Synthetic biology is a new research field that comprises many different projects, approaches and definitions, which, at first glance, do not apparently have much in common. These approaches include the generation of bioengineered bacteria, artificial protocells or synthetic genomes, as well as pure in silico models of protocells or regulatory and metabolic pathways, all of which are products of synthetic biology. This Viewpoint tries to put together the various pieces by proposing five categories or subfields of synthetic biology and looking for their common denominators. The five categories are bioengineering, synthetic genomics, protocell synthetic biology, unnatural molecular biology and in silico approaches. For societal and ethical assessments of synthetic biology, it is important to consider both the differences and the similarities between the branches of synthetic biology in order to distinguish between questions that affect the field as a whole and those that are specific to individual categories.
Bioengineering, as part of synthetic biology, is the design of entirely new signalling pathways, including multiple genes and regulatory elements…
The various definitions of synthetic biology shown in Sidebar A indicate that it is not a uniform discipline. Several commentators and scientists each have individual and varying definitions or classifications (Benner & Sismour, 2005; de Lorenzo et al, 2006; Forster & Church, 2007; O'Malley et al, 2008). The five categories proposed in this article take into consideration various aspects of these earlier classifications: in particular, the scientific disciplines from which the different branches of synthetic biology emerged, the stated goals and aims that researchers are striving for, and the techniques and strategies that they use to achieve these goals (Table 1). These categories should help to make societal assessments of synthetic biology more specific and precise by clarifying which issues concern which branches.
Table 1.
Overview of the various approaches to synthetic biology
| Approach | Scientific background | Vision | Technique | Notable societal impact |
|---|---|---|---|---|
| Bioengineering | Engineering, biotechnology | Making biology an engineering discipline | Standardized and elaborated genetic engineering | Biosafety: interaction with environment Ethics: turning organisms into machines |
| Synthetic genomics | Molecular biology, chemistry | Chassis organisms | DNA synthesis | Biosecurity: synthesis of pathogens |
| Protocell synthetic biology | Chemistry, biochemistry | Synthetic cells | Chemical synthesis of a cell | Ethics: in vitro synthesis of life |
| Unnatural molecular biology | Chemistry, biochemistry, molecular biology | ‘Parallel life' | Synthesis of unnatural genomes and biological adaptation of the cell | Ethics: in vitro synthesis of life Biosafety: resistance to viruses |
| In silico synthetic biology | Computer science, engineering | Designed organisms | Computer technology | Only as applied to other approaches |
| All synthetic-biology approaches | – | New forms of life, designed life | – | Biosafety and biosecurity depending on applications Ethics: related to the impact on society and related to dealing with life |
This categorization is clearly open to dispute and certain categories could be combined; I do not try to define clear-cut fields of synthetic biology, but rather suggest a possible framework within which to arrange individual research projects. Nonetheless, these categories, or whatever other definitions and subfields one might use and consider, are all parts of one research discipline: synthetic biology, the overarching aim of which is to produce new forms of life either de novo or by redesigning existing life forms.
The category that probably attracts most scientists and research funding at present is the subfield of bioengineering. It is driven mainly by the idea of turning biotechnology into a true engineering discipline. The term ‘bioengineering', as used to describe this branch of synthetic biology, should not be confused with traditional genetic engineering, which introduces singular transgenes into the target organism. The bioengineers of synthetic biology have a fundamentally more integral view of how to modify organisms or metabolic pathways, which has been adopted from systems biology. For example, inserting a gene that encodes human insulin into bacteria in order to produce transgenic protein is classical single-gene genetic engineering. Bioengineering, as part of synthetic biology, is the design of entirely new signalling pathways, including multiple genes and regulatory elements, such as an oscillator circuit to trigger the periodic expression of green fluorescent protein (GFP) in mammalian cells (Tigges et al, 2009).
Similar to the bioengineering approach, synthetic genomics aims to generate organisms with new ‘architectures' and takes an integral or holistic view of the organism
In contrast to systems biology, which aims to understand complex biological systems such as regulatory networks, organisms or even ecosystems, bioengineering tries to design novel biological systems by using abstract and simplified metabolic and regulatory modules and other standardized components that can be combined freely into new pathways or organisms. This approach not only generates countless possibilities for new applications, but is also expected to make bioengineering more predictable and controllable than traditional biotechnology (Andrianantoandro et al, 2006; Breithaupt, 2006; Endy, 2005; Heinemann & Panke, 2006).
The idea of synthetic biology as a bioengineering field is demonstrated nicely by the annual international genetically engineered machine competition (iGEM; http://2009.igem.org/Main_Page), during which students—instructed by synthetic biologists—engineer new metabolic pathways in bacteria or eukaryotic cells based on standardized DNA elements known as BioBricks™. Although the iGEM projects have not yet lead to scientific breakthroughs—or been developed into fully elaborated projects—some examples serve to illustrate the types of questions and problems that can be tackled by synthetic biology. Among the iGEM projects are bacteria that are able to differentiate into various cell types analogous to multicellular organisms (Peking University, 2007), to do basic mathematical addition (ETH Zurich, 2006) or to work as biosensors for arsenic (University of Edinburgh, 2006), and mammalian cells that are engineered to prevent sepsis (University of Ljubljana, 2006). Some of these projects already anticipate how ‘genetically engineered machines' could be used for bioremediation or medical applications. So far, however, there are only a few practical applications of bioengineering that are close to commercial use such as the production of artemisinic acid, which is the precursor of the anti-malaria drug artemisin (Ro et al, 2006), and the production of biofuels (Atsumi & Liao, 2008; Lee et al, 2008).
While bioengineering focuses on the design and generation of new metabolic and regulatory pathways, synthetic genomics emphasizes another aspect of synthetic biology: namely, the creation of organisms with a chemically synthesized (minimal) genome. This branch of synthetic biology has been made possible by the constant improvements in DNA-synthesis technology over the past years, which now allows the generation of DNA molecules in the range of thousands of base pairs at a competitive price. The aim is to merge these molecules into full genomes and transplant them into living cells, thereby replacing the genome of the host cell and reprogramming its metabolism to undertake new tasks.
Scientists have already shown the potential of this technology by synthesizing the genomes of several viruses and using these synthetic DNA molecules to produce infectious viruses. These huge scientific and technological breakthroughs provoked the first public discussions about the dangers of this technology (Cello et al, 2002; Check, 2002, 2005; Couzin, 2002; Smith et al, 2003).
Researchers at the J. Craig Venter Institute (Rockville, MD, USA) recently published the de novo synthesis of the full genome of Mycoplasma genitalium, which comprises more than 580,000 base pairs (Gibson et al, 2008). Although, at the time of writing, this synthetic genome has not yet been transplanted into a bacterial cell, the fully synthetic Mycoplasma genome is considered to be the first step towards the synthesis of bacteria with reduced or even minimal genomes. Such organisms, if viable, could provide important information about the minimal set of genes and/or functions that are required for life.
The aim of researchers following the protocell branch of synthetic biology is to construct artificial cells in vitro
Furthermore, a minimal genome could serve as a ‘chassis genome' that might easily be expanded by the addition of genes designed for specific functions. Such ‘chassis organisms' would be optimized for the insertion of new functions, not only through dedicated insertion sites, but also because they would have fewer biological pathways that potentially interfere with the added functions compared with natural organisms. Similar to the bioengineering approach, synthetic genomics aims to generate organisms with new ‘architectures' and takes an integral or holistic view of the organism. However, in this case, the target is not the design of metabolic or regulatory pathways based on abstract standards, but the design of chassis genomes based on essential genes and other requisite DNA sequences.
The aim of researchers following the protocell branch of synthetic biology is to construct artificial cells in vitro. Such synthetic cells can be built from lipid vesicles, which contain the essential components needed to become a fully functional system. Ultimately, these synthetic cells should fulfil the necessary criteria to be considered alive: namely, to be able to self-reproduce, self-maintain and evolve (Deamer, 2005; Luisi et al, 2006b; Sole et al, 2007). Although this is the ultimate goal of the protocell approach, there are different intermediate stages that do not fulfil all the requirements of a living cell. These are lipid vesicles containing cell extracts or more specified sets of biological macromolecules and complex structures such as enzymes, nucleic acids or ribosomes, in order to fulfil a certain function—for example, liposomes that could perform specific polymerase chain reactions or synthesize a particular protein (Oberholzer et al, 1995, 1999; Luisi et al, 2006b; Sole et al, 2007).
Compared with the synthetic genomics approach, which is based on forcing a natural cell to follow the instructions encoded by the introduced synthetic genome, protocell synthetic biology goes one step further towards a fully artificial organism, as eventually not only the genome but also all the components of the cell would be synthesized in vitro. More than in any of the other approaches, synthetic biologists in this field consider their work as basic research into the minimal requirements for, and the origin of, life (Luisi, 2006). However, the protocell approach also lends itself easily towards applications; similar to other products of synthetic biology, protocells could be used for the synthesis of biopolymers and therapeutics (Pohorille & Deamer, 2002; Sole et al, 2007).
The ‘unnatural molecular biology' approach aims to synthesize novel forms of life, which are based on a new kind of molecular biology, for example, new types of nucleic acid or a different genetic code. Alternative forms of nucleic acids could be achieved by altering different components of DNA or RNA such as the bases or the backbone sugars (Benner, 2004; Benner & Sismour, 2005; Chin et al, 2003; Wang et al, 2001), to create new types of nucleotide that can be assembled into novel nucleic acids.
Modification of the standard genetic code is being tackled by substituting some codons to encode new amino acids or by inserting quadruplet codons, which would then allow the use of non-natural amino acids with novel properties in protein synthesis (Anderson et al, 2004; Benner & Sismour, 2005; Xie & Schultz, 2006). Both strategies require the enzymatic machineries of the cell to be adapted, which is a scientific and technological challenge.
Organisms with a genome based on unnatural nucleic acids or on an entirely new coding system for unnatural amino acids would form a new type of life, which would present some advantages, but also new risks. There would be no outcrossing of genes or horizontal gene transfer with natural organisms on release into the environment. Furthermore, these types of synthetic organism could be designed to depend on non-natural substances for nucleic-acid or protein synthesis, so that they could not survive in the wild if they escaped spontaneously. Conversely, if such organisms eventually managed to survive outside a controlled environment, they could have a selective advantage over natural organisms, as they would be resistant to natural viruses or other predatory life forms, which might result in an uncontrolled propagation of the synthetic organisms.
Synthetic biology In silico is closely linked to the other approaches. One of the main challenges of the four synthetic-biology approaches discussed above is the establishment of complicated designs, whether these are metabolic pathways, basic cellular functions or chassis genomes. Similar to systems biology, synthetic biology therefore has a strong in silico branch that seeks to establish computational models for the design of standard biological components or synthetic circuits (Banga, 2008; Marchisio & Stelling, 2008; Simpson, 2006; Sprinzak & Elowitz, 2005); these are, so to speak, simulations of synthetic organisms. The long-term objective of in silico synthetic biology is the practical implementation of models and simulations through bioengineering or the other branches of synthetic biology (Meyer et al, 2007). However, many of the computational models of synthetic organisms so far have little or no direct reference to living organisms. For this reason, in silico synthetic biology is considered to be an independent category in this article.
The ‘unnatural molecular biology' approach aims to synthesize novel forms of life, which are based on a new kind of molecular biology…
Each of these categories can be associated with different scientific disciplines: bioengineering with biotechnology and engineering; synthetic genomics and the unnatural molecular biology approach with molecular biology and chemistry; the protocell approach with biochemistry and chemistry; and in silico synthetic biology with computer sciences. As a result of these different scientific backgrounds, each branch is characterized by a specific methodology, strategy and immediate goal. The end product of the bioengineering approach ought to be a fully controllable living organism; synthetic genomics aims to produce a simplified chassis organism; and the protocell approach would generate an artificial cell that, in contrast to the products of bioengineering, would be more autonomous than a human-controlled machine.
Given these differences, does it make sense to combine the five categories into the single research field of synthetic biology? It seems that it does, mainly because these five different approaches all contribute to the same goal of generating new forms of living organisms, albeit by addressing different aspects of life such as metabolic regulation, minimal components or biochemical composition. Moreover, the various approaches start from different methodological strategies, which results in the diversity of synthetic biology approaches described here.
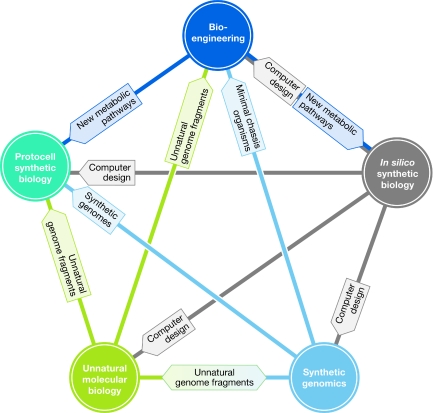
However, it is possible that the different objectives and strategies could be combined to create a completely computer-designed organism with a synthetic genome of unnatural nucleotides in a lipid vesicle with an engineered metabolism. Even before such an organism is achieved, some of these approaches can complement each other (Fig 1). The in silico approach, for example, already pervades the other categories; synthetic biology is simply not conceivable without computer-based design. Synthetic genomes, as a basic chassis to create tailor-made organisms, would of course be a powerful tool for bioengineering, whereas non-natural genomes resulting from the unnatural molecular biology approach could be used to create synthetic genomes, which again could be used to produce protocells.
Figure 1.
Schematic representation of possible connections between the categories of synthetic biology. Computer design established by the in silico approach influences all the other synthetic-biology approaches. Minimal chassis organisms synthesized by synthetic genomics could be useful for bioengineering. Synthetic genomes could be introduced into protocells and the synthetic genomics approach could be used for the synthesis of unnatural genome fragments, which could again be used in the protocell approach. Unnatural molecular biology could also be used in bioengineering independently of protocells. Metabolic pathways engineered by the bioengineering approach could be applied in protocells, and the pathways found to be required for bioengineering affect the models of the in silico approach.
Despite all the methodological and strategic differences, the idea of ‘designing life' is a common theme that underlies all aspects of synthetic biology. The term ‘designing' in this context is not only used in the sense that engineers use it, which would imply the organized assembly of prefabricated modules and standards. Rather, it refers to the design of the overall concept of an organism, which could define its metabolic pathways, genome, cellular structure or genetic code. This idea of synthesizing life is fundamentally different from, for example, the ‘creation of life' by in vitro fertilization, whereby humans so far cannot determine directly the genotype and phenotype of the organism that is generated.
Not surprisingly, synthetic biology has enormous potential implications for society. The ability to modify and create life means that scientists could tailor-make organisms to fulfil specific functions such as the production of new forms of drugs or other chemicals, the production of biofuels or hydrogen, the bioremediation of toxic chemicals and various applications in medicine. By contrast, even more than genetic engineering, synthetic biology might also raise fears about potential abuse, unintended environmental damage, health risks or ethical issues. Various stakeholders have already begun to discuss these concerns and risks, and to propose possible countermeasures.
Both the possible benefits and the possible risks of synthetic biology require broader discussion and assessment of the various ethical and societal aspects including biosafety, biosecurity, intellectual property rights, and legal and regulatory frameworks. These discussions should be guided both by the differences between the various branches of synthetic biology and by their common features.
Clearly, the various approaches imply different risks, dangers and ethical issues. A report on synthetic genomics, for example, discusses the risks related to the uncontrolled synthesis and distribution of pathogenic viruses—a biosecurity issue, which is particularly relevant for this approach (Garfinkel et al, 2007). Bioengineering, in turn, creates worries about biosafety related to the handling of such synthetic organisms. Ethical issues can also vary between different disciplines. Bioengineering might raise the question of whether it is ethical to regard and treat a living organism as a mere machine, whereas the protocell approach might raise ethical questions about creating life de novo. In addition to specific topics, issues that are particular to certain products or applications of synthetic biology require a case-by-case assessment. The broad spectrum of societal issues that seem to be raised by synthetic biology can be explained in part by the fact that this field is heterogeneous and each branch generates its own issues.
However, in some cases, the common features of synthetic biology justify a combined assessment. The fact that the products of synthetic biology are ‘alive'—and are therefore capable of reproducing and evolving—implies particular risks that are not relevant for other technological products: for example, although the release of harmful chemical substances or genetically modified organisms poses potential risks for the environment, the former do not reproduce. Similarly, unlike other technologies, synthetic biology synthesizes or substantially modifies living organisms, which might require a specific ethical assessment of the whole field (Boldt & Muller, 2008).
Finally, there are many issues related to the social impact, global distribution and access to the benefits of synthetic biology that should be addressed with a focus on the individual branches, in regard to synthetic biology as a whole and in the context of emerging technologies in general.
In summary, synthetic biology is not a homogenous discipline, but a heterogeneous assembly of different fields and approaches that draw on and are inspired by various scientific disciplines. Although synthetic biologists might use different strategies, approaches and research tools, they all share the overarching aim of designing and creating new forms of life. Any assessment of synthetic biology—whether it considers ethical, legal or safety issues—needs to take into account the fact that certain questions, risks and problems are specific to each approach, whereas in other cases it is necessary to regard synthetic biology as a whole.
Sidebar A | Definitions of synthetic biology.
“Synthetic biology is (A) the design and construction of new biological parts, devices and systems, and (B) the re-design of existing, natural biological systems for useful purposes” (http://www.syntheticbiology.org). This definition applies to the categories of bioengineering, synthetic genomics and in silico synthetic biology.
“The term synthetic biology describes, rather broadly, those avenues of research, within the life sciences, interested in the synthesis of parts of biological systems, or in the construction of models of biological systems. Synthetic biology comprises (and somehow is an extension of) biomimetic chemistry, but with the additional issue of ‘systems thinking'” (Luisi et al, 2006a). This definition describes bioengineering, in silico and protocell synthetic biology.
“Synthetic biology is an increasingly high-profile area of research that can be understood as encompassing three broad approaches towards the synthesis of living systems: DNA-based device construction, genome-driven cell engineering and protocell creation. Each approach is characterized by different aims, methods and constructs, in addition to a range of positions on intellectual property and regulatory regimes” (O'Malley et al, 2008). This definition describes unnatural molecular biology, bioengineering and protocell synthetic biology.
“Synthetic biologists come in two broad classes. One uses unnatural molecules to reproduce emergent behaviours from natural biology, with the goal of creating artificial life. The other seeks interchangeable parts from natural biology to assemble into systems that function unnaturally. Either way, a synthetic goal forces scientists to cross uncharted ground to encounter and solve problems that are not easily encountered through analysis” (Benner & Sismour, 2005). This definition describes unnatural molecular biology and bioengineering.

Acknowledgments
I thank S. Panke, A. Ganguli, S. Leidel, C. Kraft, N. Möckli and I. Zemp for their helpful comments on earlier drafts of this manuscript. I am grateful to N. Biller-Andorno, M. Huppenbauer and my collaborators from the synbiosafe team—who work on the safety and ethical aspects of synthetic biology—for many stimulating discussions. This work was supported by the University Research Priority Programme (URPP) Ethics of the University of Zurich, Switzerland.
References
- Anderson JC, Wu N, Santoro SW, Lakshman V, King DS, Schultz PG (2004) An expanded genetic code with a functional quadruplet codon. Proc Natl Acad Sci USA 101: 7566–7571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrianantoandro E, Basu S, Karig DK, Weiss R (2006) Synthetic biology: new engineering rules for an emerging discipline. Mol Syst Biol 2: 2006.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsumi S, Liao JC (2008) Metabolic engineering for advanced biofuels production from Escherichia coli. Curr Opin Biotechnol 19: 414–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banga JR (2008) Optimization in computational systems biology. BMC Syst Biol 2: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benner SA (2004) Understanding nucleic acids using synthetic chemistry. Acc Chem Res 37: 784–797 [DOI] [PubMed] [Google Scholar]
- Benner SA, Sismour AM (2005) Synthetic biology. Nat Rev Genet 6: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldt J, Muller O (2008) Newtons of the leaves of grass. Nat Biotechnol 26: 387–389 [DOI] [PubMed] [Google Scholar]
- Breithaupt H (2006) The engineer's approach to biology. EMBO Rep 7: 21–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cello J, Paul AV, Wimmer E (2002) Chemical synthesis of poliovirus cDNA: generation of infectious virus in the absence of natural template. Science 297: 1016–1018 [DOI] [PubMed] [Google Scholar]
- Check E (2002) Poliovirus advance sparks fears of data curbs. Nature 418: 265. [DOI] [PubMed] [Google Scholar]
- Check E (2005) Synthetic biologists face up to security issues. Nature 436: 894–895 [DOI] [PubMed] [Google Scholar]
- Chin JW, Cropp TA, Anderson JC, Mukherji M, Zhang Z, Schultz PG (2003) An expanded eukaryotic genetic code. Science 301: 964–967 [DOI] [PubMed] [Google Scholar]
- Couzin J (2002) Bioterrorism. A call for restraint on biological data. Science 297: 749–751 [DOI] [PubMed] [Google Scholar]
- Deamer D (2005) A giant step towards artificial life? Trends Biotechnol 23: 336–338 [DOI] [PubMed] [Google Scholar]
- de Lorenzo V, Serrano L, Valencia A (2006) Synthetic biology: challenges ahead. Bioinformatics 22: 127–128 [DOI] [PubMed] [Google Scholar]
- Endy D (2005) Foundations for engineering biology. Nature 438: 449–453 [DOI] [PubMed] [Google Scholar]
- Forster AC, Church GM (2007) Synthetic biology projects in vitro. Genome Res 17: 1–6 [DOI] [PubMed] [Google Scholar]
- Garfinkel MS, Endy D, Epstein GL, Friedman RM (2007) Synthetic Genomics, Options for Governance. Rockville, MD, USA: J. Craig Venter Institute. www.jcvi.org [DOI] [PubMed] [Google Scholar]
- Gibson DG et al. (2008) Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science 319: 1215–1220 [DOI] [PubMed] [Google Scholar]
- Heinemann M, Panke S (2006) Synthetic biology-putting engineering into biology. Bioinformatics 22: 2790–2799 [DOI] [PubMed]
- Lee SK, Chou H, Ham TS, Lee TS, Keasling JD (2008) Metabolic engineering of microorganisms for biofuels production: from bugs to synthetic biology to fuels. Curr Opin Biotechnol 19: 556–563 [DOI] [PubMed] [Google Scholar]
- Luisi PL (2006) The Emergence of Life. Cambridge, UK: Cambridge University Press [Google Scholar]
- Luisi PL, Chiarabelli C, Stano P (2006a) From never born proteins to minimal living cells: two projects in synthetic biology. Orig Life Evol Biosph 36: 605–616 [DOI] [PubMed] [Google Scholar]
- Luisi PL, Ferri F, Stano P (2006b) Approaches to semi-synthetic minimal cells: a review. Die Naturwissenschaften 93: 1–13 [DOI] [PubMed] [Google Scholar]
- Marchisio MA, Stelling J (2008) Computational design of synthetic gene circuits with composable parts. Bioinformatics 24: 1903–1910 [DOI] [PubMed] [Google Scholar]
- Meyer A, Pellaux R, Panke S (2007) Bioengineering novel in vitro metabolic pathways using synthetic biology. Curr Opin Microbiol 10: 246–253 [DOI] [PubMed] [Google Scholar]
- O'Malley MA, Powell A, Davies JF, Calvert J (2008) Knowledge-making distinctions in synthetic biology. Bioessays 30: 57–65 [DOI] [PubMed] [Google Scholar]
- Oberholzer T, Albrizio M, Luisi PL (1995) Polymerase chain reaction in liposomes. Chem Biol 2: 677–682 [DOI] [PubMed] [Google Scholar]
- Oberholzer T, Nierhaus KH, Luisi PL (1999) Protein expression in liposomes. Biochem Biophys Res Commun 261: 238–241 [DOI] [PubMed] [Google Scholar]
- Pohorille A, Deamer D (2002) Artificial cells: prospects for biotechnology. Trends Biotechnol 20: 123–128 [DOI] [PubMed] [Google Scholar]
- Ro DK et al. (2006) Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 44: 940–943 [DOI] [PubMed] [Google Scholar]
- Simpson ML (2006) Cell-free synthetic biology: a bottom-up approach to discovery by design. Mol Syst Biol 2: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HO, Hutchison CA, Pfannkoch C, Venter JC (2003) Generating a synthetic genome by whole genome assembly: phiX174 bacteriophage from synthetic oligonucleotides. Proc Natl Acad Sci USA 100: 15440–15445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sole RV, Munteanu A, Rodriguez-Caso C, Macia J (2007) Synthetic protocell biology: from reproduction to computation. Philos Trans R Soc Lond B Biol Sci 362: 1727–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzak D, Elowitz MB (2005) Reconstruction of genetic circuits. Nature 438: 443–448 [DOI] [PubMed] [Google Scholar]
- Tigges M, Marquez-Lago TT, Stelling J, Fussenegger M (2009) A tunable synthetic mammalian oscillator. Nature 457: 309–312 [DOI] [PubMed] [Google Scholar]
- Wang L, Brock A, Herberich B, Schultz PG (2001) Expanding the genetic code of Escherichia coli. Science 292: 498–500 [DOI] [PubMed] [Google Scholar]
- Xie J, Schultz PG (2006) A chemical toolkit for proteins: an expanded genetic code. Nat Rev Mol Cell Biol 7: 775–782 [DOI] [PubMed] [Google Scholar]