruitment of eIF6. Nature 447: 823–828 [DOI] [PubMed] [Google Scholar]
- Cho SH, Cho JJ, Kim IS, Vliagoftis H, Metcalfe DD, Oh CK (1998) Identification and characterization of the inducible murine mast cell gene, imc-415. Biochem Biophys Res Commun 252: 123–127 [DOI] [PubMed] [Google Scholar]
- Donadini A, Giodini A, Sanvito F, Marchisio PC, Biffo S (2001) The human ITGB4BP gene is constitutively expressed in vitro, but highly modulated in vivo. Gene 266: 35–43 [DOI] [PubMed] [Google Scholar]
- Donadini A, Giacopelli F, Ravazzolo R, Gandin V, Marchisio PC, Biffo S (2006) GABP complex regulates transcription of eIF6 (p27BBP), an essential trans-acting factor in ribosome biogenesis. FEBS Lett 580: 1983–1987 [DOI] [PubMed] [Google Scholar]
- Dong J, Lai R, Nielsen K, Fekete CA, Qiu H, Hinnebusch AG (2004) The essential ATP-binding cassette protein RLI1 functions in translation by promoting preinitiation complex assembly. J Biol Chem 279: 42157–42168 [DOI] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E (2008) GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat Struct Mol Biol 15: 346–353 [DOI] [PubMed] [Google Scholar]
- Fatica A, Tollervey D (2002) Making ribosomes. Curr Opin Cell Biol 14: 313–318 [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9: 102–114 [DOI] [PubMed] [Google Scholar]
- Gandin V, Miluzio A, Barbieri AM, Beugnet A, Kiyokawa H, Marchisio PC, Biffo S (2008) Eukaryotic initiation factor 6 is rate-limiting in translation, growth and transformation. Nature 455: 684–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groft CM, Beckmann R, Sali A, Burley SK (2000) Crystal structures of ribosome anti-association factor IF6. Nat Struct Biol 7: 1156–1164 [DOI] [PubMed] [Google Scholar]
- Grosso S, Volta V, Sala LA, Vietri M, Marchisio PC, Ron D, Biffo S (2008) PKCbetaII modulates translation independently from mTOR and through RACK1. Biochem J 415: 77–85 [DOI] [PubMed] [Google Scholar]
- Harris MN, Ozpolat B, Abdi F, Gu S, Legler A, Mawuenyega KG, Tirado-Gomez M, Lopez-Berestein G, Chen X (2004) Comparative proteomic analysis of all-trans-retinoic acid treatment reveals systematic posttranscriptional control mechanisms in acute promyelocytic leukemia. Blood 104: 1314–1323 [DOI] [PubMed] [Google Scholar]
- Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y (2008) The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci 65: 2334–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock J, Weinmann L, Ender C, Rudel S, Kremmer E, Raabe M, Urlaub H, Meister G (2007) Proteomic and functional analysis of Argonaute-containing mRNA–protein complexes in human cells. EMBO Rep 8: 1052–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kispal G et al. (2005) Biogenesis of cytosolic ribosomes requires the essential iron–sulphur protein Rli1p and mitochondria. EMBO J 24: 589–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam YW, Trinkle-Mulcahy L, Lamond AI (2005) The nucleolus. J Cell Sci 118: 1335–1337 [DOI] [PubMed] [Google Scholar]
- Lebreton A, Saveanu C, Decourty L, Rain JC, Jacquier A, Fromont-Racine M (2006) A functional network involved in the recycling of nucleocytoplasmic pre-60S factors. J Cell Biol 173: 349–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Grummt I (2006) Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene 25: 6384–6391 [DOI] [PubMed] [Google Scholar]
- Menne TF, Goyenechea B, Sanchez-Puig N, Wong CC, Tonkin LM, Ancliff PJ, Brost RL, Costanzo M, Boone C, Warren AJ (2007) The Shwachman–Bodian–Diamond syndrome protein mediates translational activation of ribosomes in yeast. Nat Genet 39: 486–495 [DOI] [PubMed] [Google Scholar]
- Nilsson J, Sengupta J, Frank J, Nissen P (2004) Regulation of eukaryotic translation by the RACK1 protein: a platform for signalling molecules on the ribosome. EMBO Rep 5: 1137–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell DW, Spremulli LL (1979) Purification and characterization of a ribosome dissociation factor (eukaryotic initiation factor 6) from wheat germ. J Biol Chem 254: 8796–8800 [PubMed] [Google Scholar]
- Sanvito F, Piatti S, Villa A, Bossi M, Lucchini G, Marchisio PC, Biffo S (1999) The beta4 integrin interactor p27(BBP/eIF6) is an essential nuclear matrix protein involved in 60S ribosomal subunit assembly. J Cell Biol 144: 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanvito F, Vivoli F, Gambini S, Santambrogio G, Catena M, Viale E, Veglia F, Donadini A, Biffo S, Marchisio PC (2000) Expression of a highly conserved protein, p27BBP, during the progression of human colorectal cancer. Cancer Res 60: 510–516 [PubMed] [Google Scholar]
- Senger B, Lafontaine DL, Graindorge JS, Gadal O, Camasses A, Sanni A, Garnier JM, Breitenbach M, Hurt E, Fasiolo F (2001) The nucle(ol)ar Tif6p and Efl1p are required for a late cytoplasmic step of ribosome synthesis. Mol Cell 8: 1363–1373 [DOI] [PubMed] [Google Scholar]
- Si K, Maitra U (1999) The Saccharomyces cerevisiae homologue of mammalian translation initiation factor 6 does not function as a translation initiation factor. Mol Cell Biol 19: 1416–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si K, Chaudhuri J, Chevesich J, Maitra U (1997) Molecular cloning and functional expression of a human cDNA encoding translation initiation factor 6. Proc Natl Acad Sci USA 94: 14285–14290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Dever TE (2003) Eukaryotic translation initiation factors and regulators. Curr Opin Struct Biol 13: 56–63 [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG (2009) Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136: 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valasek L, Hasek J, Nielsen KH, Hinnebusch AG (2001) Dual function of eIF3j/Hcr1p in processing 20 S pre-rRNA and translation initiation. J Biol Chem 276: 43351–43360 [DOI] [PubMed] [Google Scholar]
- Valenzuela DM, Chaudhuri A, Maitra U (1982) Eukaryotic ribosomal subunit anti-association activity of calf liver is contained in a single polypeptide chain protein of Mr = 25,500 (eukaryotic initiation factor 6). J Biol Chem 257: 7712–7719 [PubMed] [Google Scholar]
- Volta V, Ceci M, Emery B, Bachi A, Petfalski E, Tollervey D, Linder P, Marchisio PC, Piatti S, Biffo S (2005) Sen34p depletion blocks tRNA splicing in vivo and delays rRNA processing. Biochem Biophys Res Commun 337: 89–94 [DOI] [PubMed] [Google Scholar]
- Vreugde S, Ferrai C, Miluzio A, Hauben E, Marchisio PC, Crippa MP, Bussi M, Biffo S (2006) Nuclear myosin VI enhances RNA polymerase II-dependent transcription. Mol Cell 23: 749–755 [DOI] [PubMed] [Google Scholar]
- Wang B, Yanez A, Novina CD (2008) MicroRNA-repressed mRNAs contain 40S but not 60S components. Proc Natl Acad Sci USA 105: 5343–5348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR (1999) The economics of ribosome biosynthesis in yeast. Trends Biochem Sci 24: 437–440 [DOI] [PubMed] [Google Scholar]
- Waskiewicz AJ, Flynn A, Proud CG, Cooper JA (1997) Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J 16: 1909–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek RC, Jiang HY, Anthony TG (2006) Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans 34: 7–11 [DOI] [PubMed] [Google Scholar]
- White RJ (2008) RNA polymerases I and III, non-coding RNAs and cancer. Trends Genet 24: 622–629 [DOI] [PubMed] [Google Scholar]
- Wood LC, Ashby MN, Grunfeld C, Feingold KR (1999) Cloning of murine translation initiation factor 6 and functional analysis of the homologous sequence YPR016c in Saccharomyces cerevisiae. J Biol Chem 274: 11653–11659 [DOI] [PubMed] [Google Scholar]