Abstract
Background
NBS1 is a key DNA repair protein in the homologous recombination repair pathway and a signal modifier in the intra-S phase checkpoint that plays important roles in maintaining genomic stability. The NBS1 8360G>C (Glu185Gln) is one of the most commonly studied polymorphisms of the gene for their association with risk of cancers, but the results are conflicting.
Methods
We performed a meta-analysis using 16 eligible case-control studies (including 17 data sets) with a total of 9,734 patients and 10,325 controls to summarize the data on the association between the NBS1 8360G>C (E185Q) polymorphism and cancer risk.
Results
Compared with the common 8360GG genotype, the carriers of variant genotypes (i.e., 8360 GC/CC) had a 1.06-fold elevated risk of cancer (95% CI = 1.00–1.12, P = 0.05) in a dominant genetic model as estimated in a fixed effect model. However, the association was not found in an additive genetic model (CC vs GG) (odds ratio, OR = 0.98, 95% CI = 0.85–1.13, P = 0.78) nor in a recessive genetic model (CC vs GC +GG) (OR = 0.94, 95% CI = 0.82–1.07, P = 0.36). The effect of the 8360G>C (E185Q) polymorphism was further evaluated in stratification analysis. It was demonstrated that the increased risk of cancer associated with 8360G>C variant genotypes was more pronounced in the Caucasians (OR = 1.07, 95% CI = 1.01–1.14, P = 0.03).
Conclusion
Our meta-analysis suggests that the NBS1 E185Q variant genotypes (8360 GC/CC) might be associated with an increased risk of cancer, especially in Caucasians.
Background
DNA damage may increase cancer risk, and DNA double-strand breaks (DSBs) cause the most potentially serious damage to the genome. If unrepaired, DSBs may lead to genomic instability and thus cancer [1]. The repair of DSBs in human cells includes two different pathways, homologous recombination repair (HR) and non-homologous end joining (NHEJ) pathways [2]. The initial step in both pathways is the recognition and signaling of DNA DSBs by a protein complex containing Nijmegen breakage syndrome 1 (NBS1), meiotic recombination 11 homologue (MRE11), and human RAD50 homologue (RAD50) proteins [3]. NBS1 plays an important role as a sensor in repairing the DSBs and activates the cell-cycle checkpoint signaling; it also directly binds to the phosphorylated histone H2AX that is located around DSBs, participating in maintaining genomic stability [4], and prevents cells from telomeric fusion [2,5]. A markedly impaired DSB repair was observed in cells from patients with Nijmegen breakage syndrome [6], in which cells are characteristic of chromosome instability and sensitivity to DSB-causative agents [7,8].
The NBS1 gene is located on human chromosome 8q21 and codes for a protein termed nibrin (754-amino acid protein) [9-11]. Some NBS1 mutations and polymorphisms have been reported to be associated with risk of several cancers, including cancers of the breast, lung, bladder, ovaries, non-Hodgkin lymphoma, malignant melanoma and basal cell carcinoma of the skin [12-18]. A homozygous 5-bp deletion in exon 6 (657del5) has been reported to be associated with an elevated risk of breast cancer in Polish and Russian populations [15,19]. However, the 657del5 variant appears to be a Slavic origin with a low frequency of approximate 0.5% in Eastern Europe populations and even lower in other ethnic groups. According to the Environmental Genome Project (EGP) SNP database of the (NIEHS) (http://egp.gs.washington.edu, accessed on March 1, 2008), 249 single nucleotide polymorphisms (SNPs) are reported, of which 84 are common polymorphisms (with a minor allele frequency > 5%). Among these SNPs, the 8360G>C (Glu185Gln, E185Q, rs1805794) is one of the most commonly studied polymorphisms. However, the results of studies on association between the 8360G>C (E185Q) polymorphism and the risk of cancers are conflicting [1,20,21]. To summarize the published data, we performed a meta-analysis from all eligible case-control studies to assess the association between the NBS1 E185Q polymorphism and cancer risk.
Methods
Bioinformatics Analysis
Based on the resequencing information about the NBS1 gene provided by the NIEHS Environmental Genome Project http://egp.gs.washington.edu/data/nbs1/, we calculated the D' value and r2 coefficient by linkage disequilibrium (LD) analysis. In addition, we further analyzed SNPs in LD with 8360G>C in a 5-Mb region on chromosome 8 via the HapMap SNP database (release #36, http://www.hapmap.org/, March 1, 2008) that have genotypes for the CEPH trios with the ssSNPer web interface http://gump.qimr.edu.au/general/daleN/ssSNPer/[22].
Literature search strategy for identification of the studies
We carried out a literature search in the PubMed and SciFinder Scholar (CA web version) database (between January 2000 and February 2008) to identify all articles that investigated the association between the NBS1 E185Q polymorphism and cancer risk in all ethnic groups, using the following keywords and subject terms: 'NBS1,' 'cancer,' and 'polymorphism.' We evaluated the titles and abstracts of all relevant publications first but excluded abstracts, case reports, editorials, and review articles. Studies included in the current meta-analysis had to meet the following criteria: (a) The study used a case-control study design; (b) the report described cancer diagnoses and the sources of cases and controls; (c) the report had available genotype frequency; (d) the authors offered the size of the samples, odds ratios (ORs) and their 95% confidence intervals (CIs); (e) the definition of the exposure or risk genotypes was similar in all reports; and (f) the methods of data collection and analysis were statistically acceptable.
Data extraction
Data were collected on the NBS1 E185Q genotype for studies of different types of cancers. The first author, a year of publication, country, ethnicity of the study population, and the number of cases and controls and allele frequency were also described.
Methods for quantitative synthesis
The Hardy-Weinberg equilibrium (p2 + 2pq + q2 = 1, where p is the frequency of the variant allele and q = 1 - p), was tested by goodness-of-fit Chi-square tests to compare the observed genotype frequencies with expected genotype frequencies in cancer-free controls for all studies. The selections of published studies used for meta-analysis were further evaluated in sensitivity analyses. Odds ratio (OR) and 95% confident interval (CI) in each case-control study was used to assess the strength of association between the NBS1 8360G>C (E185Q) genotypes and the risk of cancer in dominant (GC+CC vs GG), additive (CC vs GG), and recessive (CC vs GC +GG) genetic models. The combined OR was calculated according to the method of Woolf [23]. A χ2-based Q statistic test was performed to assess the between-study heterogeneity [24]. If the P value of heterogeneity test was ≥ 0.10, a fixed effect model using the Mantel-Haenszel method was used to calculate the combined OR, which assumed the same homogeneity of the effect size across all the studies. If the P value of the heterogeneity test was <0.10, it showed that the heterogeneity between-study was statistically significant. The random effects model using the DerSimonian and Laird method was performed to calculate the combined OR [25]. If there was no between-study heterogeneity, the results from those two methods calculating the combined OR would be identical. The significance of the combined OR was determined by the Z-test, in which P< 0.05 was considered significant. Finally, combined ORs and their 95% CIs were presented. Stratification analyses for different types of cancers were conducted for breast cancer, lung cancer, bladder cancer, basal cell carcinoma, and other cancers (i.e., ovarian, prostate, or colorectal cancer) to estimate cancer-specific OR. Stratification analyses by ethnicity were also conducted for Caucasian, Asian and African Americans populations to estimate ethnic-specific ORs.
Publication bias was assessed with the funnel plot, in which the standard error of log (OR) of each study was plotted against its OR value. An asymmetric plot suggested possible publication bias by the method of the Egger's linear regression test [26]. The significance of the intercept was determined by the Student t-test as suggested by Egger. If the P-value of Egger's linear regression test was less than 0.05, it meant that there was publication bias in the meta-analysis.
The SAS/Genetics software program (Version 9.1, SAS Institute, Inc., Cary, NC, USA) was used to determine the LD of SNP pairs and Hardy-Weinberg equilibrium. Other statistical software used included SPSS12.0 for windows software (SPSS Inc., Chicago, USA), software Stata version 7.0, and Review Manager (version 4.2, the Cochrane Collaboration). All P-values were two-sided.
Results
Literature search and meta-analysis databases
We found 31 epidemiologic studies using the search by 'NBS1,' 'cancer' and 'polymorphism' through Pubmed and SciFinder Scholar (CA web version). Of these 31 studies, 15 studies were excluded, 9 studies were excluded because they were not case-control studies [27-35], 5 studies were excluded because E185Q polymorphism or its genotype frequency was not reported [14,36-39], and one studies focused on hematotoxicity but not cancer [40]. The remaining 16 case-control studies included 17 data sets (because Millian's study included two populations: African-American and whites) [1,13,16-18,20,21,41-49]. All the articles used DNA from blood samples for genotyping. We established a database for the extracted information from each article. Table 1 shows the essential information, including first author, cancer type, year of the publication, the numbers of cases and controls, and frequencies of NSB1 8360 C allele for all studies. There were six studies for breast cancer [1,20,21,41,42], three for the lung cancer [18,43,44], three for the bladder cancer [16,45,48], two for the basal cell carcinoma [17,46], one for the ovarian [13], one for prostate cancer [47], and one for colorectal cancer [49]. Among 17 eligible datasets included in the final analysis, there were 14 (82.3%) of Caucasians, two (11.8%) of Chinese, and one (5.9%) of African-Americans. Additional information is listed in the forest plots in our meta-analyses.
Table 1.
Summary of eligible studies considered in the meta-analysis
| First author(year) | Country | Ethnicity | Cancer type | Type of study | Case no. | Control no. | C Allele frequency (%) case/control |
|---|---|---|---|---|---|---|---|
| Kuschel(2002)[1] | Germany | Caucasians | Breast cancer | Population-based | 1694 | 734 | 34.3/32.2 |
| Forsti(2004)[20] | Finland | Caucasians | Breast cancer | Population-based | 223 | 319 | 35.7/38.6 |
| Millikan (2005)[41] | North Carolina | African-American | Breast cancer | Hospital-based | 726 | 681 | 25.1/23.6 |
| Millikan (2005)[41] | North Carolina | Caucasians | Breast cancer | Hospital-based | 1273 | 1136 | 31.6/32.3 |
| Lu(2006)[21] | USA | Caucasians | Breast cancer | Hospital-based | 421 | 423 | 35.9/29.7 |
| Zhang (2005)[42] | China | Chinese | Breast cancer | Hospital-based | 220 | 310 | 35.9/38.2 |
| Lan(2005)[18] | China | Chinese | Lung cancer | Population-based | 118 | 111 | 57.2/66.7 |
| Ryk (2006)[43] | Sweden | Caucasians | Lung cancer | Hospital-based | 177 | 152 | -/- |
| Zienoldding (2006)[44] | Norway and of Norwagian | Caucasians | Lung cancer | Hospital-based | 376 | 310 | 34.4/28.5 |
| Broberg (2005)[16] | Sweden | Caucasians | Bladder cancer | Hospital-based | 61 | 154 | 36.1/37.3 |
| Sanyal (2004)[45] | Sweden | Caucasians | Bladder cancer | Hospital-based | 299 | 278 | 38.5/34.2 |
| Figueroa(2007)[48] | Spanish | Caucasians | Bladder cancer | Hospital-based | 1086 | 1020 | 31.8/30.0 |
| Festa (2005)[17] | Sweden and Finland | Caucasians | Basal cell carcinoma | Hospital-based | 241 | 574 | 37.1/36.8 |
| Thirumaran (2006)[46] | Hungary, Romania, and Slovakia | Caucasians | Basal cell carcinoma | Hospital-based | 529 | 533 | 35.6/32.4 |
| Auranen (2005)[13] | Combined* | Caucasians | Ovarian cancer | Mixed§ | 1586 | 2685 | 32.7/33.3 |
| Hebbring (2006)[47] | Finland | Caucasians | Prostate cancer | Population-based | 200 | 200 | 35.5/35.8 |
| Pardini (2008)[49] | Czech Republic | Caucasians | Colorectal cancer | Hospital-based | 532 | 532 | 31.8/34.0 |
*: United Kingdom SEARCH study. Danish MALOVA study. United States FROC study. United Kingdom Royal Marsden Hospital and young ovarian cancer study (UK RMH/YOV).
§: Subjects including population and hospital source.
The frequency distributions of genotypes in control groups from all studies were in accordance with Hardy-Weinberg equilibrium (P > 0.05), except for Hebbring's study (χ2 = 3.93, P = 0.05). We performed a sensitivity analysis to for the selection of published studies in the meta-analysis. The frequencies of the 8360 C allele in the control groups are also listed in Table 1. Compared with the reported frequency from the database of HapMap http://www.hapmap.org/cgi-perl/gbrowse/hapmap20_B35/, the frequencies of 8360 C allele in the meta-analysis were not as similar as that from the International HapMap Project. There might be population diversity. We would take that into account in the meta-analysis.
Test for heterogeneity
Table 2 shows that no between-study heterogeneity was found in C vs G allele comparison for 16 datasets (P = 0.10) and in dominant genetic models for all 17 datasets (P = 0.54). However, there was between-study heterogeneity in the additive genetic model (P = 0.02) and recessive genetic model (P = 0.03) for 16 datasets. In the subgroup analyses by the type of cancers and ethnicity, as shown in Figures 1 and 2, the heterogeneity test did not show any significant difference in dominant genetic models: three lung cancer studies (P = 0.10), six breast cancer studies (P = 0.27), three bladder cancer studies (P = 0.86), two basal cell carcinoma studies (P = 0.42), other cancers (ovarian, prostate, and colorectal cancer) (P = 0.74), fourteen Caucasian population studies (P = 0.64), and two Chinese population studies (P = 0.14).
Table 2.
Summary of ORs for various comparisons
| E185Q comparison* |
Population (number of data sets)§ | Fixed-effects OR(95%CI) | Random-effects OR(95%CI) | Heterogeneity (P value Q test) |
P value (fixed) |
P value (random) |
|---|---|---|---|---|---|---|
| (GC+CC) vs GG | 17 | 1.06(1.00–1.12) | 1.06(1.00–1.12) | 0.54 | 0.05 | 0.05 |
| CC vs GG | 16 | 0.99(0.90–1.09) | 0.98(0.85–1.13) | 0.02 | 0.83 | 0.78 |
| CC vs (GC +GG) | 16 | 0.95(0.87–1.04) | 0.94(0.82–1.07) | 0.03 | 0.28 | 0.36 |
| C vs G | 16 | 1.02(0.97–1.08) | 1.02(0.98–1.07) | 0.10 | 0.45 | 0.36 |
*Additive genetic model: CC vs GG, recessive genetic model: CC vs (GC +GG), dominant genetic model: (GC+CC) vs GG
§Ryk's study was not involved in 16 studies.
Figure 1.
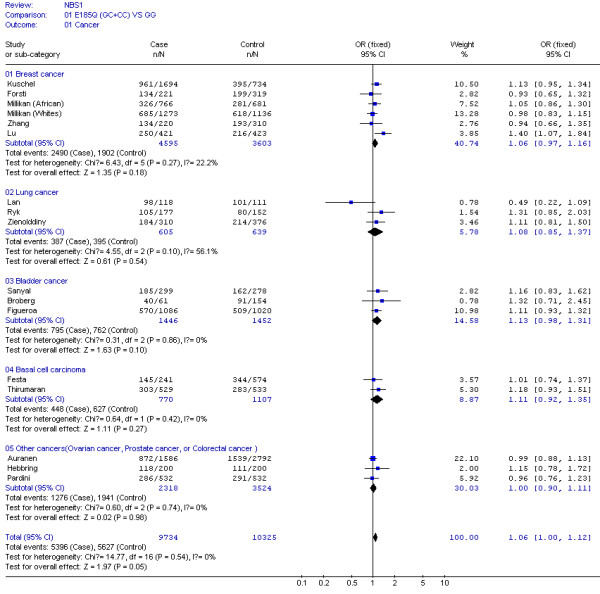
Meta-analysis for NBS1 E185Q polymorphism variant genotypes (GC and CC) vs GG in different type of cancers (breast cancer, lung cancer, bladder cancer, basal cell carcinoma, ovarian cancer, prostate cancer, and colorectal cancer).
Figure 2.
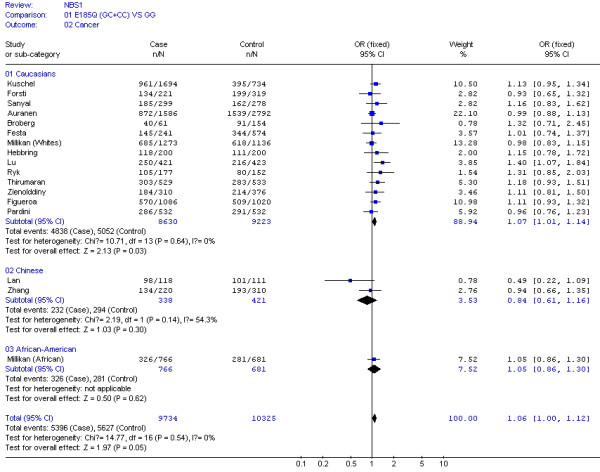
Meta-analysis for NBS1 E185Q polymorphism variant genotypes (GC and CC) vs GG in different ethnicities (Caucasians, Chinese and African Americans).
Quantitative data synthesis
For the NSB1 E185Q polymorphism, the data available for our meta-analysis were obtained from 17 datasets consisted of 9,734 cases and 10,325 controls. Associations of the NBS1 E185Q allele and genotypes with cancer risk were estimated using dominant (GC+CC vs GG), additive (CC vs GG), and recessive (CC vs GC +GG) genetic models in either fixed or random effect models according the heterogeneity Q test in Table 2. There were 16 datasets in these comparisons, except for Ryk's study that only showed data in dominant genetic models. Compared with the wild-type 8360 GG genotype, the carriers of variant genotypes (i.e., GC/CC) had a 1.06-fold elevated risk of cancer (95% CI = 1.00–1.12, P = 0.05) as estimated in a fixed effect model for dominant genetic effects. We further performed a sensitivity analysis, and found that when Hebbring's study was excluded owing to the conflict of Hardy-Weinberg equilibrium, the combined ORs of cancer risk was still 1.06 (95% CI = 1.00–1.12), and the P value of the between-study heterogeneity test was decreased significantly (from P = 0.54 to P = 0.48). However, the association between the NSB1 E185Q polymorphism and cancer risk was not significant in the additive genetic model (CC vs GG) (OR = 0.98, 95% CI = 0.85–1.13, P = 0.78) nor in the recessive genetic model (CC vs GC + GG) (OR = 0.94, 95% CI = 0.82–1.07, P = 0.36).
The effect of 8360G>C (E185Q) polymorphism was further evaluated in stratification analysis. By the types of cancer, in those three lung cancer studies consisted of 605 cases and 639 controls, the variant genotypes (387 cases and 395 controls) had a non-significantly increased risk of lung cancer (OR = 1.08, 95% CI = 0.85–1.37, P = 0.54) as estimated in a fixed effect model (Figure 1). In the six breast cancer studies of 4,595 cases and 3,603 controls, the variant genotypes (2,490 cases and 1,902 controls) had a non-significantly increased risk (OR = 1.06, 95% CI = 0.97–1.16, P = 0.18) (Figure 1). Similarly, in the three bladder cancer studies of 1,446 cases and 1,452 controls (OR = 1.13, 95% CI = 0.98–1.31, P = 0.10) and three basal cell carcinoma studies of 770 cases and 1,107 controls (OR = 1.11, 95% CI = 0.92–1.35, P = 0.27) (Figure 1).
In the stratification analyses for ethnicity, we found that the increased risk of cancer associated with 8360G>C variant genotypes was more pronounced in the Caucasians (OR = 1.07, 95% CI = 1.01–1.14, P = 0.03), but not in Chinese (OR = 0.84, 95% CI = 0.61–1.16, P = 0.30) nor in African Americans (OR = 1.05, 95% CI = 0.86–1.30, P = 0.62) (Figure 2).
Bias diagnostics
To evaluate publication biases, the NSB1 E185Q genotypes were plotted against the precision ones in a funnel plot, which is approximately symmetrical. Egger's test suggested that there was no publication bias in the current meta-analysis (t = 0.15, df = 16, P = 0.88). Furthermore, we found that the fail-safe number for the finding of NSB1 E185Q variant genotypes associated with 1.06 fold increased risk of cancer was 60, which suggests that biases from publications and other factors may not have a significant influence on the results of current meta-analysis for the association between NSB1 E185Q polymorphism and cancer risk (Figure 3).
Figure 3.
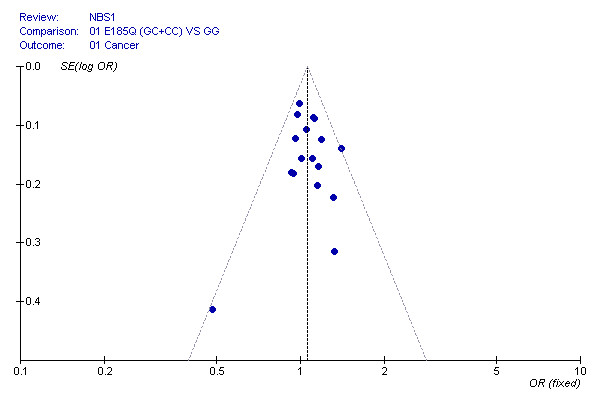
Funnel plot of the Egger's test of NBS1E185Q polymorphism for publication bias.
Discussion
In this meta-analysis consisted of 16 independent case-control studies with 17 datasets, we found that the carriers of NBS1 E185Q variant genotypes had a 1.06-fold increased risk of cancer in the dominant genetic model, and the association was more pronounced in the Caucasians. However, we did not find evidence for significant associations in the subgroup analysis for the individual type of cancers, such as lung cancer, breast cancer, basal cell carcinoma, urogenital cancers (i.e., bladder cancer, ovarian cancer, and prostate cancer), or colorectal cancer.
The NBS1 8360G>C (rs1805794) polymorphism is a non-synonymous SNP with an amino acid change (Glu185>Gln or E185Q). Either one or two missense changes in the dominant genetic model may affect the function of NBS1 and interfere protein-protein interaction [50]. The E185Q amino acid substitution was predicted to be tolerated via the SIFT prediction tool http://blocks.fhcrc.org/sift/SIFT.html. The E185Q SNP is located in a breast cancer carboxy-terminal (BRCT) domain (108–196 amino acids) of NBS1 [3], and such a domain facilitates NBS1 to interact with BRCA1 (one of two familial breast cancers mutated genes) forming BRCA1-associated genome surveillance complex (BASC), which is responsible for the recognition and repair of aberrant DNA [51].
In the LD analysis, we found that the E185Q SNP was in a completed LD (D' = 1.00, r2 = 1.00) with the loci 626G>A in the promoter of NBS1 (-1418 nt to initiation transcription code ATG), 3816G>A (Leu34Leu, rs1063045) and 40419A>G (Pro672Pro, rs1061302) [21]. In further bioinformatics analysis searching for the transcription factor binding sites with the TFSEARCH program http://mbs.cbrc.jp/research/db/TFSEARCH.html, we found that the 626G allele, but not the 626A allele, creates a new binding site of SRY that is a functional target of the tumor suppression gene WT1 [52], and that the 626G allele in LD with Glu185 may function in prohibiting carcinogenesis. Furthermore, in a region from 35309 bp upstream to 29477 bp downstream of E185Q on chromosome 8, we found that there were thirty-three polymorphisms being in completely LD (all with an r2 = 1.00) with E185Q (data not shown). Because the exact molecular mechanism involving the NBS1 E185Q variant in the etiology of cancer is still unclear, further investigations are needed to identify its LD with other unknown functional variants of cancer susceptibility candidate genes.
DNA DSBs in human cells is one of the major factors for carcinogenesis. Affected individuals when exposed to different carcinogens will have different outcomes. For example, the common NBS1 185Gln allele has been associated with an increased risk for lung cancer in a Chinese population exposed to smoky coal emissions [18]. Stable covalent BaP DNA adducts can cause single-strand breaks, resulting in DSBs during replication [53]. The established risk factors for bladder cancer include cigarette smoking, exposure to industrially related aromatic amines and drugs [54,55]. Some common, low-penetrance susceptibility genes in human populations may interact with radiation exposure to increase risk of breast cancer [56], because DNA DSBs are frequently induced by ionizing radiation, which may result in altered apoptosis or tumorigenesis [1]. For ovarian cancer, however, ovulation may play a role in ovarian cancer development [13]. Smoking and drinking habit are frequently associated with colorectal cancer risk [57]. BCCs are caused by interplay between genetic and environment factors, too [17]. The NBS1 E185Q variant genotypes (8360GC/CC) were found to be associated with a p53 mutation in lung cancer, suggesting a role in lung carcinogenesis [32]. Other report suggested that the XRCC3 interacted with NBS1 involved in the homologous recombination [58]. Therefore, it is likely that the NBS1 E185Q polymorphism may modify cancer susceptibility via gene-environment and gene-gene interactions. However, not all studies offer the information about environmental exposure.
Sensitivity analysis showed that for breast cancer no association (combined OR = 1.05, 95% CI = 0.99–1.11) was present after exclusion of the study of Lu. In Lu's study the subjects were ≤ 55 old women [21]. About 40% of NBS patients develop cancer before the age of 21 [59]. It has been suggested that there are different etiologic pathways for early-onset and late-onset types of breast cancer [60,61]. Therefore, large studies of NBS1 E185Q stratified by age are needed to investigate the inter-individual susceptibility to cancer.
There appeared to be ethnicity-specific genetic effects, because we found that the association between 8360G>C variant genotypes and increased risk of cancer was significant only in Caucasians but not in Chinese or African-Americans, suggesting genetic diversity among different ethnicities. The frequencies of 8360 C allele in the controls of selected studies were not as similar as that from the database of the International HapMap Project. We would take that into account when applying to the findings from the meta-analysis.
Potential publication bias may exist in this meta-analysis, because the studies with negative results are more likely not to be published, though there was no testable publication bias in our meta-analysis by the funnel plot. Because there were only four out of 17 datasets were population-based case-control studies, others being hospital-based case-control studies, the study subjects may not be representative of the general population and could lead to selection bias.
Conclusion
In conclusion, our meta-analysis suggests that based on the published data, the NBS1 E185Q variant genotypes (8360 GC/CC) might be associated with an increased risk of cancer, especially in Caucasians. Due to the limitations of such meta-analysis, larger association studies or multicentric case-control studies and the studies assessing gene-environment interactions are warranted to confirm these findings.
Abbreviations
NBS: Nijimegen break syndrome; NBS1: Nijimegen break syndrome mutated gene; DSBs: DNA double strand breaks; SNP: single nucleotide polymorphism; OR: odds ratio; CI: confidence interval.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
ML participated in study design and drafted the manuscript. JL carried out Bioinformatics Analysis and critically revised the manuscript. XY, MY, and HT performed the statistical analysis and participated in the critical revision of the manuscript. BY participated in collection of data and manuscript preparation. LS participated in its design. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Meixia Lu, Email: lvmeixialv@yahoo.com.cn.
Jiachun Lu, Email: jcLu@gzhmc.edu.cn.
Xiaobo Yang, Email: yxbo21021@hotmail.com.
Miao Yang, Email: y2000yangmiao@163.com.
Hao Tan, Email: tantonny@163.com.
Bai Yun, Email: henry_by@163.com.
Luyuan Shi, Email: lyshi@mails.tjmu.deu.cn.
Acknowledgements
The authors thank Xiuquan Shi, Pei Wang and Wei Gao for technical assistance, and Prof. Qingyi Wei (Department of Epidemiology, The University Texas MD Anderson Cancer Center, Houston, TX) for Science editing. This work was supported in part by the National Natural Scientific Foundation of China grants 3050416 (M. Lu), 30671813 and 30872178 (J. Lu).
References
- Kuschel B, Auranen A, McBride S, Novik KL, Antoniou A, Lipscombe JM, Day NE, Easton DF, Ponder BA, Pharoah PD, Dunning A. Variants in DNA double-strand break repair genes and breast cancer susceptibility. Hum Mol Genet. 2002;11:1399–1407. doi: 10.1093/hmg/11.12.1399. [DOI] [PubMed] [Google Scholar]
- Matsuura S, Kobayashi J, Tauchi H, Komatsu K. Nijmegen breakage syndrome and DNA double strand break repair by NBS1 complex. Adv Biophys. 2004;38:65–80. doi: 10.1016/S0065-227X(04)80076-5. [DOI] [PubMed] [Google Scholar]
- Kobayashi J. Molecular mechanism of the recruitment of NBS1/hMRE11/hRAD50 complex to DNA double-strand breaks: NBS1 binds to gamma-H2AX through FHA/BRCT domain. J Radiat Res (Tokyo) 2004;45:473–478. doi: 10.1269/jrr.45.473. [DOI] [PubMed] [Google Scholar]
- Tseng SF, Chang CY, Wu KJ, Teng SC. Importin KPNA2 is required for proper nuclear localization and multiple functions of NBS1. J Biol Chem. 2005;280:39594–39600. doi: 10.1074/jbc.M508425200. [DOI] [PubMed] [Google Scholar]
- Tauchi H, Matsuura S, Kobayashi J, Sakamoto S, Komatsu K. Nijmegen breakage syndrome gene, NBS1, and molecular links to factors for genome stability. Oncogene. 2002;21:8967–8980. doi: 10.1038/sj.onc.1206136. [DOI] [PubMed] [Google Scholar]
- Howlett NG, Scuric Z, D'Andrea AD, Schiestl RH. Impaired DNA double strand break repair in cells from Nijmegen breakage syndrome patients. DNA Repair (Amst) 2006;5:251–257. doi: 10.1016/j.dnarep.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Digweed M, Reis A, Sperling K. Nijmegen breakage syndrome: consequences of defective DNA double strand break repair. Bioessays. 1999;21:649–656. doi: 10.1002/(SICI)1521-1878(199908)21:8<649::AID-BIES4>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Digweed M, Sperling K. Nijmegen breakage syndrome: clinical manifestation of defective response to DNA double-strand breaks. DNA Repair (Amst) 2004;3:1207–1217. doi: 10.1016/j.dnarep.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Matsuura S, Tauchi H, Nakamura A, Kondo N, Sakamoto S, Endo S, Smeets D, Solder B, Belohradsky BH, Der Kaloustian VM, Oshimura M, Isomura M, Nakamura Y, Komatsu K. Positional cloning of the gene for Nijmegen breakage syndrome. Nat Genet. 1998;19:179–181. doi: 10.1038/549. [DOI] [PubMed] [Google Scholar]
- Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR 3rd, Hays L, Morgan WF, Petrini JH. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/S0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- Varon R, Vissinga C, Platzer M, Cerosaletti KM, Chrzanowska KH, Saar K, Beckmann G, Seemanova E, Cooper PR, Nowak NJ, Stumm M, Weemaes CM, Gatti RA, Wilson RK, Digweed M, Rosenthal A, Sperling K, Concannon P, Reis A. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell. 1998;93:467–476. doi: 10.1016/S0092-8674(00)81174-5. [DOI] [PubMed] [Google Scholar]
- Debniak T, Gorski B, Cybulski C, Jakubowska A, Kurzawski G, Lener M, Mierzejewski M, Masojc B, Medrek K, Kladny J, Zaluga E, Maleszka R, Chosia M, Lubinski J. Germline 657del5 mutation in the NBS1 gene in patients with malignant melanoma of the skin. Melanoma Res. 2003;13:365–370. doi: 10.1097/00008390-200308000-00005. [DOI] [PubMed] [Google Scholar]
- Auranen A, Song H, Waterfall C, Dicioccio RA, Kuschel B, Kjaer SK, Hogdall E, Hogdall C, Stratton J, Whittemore AS, Easton DF, Ponder BA, Novik KL, Dunning AM, Gayther S, Pharoah PD. Polymorphisms in DNA repair genes and epithelial ovarian cancer risk. Int J Cancer. 2005;117:611–618. doi: 10.1002/ijc.21047. [DOI] [PubMed] [Google Scholar]
- Cerosaletti KM, Morrison VA, Sabath DE, Willerford DM, Concannon P. Mutations and molecular variants of the NBS1 gene in non-Hodgkin lymphoma. Genes Chromosomes Cancer. 2002;35:282–286. doi: 10.1002/gcc.10114. [DOI] [PubMed] [Google Scholar]
- Buslov KG, Iyevleva AG, Chekmariova EV, Suspitsin EN, Togo AV, Kuligina E, Sokolenko AP, Matsko DE, Turkevich EA, Lazareva YR, Chagunava OL, Bit-Sava EM, Semiglazov VF, Devilee P, Cornelisse C, Hanson KP, Imyanitov EN. NBS1 657del5 mutation may contribute only to a limited fraction of breast cancer cases in Russia. Int J Cancer. 2005;114:585–589. doi: 10.1002/ijc.20765. [DOI] [PubMed] [Google Scholar]
- Broberg K, Bjork J, Paulsson K, Hoglund M, Albin M. Constitutional short telomeres are strong genetic susceptibility markers for bladder cancer. Carcinogenesis. 2005;26:1263–1271. doi: 10.1093/carcin/bgi063. [DOI] [PubMed] [Google Scholar]
- Festa F, Kumar R, Sanyal S, Unden B, Nordfors L, Lindholm B, Snellman E, Schalling M, Forsti A, Hemminki K. Basal cell carcinoma and variants in genes coding for immune response, DNA repair, folate and iron metabolism. Mutat Res. 2005;574:105–111. doi: 10.1016/j.mrfmmm.2005.01.026. [DOI] [PubMed] [Google Scholar]
- Lan Q, Shen M, Berndt SI, Bonner MR, He X, Yeager M, Welch R, Keohavong P, Donahue M, Hainaut P, Chanock S. Smoky coal exposure, NBS1 polymorphisms, p53 protein accumulation, and lung cancer risk in Xuan Wei, China. Lung Cancer. 2005;49:317–323. doi: 10.1016/j.lungcan.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Steffen J, Varon R, Mosor M, Maneva G, Maurer M, Stumm M, Nowakowska D, Rubach M, Kosakowska E, Ruka W, Nowecki Z, Rutkowski P, Demkow T, Sadowska M, Bidzinski M, Gawrychowski K, Sperling K. Increased cancer risk of heterozygotes with NBS1 germline mutations in Poland. Int J Cancer. 2004;111:67–71. doi: 10.1002/ijc.20239. [DOI] [PubMed] [Google Scholar]
- Forsti A, Angelini S, Festa F, Sanyal S, Zhang Z, Grzybowska E, Pamula J, Pekala W, Zientek H, Hemminki K, Kumar R. Single nucleotide polymorphisms in breast cancer. Oncol Rep. 2004;11:917–922. [PubMed] [Google Scholar]
- Lu J, Wei Q, Bondy ML, Li D, Brewster A, Shete S, Yu TK, Sahin A, Meric-Bernstam F, Hunt KK, Singletary SE, Ross MI, Wang LE. Polymorphisms and haplotypes of the NBS1 gene are associated with risk of sporadic breast cancer in non-Hispanic white women <or = 55 years. Carcinogenesis. 2006;27:2209–2216. doi: 10.1093/carcin/bgl077. [DOI] [PubMed] [Google Scholar]
- Nyholt DR. ssSNPer: identifying statistically similar SNPs to aid interpretation of genetic association studies. Bioinformatics. 2006;22:2960–2961. doi: 10.1093/bioinformatics/btl518. [DOI] [PubMed] [Google Scholar]
- Woolf B. On estimating the relation between blood group and disease. Ann Hum Genet. 1955;19:251–253. doi: 10.1111/j.1469-1809.1955.tb01348.x. [DOI] [PubMed] [Google Scholar]
- Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997;127:820–826. doi: 10.7326/0003-4819-127-9-199711010-00008. [DOI] [PubMed] [Google Scholar]
- Petitti D. Meta-analysis, Decision Analysis, and Cost-effectiveness. New York: Oxford University Press; 1994. [Google Scholar]
- Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. Bmj. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varon R, Schoch C, Reis A, Hiddemann WC, Sperling K, Schnittger S. Mutation analysis of the Nijmegen breakage syndrome gene (NBS1) in nineteen patients with acute myeloid leukemia with complex karyotypes. Leuk Lymphoma. 2003;44:1931–1934. doi: 10.1080/1042819031000099724. [DOI] [PubMed] [Google Scholar]
- Laczmanska I, Gil J, Karpinski P, Stembalska A, Kozlowska J, Busza H, Trusewicz A, Pesz K, Ramsey D, Schlade-Bartusiak K, Blin N, Sasiadek MM. Influence of polymorphisms in xenobiotic-metabolizing genes and DNA-repair genes on diepoxybutane-induced SCE frequency. Environ Mol Mutagen. 2006;47:666–673. doi: 10.1002/em.20253. [DOI] [PubMed] [Google Scholar]
- Hsu HM, Wang HC, Chen ST, Hsu GC, Shen CY, Yu JC. Breast cancer risk is associated with the genes encoding the DNA double-strand break repair Mre11/Rad50/Nbs1 complex. Cancer Epidemiol Biomarkers Prev. 2007;16:2024–2032. doi: 10.1158/1055-9965.EPI-07-0116. [DOI] [PubMed] [Google Scholar]
- Popanda O, Tan XL, Ambrosone CB, Kropp S, Helmbold I, von Fournier D, Haase W, Sautter-Bihl ML, Wenz F, Schmezer P, Chang-Claude J. Genetic polymorphisms in the DNA double-strand break repair genes XRCC3, XRCC2, and NBS1 are not associated with acute side effects of radiotherapy in breast cancer patients. Cancer Epidemiol Biomarkers Prev. 2006;15:1048–1050. doi: 10.1158/1055-9965.EPI-06-0046. [DOI] [PubMed] [Google Scholar]
- French D, Wilkinson MR, Yang W, de Chaisemartin L, Cook EH, Das S, Ratain MJ, Evans WE, Downing JR, Pui CH, Relling MV. Global gene expression as a function of germline genetic variation. Hum Mol Genet. 2005;14:1621–1629. doi: 10.1093/hmg/ddi170. [DOI] [PubMed] [Google Scholar]
- Medina PP, Ahrendt SA, Pollan M, Fernandez P, Sidransky D, Sanchez-Cespedes M. Screening of homologous recombination gene polymorphisms in lung cancer patients reveals an association of the NBS1-185Gln variant and p53 gene mutations. Cancer Epidemiol Biomarkers Prev. 2003;12:699–704. [PubMed] [Google Scholar]
- Plisiecka-Halasa J, Dansonka-Mieszkowska A, Rembiszewska A, Bidzinski M, Steffen J, Kupryjanczyk J. Nijmegen breakage syndrome gene (NBS1) alterations and its protein (nibrin) expression in human ovarian tumours. Ann Hum Genet. 2002;66:353–359. doi: 10.1046/j.1469-1809.2002.00122.x. [DOI] [PubMed] [Google Scholar]
- Goode EL, Dunning AM, Kuschel B, Healey CS, Day NE, Ponder BA, Easton DF, Pharoah PP. Effect of germ-line genetic variation on breast cancer survival in a population-based study. Cancer Res. 2002;62:3052–3057. [PubMed] [Google Scholar]
- Hama S, Matsuura S, Tauchi H, Sawada J, Kato C, Yamasaki F, Yoshioka H, Sugiyama K, Arita K, Kurisu K, Kamada N, Heike Y, Komatsu K. Absence of mutations in the NBS1 gene in B-cell malignant lymphoma patients. Anticancer Res. 2000;20:1897–1900. [PubMed] [Google Scholar]
- Ziolkowska I, Mosor M, Wierzbicka M, Rydzanicz M, Pernak-Schwarz M, Nowak J. Increased risk of larynx cancer in heterozygous carriers of the I171V mutation of the NBS1 gene. Cancer Sci. 2007;98:1701–1705. doi: 10.1111/j.1349-7006.2007.00594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laczmanska I, Gil J, Karpinski P, Stembalska A, Trusewicz A, Pesz K, Ramsey D, Schlade-Bartusiak K, Blin N, Sasiadek MM. Polymorphism in nucleotide excision repair gene XPC correlates with bleomycin-induced chromosomal aberrations. Environ Mol Mutagen. 2007;48:666–671. doi: 10.1002/em.20333. [DOI] [PubMed] [Google Scholar]
- Meyer P, Stapelmann H, Frank B, Varon R, Burwinkel B, Schmitt C, Boettger MB, Klaes R, Sperling K, Hemminki K, Kammerer S. Molecular genetic analysis of NBS1 in German melanoma patients. Melanoma Res. 2007;17:109–116. doi: 10.1097/CMR.0b013e3280dec638. [DOI] [PubMed] [Google Scholar]
- Allen-Brady K, Camp NJ. Characterization of the linkage disequilibrium structure and identification of tagging-SNPs in five DNA repair genes. BMC Cancer. 2005;5:99. doi: 10.1186/1471-2407-5-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M, Lan Q, Zhang L, Chanock S, Li G, Vermeulen R, Rappaport SM, Guo W, Hayes RB, Linet M, Yin S, Yeager M, Welch R, Forrest MS, Rothman N, Smith MT. Polymorphisms in genes involved in DNA double-strand break repair pathway and susceptibility to benzene-induced hematotoxicity. Carcinogenesis. 2006;27:2083–2089. doi: 10.1093/carcin/bgl061. [DOI] [PubMed] [Google Scholar]
- Millikan RC, Player JS, Decotret AR, Tse CK, Keku T. Polymorphisms in DNA repair genes, medical exposure to ionizing radiation, and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2005;14:2326–2334. doi: 10.1158/1055-9965.EPI-05-0186. [DOI] [PubMed] [Google Scholar]
- Zhang L, Zhang Z, Yan W. Single nucleotide polymorphisms for DNA repair genes in breast cancer patients. Clin Chim Acta. 2005;359:150–155. doi: 10.1016/j.cccn.2005.03.047. [DOI] [PubMed] [Google Scholar]
- Ryk C, Kumar R, Thirumaran RK, Hou SM. Polymorphisms in the DNA repair genes XRCC1, APEX1, XRCC3 and NBS1, and the risk for lung cancer in never- and ever-smokers. Lung Cancer. 2006;54:285–292. doi: 10.1016/j.lungcan.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Zienolddiny S, Campa D, Lind H, Ryberg D, Skaug V, Stangeland L, Phillips DH, Canzian F, Haugen A. Polymorphisms of DNA repair genes and risk of non-small cell lung cancer. Carcinogenesis. 2006;27:560–567. doi: 10.1093/carcin/bgi232. [DOI] [PubMed] [Google Scholar]
- Sanyal S, Festa F, Sakano S, Zhang Z, Steineck G, Norming U, Wijkstrom H, Larsson P, Kumar R, Hemminki K. Polymorphisms in DNA repair and metabolic genes in bladder cancer. Carcinogenesis. 2004;25:729–734. doi: 10.1093/carcin/bgh058. [DOI] [PubMed] [Google Scholar]
- Thirumaran RK, Bermejo JL, Rudnai P, Gurzau E, Koppova K, Goessler W, Vahter M, Leonardi GS, Clemens F, Fletcher T, Hemminki K, Kumar R. Single nucleotide polymorphisms in DNA repair genes and basal cell carcinoma of skin. Carcinogenesis. 2006;27:1676–1681. doi: 10.1093/carcin/bgi381. [DOI] [PubMed] [Google Scholar]
- Hebbring SJ, Fredriksson H, White KA, Maier C, Ewing C, McDonnell SK, Jacobsen SJ, Cerhan J, Schaid DJ, Ikonen T, Autio V, Tammela TL, Herkommer K, Paiss T, Vogel W, Gielzak M, Sauvageot J, Schleutker J, Cooney KA, Isaacs W, Thibodeau SN. Role of the Nijmegen breakage syndrome 1 gene in familial and sporadic prostate cancer. Cancer Epidemiol Biomarkers Prev. 2006;15:935–938. doi: 10.1158/1055-9965.EPI-05-0910. [DOI] [PubMed] [Google Scholar]
- Figueroa JD, Malats N, Rothman N, Real FX, Silverman D, Kogevinas M, Chanock S, Yeager M, Welch R, Dosemeci M, Tardon A, Serra C, Carrato A, Garcia-Closas R, Castano-Vinyals G, Garcia-Closas M. Evaluation of genetic variation in the double-strand break repair pathway and bladder cancer risk. Carcinogenesis. 2007;28:1788–1793. doi: 10.1093/carcin/bgm132. [DOI] [PubMed] [Google Scholar]
- Pardini B, Naccarati A, Novotny J, Smerhovsky Z, Vodickova L, Polakova V, Hanova M, Slyskova J, Tulupova E, Kumar R, Bortlik M, Barale R, Hemminki K, Vodicka P. DNA repair genetic polymorphisms and risk of colorectal cancer in the Czech Republic. Mutat Res. 2008;638:146–153. doi: 10.1016/j.mrfmmm.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Tauchi H. Positional cloning and functional analysis of the gene responsible for Nijmegen breakage syndrome. J Radiat Res (Tokyo) 2000;41:9–17. doi: 10.1269/jrr.41.9. [DOI] [PubMed] [Google Scholar]
- Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- Hossain A, Saunders GF. The human sex-determining gene SRY is a direct target of WT1. J Biol Chem. 2001;276:16817–16823. doi: 10.1074/jbc.M009056200. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- Steineck G, Wiholm BE, Gerhardsson de Verdier M. Acetaminophen, some other drugs, some diseases and the risk of transitional cell carcinoma. A population-based case-control study. Acta Oncol. 1995;34:741–748. doi: 10.3109/02841869509127181. [DOI] [PubMed] [Google Scholar]
- Kellen E, Hemelt M, Broberg K, Golka K, Kristensen VN, Hung RJ, Matullo G, Mittal RD, Porru S, Povey A, Schulz WA, Shen J, Buntinx F, Zeegers MP, Taioli E. Pooled analysis and meta-analysis of the glutathione S-transferase P1 Ile 105Val polymorphism and bladder cancer: a HuGE-GSEC review. Am J Epidemiol. 2007;165:1221–1230. doi: 10.1093/aje/kwm003. [DOI] [PubMed] [Google Scholar]
- Ronckers CM, Erdmann CA, Land CE. Radiation and breast cancer: a review of current evidence. Breast Cancer Res. 2005;7:21–32. doi: 10.1186/bcr970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrubsole MJ, Wu H, Ness RM, Shyr Y, Smalley WE, Zheng W. Alcohol drinking, cigarette smoking, and risk of colorectal adenomatous and hyperplastic polyps. Am J Epidemiol. 2008;167:1050–1058. doi: 10.1093/aje/kwm400. [DOI] [PubMed] [Google Scholar]
- Forget AL, Bennett BT, Knight KL. Xrcc3 is recruited to DNA double strand breaks early and independent of Rad51. J Cell Biochem. 2004;93:429–436. doi: 10.1002/jcb.20232. [DOI] [PubMed] [Google Scholar]
- Nijmegen breakage syndrome. The International Nijmegen Breakage Syndrome Study Group. Arch Dis Child. 2000;82:400–406. doi: 10.1136/adc.82.5.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson WF, Matsuno RK, Sherman ME, Lissowska J, Gail MH, Brinton LA, Yang XR, Peplonska B, Chen BE, Rosenberg PS, Chatterjee N, Szeszenia-Dabrowska N, Bardin-Mikolajczak A, Zatonski W, Devesa SS, Garcia-Closas M. Estimating age-specific breast cancer risks: a descriptive tool to identify age interactions. Cancer Causes Control. 2007;18:439–447. doi: 10.1007/s10552-006-0092-9. [DOI] [PubMed] [Google Scholar]
- Anderson WF, Chen BE, Brinton LA, Devesa SS. Qualitative age interactions (or effect modification) suggest different cancer pathways for early-onset and late-onset breast cancers. Cancer Causes Control. 2007;18:1187–1198. doi: 10.1007/s10552-007-9057-x. [DOI] [PubMed] [Google Scholar]


