SUMMARY
Helicobacter pylori colonization of the human stomach is characterized by profound disease-causing inflammation. Bacterial proteins that detoxify reactive oxygen species or recognize damaged DNA adducts promote infection, suggesting that H. pylori requires DNA damage-repair for successful in vivo colonization. The molecular mechanisms of repair remain unknown. We identified homologs of the AddAB class of helicase-nuclease enzymes, related to the Escherichia coli RecBCD enzyme, which, with RecA, is required for repair of DNA breaks and homologous recombination. H. pylori mutants lacking addA or addB genes lack detectable ATP-dependent nuclease activity, and the cloned H. pylori addAB genes restore both nuclease and helicase activities to an E. coli recBCD deletion mutant. H. pylori addAB and recA mutants have a reduced capacity for stomach colonization. These mutants are sensitive to DNA damaging agents and have reduced frequencies of apparent gene conversion between homologous genes encoding outer membrane proteins. Our results reveal requirements for double-strand break repair and recombination during both acute and chronic phases of H. pylori stomach infection.
Keywords: Helicobacter pylori, stomach infection, recombination, DNA repair, gene conversion
INTRODUCTION
The Gram(−) bacterium Helicobacter pylori chronically infects the stomach of half of the world’s human population, causing inflammation in the stomach that can lead to peptic ulcer disease and gastric cancers (Kusters et al., 2006). The host immune system wards off these and other bacteria by exposing them to DNA damaging agents. Bacteria overcome this damage in part by repairing their damaged DNA using homologous recombination. Homologous recombination involves three steps. First, a presynaptic step processes the damaged DNA to produce single-stranded DNA coated with RecA protein. The second, synaptic step involves homology searching and strand exchange promoted by RecA to produce a joint DNA molecule between the damaged DNA and intact DNA. A third, post-synaptic step results in resolution of the joint molecule or priming of new DNA synthesis.
In Escherichia coli the presynaptic step can be catalyzed by two distinct sets of proteins, the heterotrimeric RecBCD complex and the RecFOR proteins, which convert a DNA lesion into a RecA-coated filament (Amundsen and Smith, 2003). It has been proposed that the choice of repair complex depends on whether a double-strand (ds) break (by RecBCD) or a single-strand (ss) break or gap (by RecFOR) must be repaired. Curiously, H. pylori and other bacteria in the epsilon branch of Proteobacteria with sequenced genomes were thought to lack many or all components of these complexes due to a failure to identify homologs in their genomes (Rocha et al., 2005). In H. pylori, the only convincing, annotated homologs of presynaptic proteins are RecJ and RecR. This is in spite of the facts that H. pylori contains RecA (Schmitt et al., 1995, Thompson and Blaser, 1995) and post-synaptic proteins (Tomb et al., 1997, Loughlin et al., 2003), has a population genetic structure indicative of a high amount of recombination between strains (Suerbaum et al., 1998, Falush et al., 2003), and uses gene conversion to vary expression of surface proteins during the course of infection (Solnick et al., 2004).
Recombination and DNA repair initiated at DNA ds breaks in E. coli requires the RecBCD enzyme, a heterotrimer composed of one copy of the products of the recB, recC, and recD genes (Taylor and Smith, 1995). The enzyme is an ATP-dependent ds and ss exonuclease, a ss endonuclease, an ATPase, and a highly processive helicase. RecBCD binds tightly to ds DNA ends and initiates unwinding using the fast RecD helicase and the slower RecB helicase (Taylor and Smith, 2003). Although the single nuclease domain resides in RecB (Yu et al., 1998), all three subunits and ATP are required for substantial nuclease activity, because DNA hydrolysis occurs during ATP-dependent DNA unwinding (Amundsen et al., 1986). When the enzyme interacts with a Chi site (5′ GCTGGTGG 3′) during unwinding, a 3′-terminated ss DNA end is produced, onto which RecBCD loads RecA protein (Smith et al., 1981, Ponticelli et al., 1985, Anderson and Kowalczykowski, 1997). This presynaptic filament is the substrate for subsequent steps in recombination or repair with an intact duplex.
Activities similar to those of RecBCD enzyme are also found in the two subunit AddAB bacterial enzyme, most extensively studied from Bacillus subtilis. Like RecBCD, AddAB has nuclease and helicase activities, both of which are ATP-dependent (Kooistra et al., 1988) and can produce the 3′-terminated ss DNA end required for presynaptic filament formation and recombination (Chedin and Kowalczykowski, 2002). Both the AddA and AddB proteins have nuclease domains, while apparently active helicase motifs are found only in AddA (Kooistra et al., 1997, Yeeles and Dillingham, 2007). Thus, although the structure of AddAB differs from that of RecBCD, both enzymes contain ATP-dependent nuclease and helicase activities.
Here we demonstrate that H. pylori and, by homology, other episilon Proteobacteria do have ATP-dependent nuclease and helicase activities, which, as in most Gram(+) bacteria and some Gram(−) bacteria (Rocha et al., 2005), are encoded by addA and addB genes. We show that AddAB and RecA are required for efficient colonization of the stomach of mice, RecA more than AddAB. We also demonstrate that AddAB and RecA promote a gene conversion-like event that modulates surface expression of a bacterial adhesin. These results suggest that recombination plays multiple roles during infection: recombination-related repair of DNA damage encountered during infection as well as remodeling of the bacterial surface that may allow evasion of adaptive immune responses or altered bacterial tropism.
RESULTS
Identification of H. pylori addAB Homologs
No genes in either of two sequenced H. pylori genomes available on the NCBI website (http://www.ncbi.nlm.nih.gov/) were annotated as either recB or addA. Two BLAST searches against H. pylori using the RecB sequence of Escherichia coli K12 or and the AddA sequence of Bacillus subtilis subtilis identified a group of three proteins with significant E values (from 8 × 10−7 to 4 × 10−16) in H. pylori strains J99 and 26695. Both AddA and RecB consist of a highly conserved helicase domain and a highly conserved nuclease domain. The helicase domain is also found in a group of related helicases including Rep, UvrD, and PcrA. In neither H. pylori strain J99 nor strain 26695 did the highest scoring alignments to RecB or AddA include their nuclease domains, suggesting that these H. pylori proteins might not be homologs of AddA or RecB but instead might be related helicases.
To identify more likely H. pylori AddA or RecB proteins, the group of three high-scoring helicases from each strain was searched for conserved domains against the NCBI Conserved Domain Database (CDD). For both of strains J99 and 26695 only one protein showed a significant alignment to the RecB profile (COG1074); these alignments included both the helicase and nuclease domains of RecB. These two proteins (HP1553 from strain 26695 and jhp1446 from strain J99) were not those with the maximum scoring BLAST results but were 93% identical to each other. The J99 protein was previously annotated as PcrA. Each of the two protein sequences bears a RecB domain corresponding to essentially the full length of the protein sequence. To classify these proteins as AddA or RecB they were BLASTed against the TIGR profile database (http://tigrblast.tigr.org/web-hmm/), which identified them as AddA, rather than RecB, sequences.
No significant hits were obtained in a BLAST search using the Bacillus subtilis AddB sequence against Helicobacter species. This is likely because AddB proteins consist of a large, poorly-conserved RecC-like “inactivated helicase” domain and a short, well-conserved, RecB-like nuclease domain. Therefore, the highly conserved AddB nuclease motif “GRIDRID” was used to identify the H. pylori AddB homologs. One perfect match to this sequence was identified in both strains 26695 and J99 (proteins HP1089 and jhp0336, respectively). These proteins are 94% identical and include an “inactivated superfamily I helicase” domain. Both were described as hypothetical proteins, without an assigned function. Alignment of these proteins to B. subtilis AddB showed conservation of the nuclease domain (Figure 1). Interestingly, the HP0275 protein from strain 26695, which was annotated as addB, lacks the inactivated helicase domain annotation and does not show significant alignment to the conserved nuclease sequence.
Figure 1.
Epsilon Proteobacteria AddA and AddB homologs with conserved nuclease domains. Nuclease domains of AddA (A) and AddB (B) proteins from epsilon Proteobacteria with fully sequenced genomes show highly conserved residues indicated by shading. Sites where mutations in B. subtilis addA and addB abolish nuclease activity (Kooistra et al., 1997) are indicated by an asterisk. Species names, and first and last residue numbers in the displayed alignment are shown on the left. Alignment to the nuclease domains of E. coli RecB and B. subtilis AddA (A) and E. coli RecB and B. subtilis AddB (B) are shown for comparison (bottom of each alignment). The corresponding locus tags and (GI numbers) for the epsilon Proteobacterial AddA proteins are (top to bottom) CCV52592_0910 (154174808), CFF8240_0386 (118474774), CHAB381_1391 (154148488), CJE1654 (57238504), JJD26997_1828 (153951355), CJJ81176_1474 (121612796), Cj1481c (15792796), CLA0743 (57240914), CUP1844 (57242577), Hac_0287 (109946901), HH1643 (32267142), HP1553 (15646160), HPAG1_1502 (108563927), jhp1446 (15612511), NIS_0366 (152990115), SUN_0187 (152991783), Tmden_0090 (78776291), WS1252 (34557616). The corresponding locus tags and GI numbers for the epsilon Proteobacterial AddB proteins are (top to bottom) Ccur5_02000280 (145956769), CFF8240_0385 (118475773), CHAB381_1392 (154149234), CJE1655 (57238505), Cjejd_02001773 (145960086), CJJ81176_1475 (121613506), Cj1482c (15792797), CLA0742 (57240913), CUP1843 (57242576), Hac_0793 (109947353), HH0025 (=(32265524), HP1089 (15645703), HPAG1_0358 (108562783), jhp0336 (15611404), NIS_0365 (152990114), SUN_0185 (152991781), Tmden_0088 (78776289), WS0562 (34556981).
Using the criterion of reciprocal best hit by BLAST with the H. pylori 26695 AddAB sequences, we identified highly related proteins in all of the sequenced epsilon Proteobacteria, suggesting that these bacteria all contain AddAB. As shown in Figure 1, the nuclease domains of the epsilon Proteobacteria homologs of both AddA and AddB are highly conserved. Unlike B. subtilis AddB (Kooistra and Venema, 1991), H. pylori AddB does not contain a detectable Walker A box, which is often involved in ATP hydrolysis. This motif is found in AddB of some, but not all, firmicute species. Many Gram (+) and essentially all Gram (−) AddB sequences lack this motif. While addA and addB are adjacent in the chromosome in most bacteria, including other epsilon Proteobacteria, this is not the case in H. pylori. Both genes that we identified as addA and addB are considered core genes that are not strain variable, since they were observed in 56 H. pylori clinical isolates from around the world (Gressmann et al., 2005).
H. pylori AddAB Has ATP-dependent Nuclease and Helicase Activity
The defining characteristic of AddAB and RecBCD enzymes is ATP-dependent DNA exonuclease activity; this nuclease is apparently active only during DNA unwinding, which requires ATP-hydrolysis. Thus, we measured this activity in wild-type, mutant, and complemented H. pylori NSH57 strains; the latter have the addA+ or addB+ gene inserted into the chromosomal rdxA locus, often used for this purpose (Smeets et al., 2000). As shown in Table 1, cytosolic extracts from wild-type bacteria showed detectable ATP-dependent nuclease activity with ds DNA substrate under conditions optimized for the H. pylori enzyme (supplemental Figure S1). Replacement of either the addA or addB gene with an antibiotic resistance cassette to create deletion (null) alleles abolished activity. ATP-dependent nuclease activity could be restored in these strains by complementation with addA+ or addB+. As a further control, we showed that disruption of two other recombination genes, recA and ruvC, had no effect on ATP-dependent nuclease activity.
Table 1.
H. pylori addA and addB Mutants Lack ATP-dependent ds DNA Exonuclease Activity
| Relevant genotypea | Allele at rdxb | ATP-dependent ds DNA exonuclease (Units/mg extract protein) |
|
|---|---|---|---|
| Meanc | Range | ||
| WT | - | 5.3 ± 0.9 (6) | 3.4 – 6.8 |
| ΔaddA::cat | - | 0.009 ± 0.0007 (6) | 0.009 – 0.010 |
| ΔaddA::cat | addA+ | 6.0 (2) | 5.7 – 6.3 |
| ΔaddB::cat | - | 0.02 ± 0.017 (6) | 0.010 – 0.040 |
| ΔaddB::cat | addB+ | 7.1 (2) | 7.0 – 7.2 |
| ΔrecA::cat | - | 6.1 ± 0.25 (3) | 5.9 – 6.4 |
| ΔruvC::cat | - | 4.8 ± 0.4 (3) | 4.3 – 5.1 |
These strains are derivatives of H. pylori strain NSH57 with the indicated deletion allele on the chromosome.
The indicated allele is at the rdx chromosomal locus. “-“ indicates wild-type rdx.
The values are the means and standard deviations of the number of extracts assayed in parentheses. For those experiments where two extracts were assayed only the mean is indicated.
To determine if addA and addB are the structural genes sufficient to confer ATP-dependent nuclease activity, we expressed these proteins in an E. coli strain deleted for recBCD. Cytosolic extracts of this strain without the addA and addB genes showed a very low level of ATP-dependent nuclease activity that was not enhanced by introduction of the vector control (Table 2). Introduction of an addAB co-expression construct (pETDuet-1 addA addB) resulted in a >40-fold increase in ATP-dependent nuclease activity. The non-inducible recBCD+ control plasmid (pMR3) resulted in a 10-fold increase in activity.
Table 2.
Cloned H. pylori addA and addB Genes Express ATP-dependent Exonuclease in E. coli Cells
| Plasmid | ATP-dependent ds DNA exonuclease (Units/mg extract protein) a | Efficiency of platingb |
|
|---|---|---|---|
| T4 | T4 2− | ||
| pBR322 | 18 ± 4 (4) | 1.0 | 1.0 |
| pMR3 (recBCD) | 140 ± 11 (6) | 0.94 | 8.3 × 10−7 |
| pETDuet-1 | 24 ± 5 (6) | 0.97 | 0.91 ± 0.04 |
| pSA405 (addA addB) | 880 ± 12(6) | 0.91 | 6.2 × 10−6 |
Extracts were prepared from transformants of E. coli strain V2831 (ΔrecBCD2731 <kan>) with pBR322 or pMR3 or transformants of E. coli strain V3060 (ΔrecBCD2731 <kan> DE3) with pETDuet-1 or its derivatives. Cells containing pETDuet-1 were harvested 3 hr after induction with 1 mM IPTG. Data are the mean ± SEM from the indicated number of extracts (n) from separate cultures.
Phage titer on E. coli strain V3060 (ΔrecBCD2731 <kan> DE3) with the indicated plasmid divided by the phage titer on strain V3060 with pBR322. Data are the mean from 3 separate experiments.
For an intracellular measure of H. pylori AddAB nuclease activity in these E. coli cells, we examined the ability of the addAB genes to restrict T4 phage infection. The T4 gene 2 protein blocks RecBCD-dependent degradation of the phage DNA upon infection, perhaps by binding to the DNA ends in the virion (Oliver and Goldberg, 1977). A T4 gene 2 mutant can productively infect an E. coli strain lacking RecBCD nuclease activity (Amundsen et al., 1990) but shows a six-log reduction in plating efficiency in recBCD+ E. coli (Table 2). Similarly, expression of addAB in the recBCD deletion strain efficiently restricted T4 2− infection. Thus, the H. pyori addAB genes confer nuclease activity both in cell-free extracts and in intact cells.
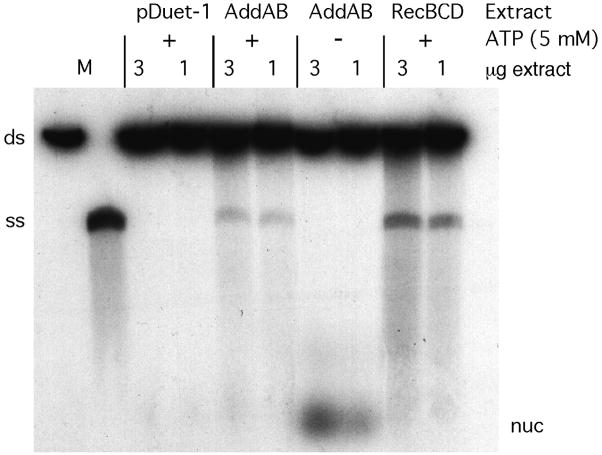
In addition to being an ATP-dependent ds DNA exonuclease, RecBCD enzyme is a highly processive DNA helicase (Taylor and Smith, 1980). We tested extracts of E. coli recBCD mutant cells expressing H. pylori addAB or E. coli recBCD for unwinding activity using linearized, 5′end-labeled pBR322 DNA as substrate. These extracts unwound linear DNA, but extracts with the vector (pETDuet-1) lacking the addA and addB genes did not (Figure 2). Unwinding activity by AddAB was ATP-dependent but slightly weaker than that by RecBCD. These results confirm that the H. pylori addA and addB genes encode an ATP-dependent helicase-nuclease similar to RecBCD enzyme of E. coli. In the absence of ATP the effective Mg2+ concentration is elevated and ATP-independent nucleases present in the extract degraded some of the substrate to oligonucleotides (Figure 2).
Figure 2.
AddAB promotes ATP-dependent DNA unwinding. Extracts were prepared from strain V3060 (ΔrecBCD2731 DE3) carrying vector pETDuet-1 with or without insertion of the H. pylori addA and addB genes or pBR322 with recBCD (pMR3). The indicated amount of extract protein was incubated with pBR322 linearized and 5′ end-labeled with 32P. Reactions contained ATP (5 mM) as indicated. The positions of ds DNA substrate (ds), unwound ss DNA (ss) produced by boiling or enzymatic reaction, and degraded DNA (nuc) are shown.
addAB and recA Mutants Are Hypersensitive to DNA Damaging Agents
Mutants lacking E. coli RecBCD or B. subtilis AddAB show increased sensitivity to several antibiotics that damage DNA (Alonso et al., 1993), as do recA mutants of several species including H. pylori (Schmitt et al., 1995, Thompson and Blaser, 1995). As expected, H. pylori addA and addB mutant strains also showed heightened sensitivity to the alkylating agent mitomycin C and the DNA gyrase inhibitor ciprofloxacin (Table 3), both of which lead to DNA ds breaks (Iyer and Szybalski, 1963, Wolfson and Hooper, 1985, Sioud and Forterre, 1989). The sensitivity observed for the addA and addB mutants was similar to that seen for a recA mutant and could be complemented by expression of the corresponding gene. We conclude that H. pylori AddAB is required for repair of intracellular ds breaks.
Table 3.
Antibiotic Sensitivity of Recombination Mutants
| Minimal inhibitory concentration |
||
|---|---|---|
| Genotypea | Mitomycin C (ng/ml) | Ciprofloxacin (μg/ml) |
| WT | 6.0 | 0.5 |
| ΔrecA::cat | 1.8 | 0.1 |
| ΔaddA::cat | 1.8 | 0.1 |
| ΔaddB::cat | 1.8 | 0.1 |
| ΔaddA:cat, rdxA::addA | 6.0 | 0.5 |
| ΔaddB::cat rdxA::addB | 6.0 | 0.5 |
NSH57 strain background.
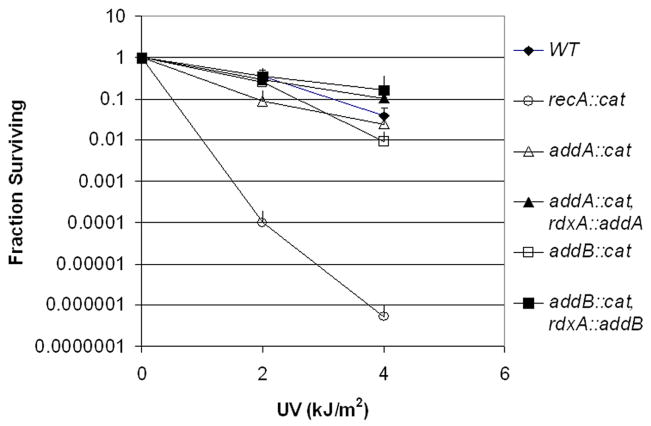
In contrast, when we examined UV sensitivity, addA and addB mutants were markedly less sensitive than a recA mutant (Figure 3), suggesting that ds break repair does not play a major role in repair of this damage in H. pylori. The modest UV sensitivity of the addA and addB mutants could be complemented by the corresponding genes. This complementation was particularly evident at the 4 kJ/m2 exposure, where the complemented strains were slightly more resistant than wild type. This enhanced resistance may result from a higher-than-wild-type level of expression when the genes are at the rdxA locus, which shows constitutive high level expression of several proteins (D. M. Pinto-Santini, L. K. Sycuro, N. R. Salama, unpublished observations).
Figure 3.
AddAB confers modest protection against UV damage. The indicated bacterial strains (wild-type, mutant, and complemented mutant strains) were exposed to UV irradiation, and the fraction of bacteria surviving was determined. The data plotted represent averages of three separate experiments, and bars indicate 1 standard deviation.
We also queried the role that H. pylori AddAB might play in homologous recombination during natural transformation with a chromosomal marker. While the recA mutant completely lost the ability to undergo natural transformation, as reported previously (Schmitt et al., 1995, Thompson and Blaser, 1995), there was no measurable difference in transformation efficiency for either the addA or addB mutant (unpublished data). Thus, while H. pylori AddAB does appear to play an important role in repair of certain types of DNA damage, it does not appear to participate in homologous recombination during natural transformation.
AddAB Enzyme and RecA Protein Are Required for Optimal Stomach Colonization
H. pylori proteins that neutralize reactive oxygen species, such as superoxide dismutase (Seyler et al., 2001) and catalase (Harris et al., 2003), promote stomach colonization. Similarly endonuclease III, a protein involved in recognition and processing of oxidized DNA, promotes stomach colonization (O’Rourke et al., 2003), suggesting that H. pylori DNA experiences oxidative damage during infection. A possible role for recombination-based repair during infection was suggested by the observation that a mutant lacking a Holliday junction resolvase homolog RuvC had persistence defects during stomach colonization (Loughlin et al., 2003). Therefore, we investigated the role that AddAB and RecA, proteins whose homologs promote the early steps of recombination-based repair, might play in stomach colonization.
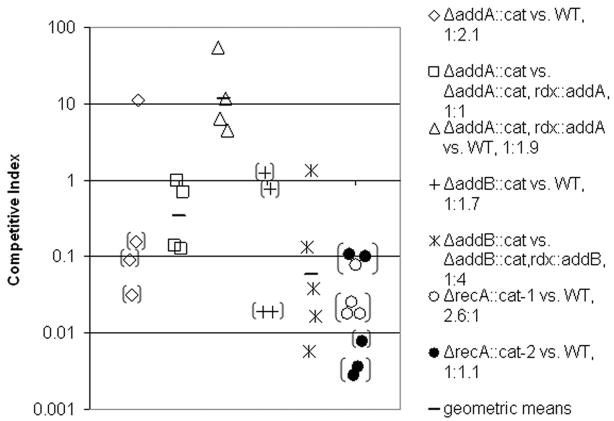
We first performed competition experiments by oral infection with 1:1 mixtures of mutant and either wild type or complemented mutant in the NSH57 strain background. We allowed the infection to continue for one week and then harvested the bacteria from the stomachs. Plating on selective and non-selective media allowed enumeration of mutant and total bacteria. A competitive index was computed for each animal as the ratio of mutant to wild-type (or complemented mutant) bacteria recovered after one week, adjusted for the ratio of strains in the inocula. While there is considerable mouse-to-mouse variation in the assay, the average competitive index of either addA or addB mutants in competition with either wild type or the complemented mutant was below 1, indicating a colonization defect for the mutants (Figure 4). Curiously, when the addA complemented strain was competed with wild type, the competitive index was above 1. As described above, expression of addA from the rdxA locus may result in a higher expression level that is protective under some circumstances. We were unable to complement the recA mutant clone because of the requirement of RecA for natural transformation. However, two independently generated recA mutant clones both yielded even lower competitive indices than addA or addB mutants (Figure 4). While we recovered at least some mutant bacteria in 5 of 8 addA mutant and 5 of 9 addB mutant competitions with wild type or complemented strains, we never recovered recA mutant bacteria in competition experiments. All three mutants (recA, addA, and addB) showed comparable growth to wild type during in vitro culture (supplemental Figure S2).
Figure 4.
AddAB and RecA promote stomach colonization. Mice were orally infected with a 1:1 mixture of the indicated strains. The actual ratio in the inoculum of mutant to wildtype or complemented mutants is indicated. After one week the bacteria colonizing the stomach were harvested and the competitive index determined (ratio of mutant to wild type in the output corrected for the input ratio) as described in the Experimental Procedures. Each data point is from one mouse, and the geometric means are indicated by horizontal bars. Infections with two independent ΔrecA::cat clones are indicated by open and filled circles. Bracketed data points represented an upper limit on the competitive index with 95% confidence because of failure to isolate mutant clones from the stomach as described in the Experimental Procedures. Competitions of mutant clones with wild type were repeated with similar results (data not shown). A colonization defect is indicated by a competitive index of less than one.
In order to gain further insight into the infection potential of our H. pylori strains we infected groups of five animals with decreasing titers of individual strains to determine the dose required for detectable infection of 50% of the animals (ID50) (supplemental table S4). In this experiment the ID50 was 2.3 × 104 bacteria for the wild-type strain, 2.0 × 107 for the addA mutant strain, and greater than 2.4 × 1010 for the recA mutant strain. We recovered bacteria from only a single animal at a high infecting dose of recA mutant bacteria. These results mirror the data from the competition experiments showing significantly attenuated colonization by strains lacking AddAB activity and essentially no colonization by strains lacking RecA.
Recombination Proteins Promote Apparent Gene Conversion at the babA Locus
As mentioned above, previous work demonstrated that mutants lacking RuvC, a protein required for resolving recombination intermediates, also have partially attenuated stomach persistence (Loughlin et al., 2003). Interestingly, further studies suggested that RuvC function and, by inference, recombination facilitate bacterial immune evasion by altering the adaptive immune response (Robinson et al., 2005). The mechanisms by which the immune system becomes redirected remain obscure. We previously showed that H. pylori can abolish babA-dependent adhesion by a gene conversion-like event between babA and a related locus, babB, and that this event is selected during infection of some hosts (Solnick et al., 2004). We investigated whether this gene conversion event requires recombination protein activity. To do this, we created null alleles of recA, addA, and ruvC in the J166 strain background where we had previously measured spontaneous babA to babB gene conversion during in vitro culture (Solnick et al., 2004). Real-time PCR was used to quantify the frequency of babB at the babA locus in bacterial populations expanded from single colonies in vitro. The wild-type J166 strains showed a frequency of 3 × 10−5, corresponding to an approximate rate of gene conversion of 3 × 10−6 per cell division (Lea and Coulson, 1949). As shown in Figure 5, loss of AddA or RecA significantly reduced the frequency of the babA to babB gene conversion. In contrast loss of RuvC only slightly reduced the frequency of gene conversion, and this difference was not statistically significant. The limit of detection of our assay for the frequency of convertants is approximately 4 × 10−7. These results suggest that this gene conversion event frequently occurs by a RecA-dependent mechanism that may involve a ds break, since it is AddAB-dependent.
Figure 5.
AddA and RecA promote babA to babB gene conversion. Quantitative PCR was used to determine the number of copies of babA and babB at the babA locus for genomic DNA prepared from each of three cultures started from individual colonies as described in the Experimental Procedures. The geometric mean of the frequency of babB/babA at the babA locus is reported and the bars indicate 1 standard deviation. The difference in means between groups was considered significant (one-way analysis of variance, p = 0.0002). Significant P values for pair-wise comparisons between groups are indicated **p <0.001, *p <0.01.
DISCUSSION
We report here that the DNA double-strand (ds) break repair enzyme AddAB and the homologous recombination-promoting protein RecA are required for high-level infection by H. pylori. AddAB is a functional homolog of the RecBCD enzyme of E. coli, Salmonella enterica serovar Typhimurium, and Neisseria gonorrhoea. The E. coli enzyme is crucial for repair of DNA breaks and genetic recombination involving linear DNA (Smith, 2001), and the recC gene appears to be under positive selection in uropathogenic strains of E. coli (Chen et al., 2006). N. gonorrhoeae recB, recC, and recD mutants are more sensitive than wild-type strains to hydrogen peroxide (Stohl and Seifert, 2006). We discuss below the properties of H. pylori AddAB enzyme and its role in colonization of the stomach of mice and compare its role with that of RecA protein, which is also required for DNA repair as well as homologous recombination.
We identified the addA and addB genes using BLAST searches that started with a consensus sequence for RecB. This polypeptide contains both the canonical seven helicase motifs (Bork and Koonin, 1993) and a nuclease domain (Yu et al., 1998). Although the helicase motifs are highly conserved among a large group of helicases with highly divergent cellular functions, the nuclease domain appears to be unique to RecB-related polypeptides. This dual criterion in our searches may account for our finding the RecB-related polypeptide AddA of H. pylori, whereas previous searches were not successful. The AddB polypeptide also contains a closely related nuclease domain and a large region with only scant similarity to helicases. These properties allowed us to identify H. pylori AddB. Identification of the H. pylori addA and addB genes allowed us to discover further that AddAB proteins are in fact well conserved among all the sequenced epsilon Proteobacteria in spite of the fact they have not been annotated in most sequencing projects (Rocha et al., 2005)
Although the addA and addB genes of many groups of bacteria are adjacent and appear to form an operon, the H. pylori addA and addB genes are not; they are separated by approximately 500 kb. We suppose that the AddA and AddB polypeptides act together in a complex, as do the RecBCD polypeptides and AddAB polypeptides of other bacteria investigated (Kooistra and Venema, 1991, Taylor and Smith, 1995). As noted below, the phenotypes of H. pylori addA and addB mutants are indistinguishable, as expected if the polypeptides act in a complex. If so, the control of the unlinked addA and addB genes to maintain the proper stoichiometry of the two polypeptides remains an interesting question.
By assaying extracts of H. pylori, we detected an ATP-dependent nuclease (Table 1 and Figure S1), the defining characteristic of the RecBCD class of enzymes, also called exonuclease V. The ATP-dependence of the nuclease activity is a consequence of DNA degradation occurring only during unwinding, which requires the energy of ATP hydrolysis. ATP-dependent nuclease activity was undetectable in addA and addB mutants; as expected, this loss was complemented by insertions of the corresponding genes at a distant locus (Table 1). These genes also conferred ATP-dependent nuclease and ATP-dependent DNA unwinding activity to an E. coli recBCD deletion mutant (Table 2 and Figure 2), indicating that they are the structural genes for this enzyme. Activity was detected both in extracts and in intact cells, which blocked the growth of a phage T4 mutant lacking a protein that protects linear DNA from nuclease digestion. Expression of these genes in E. coli provides a way to make large amounts of H. pylori AddAB enzyme for further analysis.
Our phenotypic analyses suggest that, like the E. coli RecBCD enzyme, H. pylori AddAB functions to repair DNA damage that results in ds breaks; both addA and addB mutants were highly sensitive in vitro to the alkalating agent mitomycin C and the topoisomerase inhibitor ciprofloxacin but only slightly sensitive to UV-induced damage (Table 3). While addA and addB mutants grow well in vitro in the absence of overt DNA damage (Figure S2), these strains have a lower colonization potential in both competition experiments and during single-strain infections (Figure 4, supplementary table S4 and data in Results). This may result from DNA damage specifically encountered in the host environment.
Previous studies of enteric pathogens revealed a role for recombinational repair proteins but apparently not recombination per se during infection. Salmonella enterica serovar Typhimurium recBC mutants are severely attenuated for infection and killing, but suppressors that restore recombinational repair by activating the RecFOR pathway of homologous recombination do not suppress the in vivo defects (Buchmeier et al., 1993, Cano et al., 2002). Moreover, recA mutants, which essentially lack homologous recombination, show a milder phenotype than recBC Salmonella mutants, and loss of RecA does not impact colonization by the E. coli extracellular pathogens EHEC or UPEC (Fuchs et al., 1999). Based on double mutant analyses, the essential in vivo function of RecBCD enzyme during Salmonella infection appears to be restoration of stalled replication forks (Schapiro et al., 2003), not recombination. In contrast, our results with H. pylori, reported here, indicate a direct role for homologous recombination in stomach infection.
Our results show that two enzymes, AddAB and RecA, which process damaged DNA, enhance the ability of H. pylori strains to colonize the stomach (Figure 4 and lower ID50). In contrast to the Salmonella results, loss of H. pylori RecA causes even more severe attenuation of stomach colonization than loss of AddAB. This result suggests that H. pylori experiences a different spectrum of DNA damage during infection than that encountered by Salmonella, and that this spectrum includes ds breaks. Furthermore, recombination functions are essential either to repair DNA damage or for other recombination protein-mediated processes early in H. pylori infection.
Unlike Salmonella, H. pylori primarily remains extracellular where it is not exposed to phagosome-specific host defenses, although recent studies suggest that a small sub-population of H. pylori do reside in an intracellular niche primarily in epithelial cells (Amieva et al., 2002, Aspholm et al., 2006, Necchi et al., 2007). The different cellular and tissue tropisms of these bacteria may account for the different recombination-protein requirements for successful infection of Salmonella and H. pylori. Several lines of evidence suggest that even in its extracellular niche, H. pylori is exposed to oxidative damage soon after infection. Recent work using the mouse model showed H. pylori-dependent infiltration of neutrophils and macrophages one and two days post-infection which then decreased to low levels at three and ten days post-infection, suggesting a rapid innate immune response to infection that is then down-regulated (Algood et al., 2007). Because neutrophil infiltration is a hallmark of human H. pylori infection, the interaction of H. pylori and cultured neutrophils has been studied in some detail. Interestingly, while H. pylori is readily taken up by neutrophils and is a potent activator of the phagosome NADPH oxidase, active flavocytochrome b558 complex does not assemble on the H. pylori-containing phagosome and instead is redirected to the plasma membrane, leading to extracellular superoxide accumulation (Allen et al., 2005). H. pylori also induce both macrophages (Chaturvedi et al., 2004) and epithelial cells (Xu et al., 2004) to produce extracellular hydrogen peroxide by stimulating polyamine oxidase. Finally, isolated gastric pit cells express the phagosome NADPH oxidase components at the plasma membrane and show measurable constitutive extracellular superoxide production that is further induced after exposure to H. pylori-derived LPS (Teshima et al., 1999). These innate immune responses likely contribute to host cellular damage that may benefit H. pylori but also necessitates bacterial mechanisms to combat oxidative damage to its DNA, proteins and lipids. AddAB appears to play such a role for H. pylori.
Loss of several H. pylori proteins shown or annotated to recognize DNA damage cause lower colonization loads or decreased persistence of H. pylori strains in the mouse model, suggesting that the bacteria experience DNA damage stresses during infection. These proteins include HP0585, a homolog of E. coli endonuclease III, which repairs oxidized pyrimidine residues (O’Rourke et al., 2003), MutS2, which in H. pylori recognizes and binds 8-oxoguanine (Wang et al., 2005), two DNA glycosylases (Baldwin et al., 2007), and a putative RecN homolog (Wang and Maier, 2008). In E. coli and B. subtillis, RecN promotes RecBCD (AddAB)-dependent ds break repair under some stress conditions and is recruited to large damage foci in the absence of ds break repair (Meddows et al., 2005, Sanchez et al., 2006) Thus, H. pylori RecN may interact with AddAB to promote ds break repair during stomach colonization.
Our in vitro data suggest that H. pylori AddAB, like E. coli RecBCD, functions specifically at ds breaks. Mutants with loss of AddAB or RecA show equivalent sensitivity to chemicals leading to ds breaks, but the recA mutant is much more sensitive to UV exposure than addAB mutants (Table 3, Figure 3). These results suggest that an additional RecA-dependent pathway operates in H. pylori to repair damage induced by UV. A likely candidate is an analog of the E. coli RecFOR pathway. A recR homolog has been annotated in the H. pylori genome (Tomb et al., 1997). The more severe stomach colonization phenotype of recA mutants than of addAB mutants (supplemental table S4) may result from an additional requirement for the RecFOR DNA repair pathway during infection. The role of RecA in competence for natural transformation may also contribute to the more severe attenuation of this mutant. Unlike other bacteria, such as Rhizobium, Bacillus, and Neisseria (Haijema et al., 1996, Mehr and Seifert, 1998, Zuniga-Castillo et al., 2004), H. pylori AddAB does not appear to contribute to competence, while RecA is absolutely required (Schmitt et al., 1995). Two studies have suggested that competence contributes to stomach colonization, even at early time points (Kavermann et al., 2003, Baldwin et al., 2007). Natural transformation is thought to contribute to genetic iversification of the H. pylori population later in infection by allowing new alleles to spread through the population via recombination (Suerbaum and Josenhans, 2007). Multiple mutant analyses involving addAB, recR, and com (DNA transformation competence) genes should begin to address the relative importance of these pathways during infection.
RecA and AddAB may also contribute to long-term adaptation to the host environment. We discovered a role for RecA and AddAB in promoting gene conversion between two outer membrane protein (OMP) genes, babA and babB (Figure 5). H. pylori genomes encode a large number of OMPs (60), some of which have been annotated as porins, adhesins or outer membrane transporters. Subsets of OMPs have been grouped into paralogous families suggested to be at least partially redundant (Alm et al., 2000). The largest family of OMPs is the Hops which include the Lewis B blood group antigen-binding adhesin BabA and two highly related Hops BabB and BabC (Alm et al., 2000, Hennig et al., 2006). Gene conversion via conserved 5′- and 3′-terminal sequences in babB or babC can eliminate babA adhesin-gene expression. Loss of babA expression has been observed in a majority of clones from the infecting bacterial population after the initial colonization event (between four and eight weeks post infection) during experimental monkey infection (Solnick et al., 2004), and BabA can be encoded by one or more of the three bab loci (Solnick et al., 2004, Colbeck et al., 2006, Hennig et al., 2006). Loss of BabA protein from the cell surface may result in an altered immune response or, alternatively, modify bacterial tropism by changing host receptor-binding interactions. In Neisseria, pilin antigenic variation is mediated by a RecA-dependent gene conversion event that also requires the RecFOR complex (Mehr and Seifert, 1998) or the RecBCD complex (Hill et al., 2007), depending on the strain background. While an engineered gene conversion event in H. pylori is RecA-dependent (Pride and Blaser, 2002), the requirement for RecA and other recombination proteins for bab locus conversion had not been examined. We find that the babA to babB gene conversion event significantly depends on RecA and AddAB (Figure 5). If AddAB, like RecBCD, depends on a DNA end for activity, this result suggests that the mechanism of this conversion involves a ds break.
Colonization by H. pylori of its host for decades is required for development of disease. The data presented here suggest that recombinational repair proteins, including AddAB and RecA, play multiple roles during infection. The in vivo phenotypes we report here after one week of infection likely result from the requirement for these proteins to combat DNA damage stress induced soon after infection. Interestingly, while recA mutants have a very severe phenotype, addAB mutants can still colonize. This outcome will allow study of genetic diversification during long-term colonization to further dissect additional roles of recombinational repair proteins during infection. Recombinational repair has been suggested as a target that could enhance the efficacy of other antibiotics that lead to intracellular oxidative stress (Kohanski et al., 2007). Our work suggests that for H. pylori and perhaps other bacteria AddAB could be a promising direct target for a novel antimicrobial drug, since this class of enzymes is widely distributed in prokaryotes but not in eukaryotes.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth
E. coli strains (Table S1) were grown in media containing Difco tryptone and yeast extract (LB), Terrific Broth (Fisher), or Difco tryptone (TB). These media, phage suspension medium (SM), and top agar have been described (Cheng and Smith, 1989). H. pylori strains (Table S1) were grown on solid horse blood agar (HB) plates containing 4% Columbia agar base (Oxoid), 5% defibrinated horse blood (HemoStat Labs), 0.2% β-cyclodextrin (Sigma), vancomycin (Sigma; 10 μg/ml), cefsulodin (Sigma; 5 μg/ml), polymyxin B (Sigma; 2.5 U/ml), trimethoprim (Sigma; 5 μg/ml), and amphotericin B (Sigma; 8 μg/ml) at 37°C either under a microaerobic atmosphere generated using a CampyGen sachet (Oxoid) in a gas pack jar or in an incubator equilibrated with 14% CO2 and 86% air. For liquid culture, H. pylori was grown in Brucella broth (Difco) containing 10% fetal bovine serum (BB10; Invitrogen) with shaking in a gas pack jar with a CampyGen sachet. For antibiotic resistance marker selection, bacterial media were additionally supplemented with ampicillin (100 μg/ml), kanamycin (50 μg/ml), chloramphenicol (Cm; 15 μg/ml) or metronidazole (Mtz; 36 μg/ml). When culturing bacteria from mouse stomachs, Bacitracin (Bac; 200 μg/ml) was added to eliminate normal mouse microbiota contamination.
DNA Manipulations
DNA manipulations, such as restriction digestion, PCR, and agarose gel electrophoresis, were performed according to standard procedures (Ausubel et al., 1997). Genomic DNA was prepared from H. pylori by Wizard genomic DNA preparation kits (Promega). Primers used for PCR and sequencing are in Table S2, and plasmids used in this study are in Table S3.
Generation of H. pylori Isogenic Knockout Mutants
In the NSH57 strain background, null alleles of addA, addB, recA, and ruvC were constructed using a vector-free allelic replacement strategy to generate alleles in which a chloramphenicol acetyl transferase resistance cassette replaced 80–90% of the coding sequence of the gene while preserving the start and stop codons (Chalker et al., 2001, Salama et al., 2004). The primers used for this procedure are in Table S2. The resistance cassette contains its own promoter but lacks a transcriptional terminator and in all cases was inserted in the same direction of transcription as that of the native gene. After natural transformation (Wang et al., 1993) with the appropriate PCR product and selection on Cm-containing media, four to eight clones were evaluated by PCR to confirm replacement of the wild-type allele with the null allele; urease activity and flagella-based motility were also confirmed. Single clones were used for infection experiments and phenotypic characterization. J166 deletion mutants of recA, addA, and ruvC were constructed by allelic exchange using methods similar to those previously described (Akopyants et al., 1998). Briefly, a non-polar kanamycin resistance (aphA) cassette (Menard et al., 1993) and ~1 kb upstream and downstream “arms” for each gene were PCR amplified using primers (Table 3) with compatible 5′ restriction sites. All three fragments were restricted with the appropriate enzymes, ligated with pBluescript SK(-) (Stratagene, La Jolla, CA) plasmid DNA which was previously digested with XhoI/NotI, and transformed into One Shot TOP10 competent E. coli (Invitrogen, Carlsbad, CA) with kanamycin selection. Each shuttle plasmid was verified by sequencing and transferred into H. pylori J166 using natural transformation with kanamycin selection, thereby deleting most of the coding region of each gene.
Generation of E. coli Expression Constructs and H. pylori Complementation Constructs
The E. coli expression construct pJF30 containing both AddA and AddB in pETDuet-1 (Novagen) was made by separately amplifying addA (HP1553) and addB (HP1089) from H. pylori strain 26695 using primers AddA-C1(SalI), AddA-N1(NcoI), AddB N1 (NdeI), and AddB C1 (AvrII). The reaction conditions included six cycles with 54°C annealing temperature followed by 24 cycles with 62°C annealing temperature and used High Fidelity Taq and Supermix (Invitrogen). Each gene was separately cloned into pETDuet-1 after digestion of the PCR product and the vector with the indicated restriction enzymes using standard procedures to generate pJF25 (addA), and pJF22 (addB). pJF30 was made by subcloning addB from pJF22 into pJF25 using NdeI and AvrII. Vectors for complementation were made by subcloning each gene individually into pRdxA and introduced into H. pylori NSH57 by natural transformation and selection on Mtz-containing media (Smeets et al., 2000). The addA complementing vector pJF29 was made by subcloning addA from pJF31 using XbaI and SalI. The addB complementing vector pJF27 was made by subcloning addB from pJF22 using AvrII and XbaI. All inserted genes contained the expected nucleotide sequences, except for addB in pJF22, pJF30, pJF31, and pJF27, which contained a single point mutation [T2311 → C2311] changing serine 771 to proline (S771P). This residue is not part of any conserved domain of AddB, and the S771P-containing clone fully complemented drug sensitivity and animal infectivity in H. pylori (Table 3 and Figure 4). For assaying enzymatic activities, this mutation was repaired using QuikChange (Strategene) on the pJF30 template to generate pSA405.
Preparation of Cell-free Extracts and Enzymatic Assays
Extracts were prepared as described by (Tomizawa and Ogawa, 1972). For H. pylori extracts bacteria were harvested from 24 – 48 hour plate-grown cultures. For E. coli extracts bacteria were harvested 3 hr after addition of 1 mM IPTG to induce expression of AddA and AddB. ds exonuclease activity was assayed as ATP-dependent solubilization of uniformly [3H]-labeled T7 DNA (specific activity 2×104 cpm/μg; Eichler and Lehman, 1977). Each assay included two or three protein concentrations that gave a linear relationship between acid-solubilized DNA and protein assayed. Assays of haploid H. pylori extracts contained 10 mM MgCl2 and 1 mM ATP (see Figure S1). Assays of H. pylori AddAB in E. coli extracts contained 50 μM ATP (Table 2).
The substrate for DNA unwinding was plasmid pBR322 digested with HindIII (New England Biolabs), treated with shrimp alkaline phosphatase (US Biochemicals), and labeled at the 5′ ends with [γ-32P] ATP (GE Biosciences). Unincorporated nucleotides were separated from the DNA substrate by passage through an SR200 minicolumn (GE Biosciences).
DNA unwinding assay mixtures contained 4.0 nM DNA substrate in 15 μl of buffer containing 25 mM Tris acetate (pH 7.5), 2 mM magnesium acetate, 5 mM ATP, 1 μM DTT and 1 μM single-stranded DNA binding protein (Promega). Reactions were for 2 min at 37°C with the amount of extract protein indicated in Figure 1. Reactions were terminated by the addition of 5 μl of buffer containing 0.1 M EDTA, 2.5% SDS, 0.125% bromophenol blue, 0.125% xylene cyanol, and 10% ficoll. Reaction products were separated on a 0.7% agarose gel (22 cm long) in Tris acetate electrophoresis (TAE) buffer (Sambrook et al, 1989) at 100 V for 2 hr and visualized by autoradiography.
Efficiency of Plaque-formation by Phage T4 and T4 2−
E. coli strain V3060 (chromosomal ΔrecBCD) containing plasmids bearing the E. coli recBCD or H. pylori addAB genes was grown in TB broth containing ampicillin (100 ug/ml) to about 2 × 108 cells/ml. Phage T4 or T4 2− in SM (50 μl) were added to 0.1 ml of bacteria and incubated for 15 min at 37°C. Top agar (2.5 ml) was added and the mixture poured onto a TB agar plate. Plates were incubated at 37°C overnight, after which plaques were enumerated.
Antibiotic Resistance Testing
Bacteria taken from fresh plates (incubated for 18 – 36 hr) were grown in liquid culture to an optical density at 600 nm (OD600) between 0.1 and 1 and examined for spiral shape and motility. Five fold serial dilutions were spotted onto plates containing increasing concentrations of mitomycin C or ciprofloxacin (Sigma). The minimal inhibitory concentration was determined when drug prevented growth of at least two five-fold serial dilutions.
UV Sensitivity
Bacteria were grown as described for antibiotic resistance testing. Dilutions were plated in duplicate, each on one half of a plate, and exposed to UV using a UV Stratalinker 2400 (Stratagene) on the energy setting. After UV exposure, plates were incubated for 3 – 4 days until single colonies could be counted. The percent survival was calculated in comparison to plates that were mock UV treated. The data presented are the average from three experiments.
Mouse Infections
Female C57BL/6 mice 24 – 28 days old were obtained from Charles River Laboratories and certified free of endogenous Helicobacter infection by the vendor. The mice were housed in sterilized microisolator cages with irradiated PMI 5053 rodent chow, autoclaved corn cob bedding, and acidified, reverse-osmosis purified water provided ad libitum. All studies were done under practices and procedures of Animal Biosafety Level 2. The facility is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care, International, and all activities were approved by the FHCRC Institutional Animal Care and Use Committee.
For competition experiments each indicated null mutant strain and the parental wild-type strain or complemented mutant were grown from frozen stock in liquid culture to mid-to-late logarithmic growth phase. The wild-type and mutant bacteria were combined to give approximately 5.0 × 108 bacteria of each in 2.5 ml, and 0.5 ml inoculated by oral gavage into each of five mice. After inoculation, a portion of the inoculum was plated on HB-Bac plates and HB-Cm plates to determine the number of wild-type and mutant bacteria, respectively, in the inocula. After 1 week the mice were euthanized by inhalation of CO2. The glandular stomach was removed and cut along the greater curvature, and any food was removed. The stomach was then cut in half longitudinally, and one half stomach placed in 0.5 mL of BB10 broth for homogenization with disposable pellet pestles (Fisher Scientific). Dilutions of the homogenate were plated on HB-Bac to enumerate total bacteria and on HB-Cm plates to enumerate mutant bacteria. If no mutant bacteria were recovered we set the number of colonies on the lowest dilution plated to three. From the Poisson distribution we estimate with 95% confidence that a plate containing zero colonies corresponds to three or less colony forming units (CFU). The average bacterial colonization load for wild type in these experiments was 1.3 × 105 CFU/g and the range was 3.3 × 103 – 9.6 × 105 CFU/g. The competitive index (CI) was computed as follows: CI = (output mutant CFU/output wild-type CFU)/(input mutant CFU/input wild-type CFU).).
To determine the dose for fifty percent infection (ID50), the indicated strains were grown in liquid culture to mid-to-late logarithmic growth phase and concentrated to give 5 × 109 – 5 × 1010 bacteria per ml. Serial ten-fold dilutions were prepared and inoculated into each of five mice. After 7 – 10 days mice were euthanized and processed as described above; bacteria were enumerated on HB-Bac plates. The number of infected mice at each titer was determined (supplemental table S4), and the ID50 was calculated using the method of Reed and Muench (Reed and Muench, 1938).
babA Gene Conversion Assay
Real-time quantitative PCR was used to determine the frequency of apparent gene conversion of babA to babB, using methods similar to those described (Solnick et al., 2004). Briefly, genomic DNA was prepared from individual colonies of wild-type H. pylori J166, and from isogenic mutants with deletions of recA, addA, or ruvC. Each 20 μl reaction contained 10 μl of iQ SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA), 5 μl of primer pair (1 μM each) containing a primer (Table S3) upstream of the babA locus (F14) and a primer in either babA (160R) or babB (178R), and 5 μl of DNA template (0.0375 ng/μl for babA; 5 ng/μl for babB). Amplification was carried out in a Bio-Rad iCycler (3 min at 95°C; 45 cycles of 30 sec at 95°C, 30 sec at 58°C, 1 min at 72°C) and the cycle threshold (Ct) was determined. Standard curves were constructed for babA and babB using plasmids (Table S2) in which babA (pJ150) or babB (pJ151) was amplified and cloned into pGEM-T Easy (Promega, Madision, WI) using TA cloning. Template for amplification of the babA and babB fragments was chromosomal DNA from wild-type H. pylori J166 (babA) or H. pylori J166 passaged through rhesus macaques (babB), in which the babA gene was converted by babB. Primers for amplification of babA and babB (Table S3) were therefore the same, since the upstream primer (HP0898F) was not in babA/babB and the downstream primer (AN5954) was in a region in which babA and babB are identical.
Acknowledgments
This project was supported by grants AI054423 (NRS), GM031693 (GRS), and AI42081 (JVS) from the National Institutes of Health of the United States of America and a grant from the FHCRC Synergy Fund (NRS and GRS). We thank Marion Dorer for helpful discussions.
References
- Akopyants NS, Clifton SW, Kersulyte D, Crabtree JE, Youree BE, Reece CA, et al. Analyses of the cag pathogenicity island of Helicobacter pylori. Mol Microbiol. 1998;28:37–53. doi: 10.1046/j.1365-2958.1998.00770.x. [DOI] [PubMed] [Google Scholar]
- Algood HM, Gallo-Romero J, Wilson KT, Peek RM, Jr, Cover TL. Host response to Helicobacter pylori infection before initiation of the adaptive immune response. FEMS Immunol Med Microbiol. 2007;51:577–586. doi: 10.1111/j.1574-695X.2007.00338.x. [DOI] [PubMed] [Google Scholar]
- Allen LA, Beecher BR, Lynch JT, Rohner OV, Wittine LM. Helicobacter pylori disrupts NADPH oxidase targeting in human neutrophils to induce extracellular superoxide release. J Immunol. 2005;174:3658–3667. doi: 10.4049/jimmunol.174.6.3658. [DOI] [PubMed] [Google Scholar]
- Alm RA, Bina J, Andrews BM, Doig P, Hancock RE, Trust TJ. Comparative genomics of Helicobacter pylori: analysis of the outer membrane protein families. Infect Immun. 2000;68:4155–4168. doi: 10.1128/iai.68.7.4155-4168.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JC, Stiege AC, Luder G. Genetic recombination in Bacillus subtilis 168: effect of recN, recF, recH and addAB mutations on DNA repair and recombination. Mol Gen Genet. 1993;239:129–136. doi: 10.1007/BF00281611. [DOI] [PubMed] [Google Scholar]
- Amieva MR, Salama NR, Tompkins LS, Falkow S. Helicobacter pylori enter and survive within multivesicular vacuoles of epithelial cells. Cell Microbiol. 2002;4:677–690. doi: 10.1046/j.1462-5822.2002.00222.x. [DOI] [PubMed] [Google Scholar]
- Amundsen SK, Neiman AM, Thibodeaux SM, Smith GR. Genetic dissection of the biochemical activities of RecBCD enzyme. Genetics. 1990;126:25–40. doi: 10.1093/genetics/126.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amundsen SK, Smith GR. Interchangeable parts of the Escherichia coli recombination machinery. Cell. 2003;112:741–744. doi: 10.1016/s0092-8674(03)00197-1. [DOI] [PubMed] [Google Scholar]
- Amundsen SK, Taylor AF, Chaudhury AM, Smith GR. recD: the gene for an essential third subunit of exonuclease V. Proc Natl Acad Sci U S A. 1986;83:5558–5562. doi: 10.1073/pnas.83.15.5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DG, Kowalczykowski SC. The recombination hot spot chi is a regulatory element that switches the polarity of DNA degradation by the RecBCD enzyme. Genes Dev. 1997;11:571–581. doi: 10.1101/gad.11.5.571. [DOI] [PubMed] [Google Scholar]
- Aspholm M, Olfat FO, Norden J, Sonden B, Lundberg C, Sjostrom R, et al. SabA is the H. pylori hemagglutinin and is polymorphic in binding to sialylated glycans. PLoS Pathog. 2006;2:e110. doi: 10.1371/journal.ppat.0020110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Short Protocols in Molecular Biology. New York: John Wiley & Sons; 1997. [Google Scholar]
- Baldwin DN, Shepherd B, Kraemer P, Hall MK, Sycuro LK, Pinto-Santini DM, Salama NR. Identification of Helicobacter pylori genes that contribute to stomach colonization. Infect Immun. 2007;75:1005–1016. doi: 10.1128/IAI.01176-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P, Koonin EV. An expanding family of helicases within the ‘DEAD/H’ superfamily. Nucleic Acids Res. 1993;21:751–752. doi: 10.1093/nar/21.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchmeier NA, Lipps CJ, So MY, Heffron F. Recombination-deficient mutants of Salmonella typhimurium are avirulent and sensitive to the oxidative burst of macrophages. Mol Microbiol. 1993;7:933–936. doi: 10.1111/j.1365-2958.1993.tb01184.x. [DOI] [PubMed] [Google Scholar]
- Cano DA, Pucciarelli MG, Garcia-del Portillo F, Casadesus J. Role of the RecBCD recombination pathway in Salmonella virulence. J Bacteriol. 2002;184:592–595. doi: 10.1128/JB.184.2.592-595.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker AF, Minehart HW, Hughes NJ, Koretke KK, Lonetto MA, Brinkman KK, et al. Systematic identification of selective essential genes in Helicobacter pylori by genome prioritization and allelic replacement mutagenesis. J Bacteriol. 2001;183:1259–1268. doi: 10.1128/JB.183.4.1259-1268.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi R, Cheng Y, Asim M, Bussiere FI, Xu H, Gobert AP, et al. Induction of polyamine oxidase 1 by Helicobacter pylori causes macrophage apoptosis by hydrogen peroxide release and mitochondrial membrane depolarization. J Biol Chem. 2004;279:40161–40173. doi: 10.1074/jbc.M401370200. [DOI] [PubMed] [Google Scholar]
- Chedin F, Kowalczykowski SC. A novel family of regulated helicases/nucleases from Gram-positive bacteria: insights into the initiation of DNA recombination. Mol Microbiol. 2002;43:823–834. doi: 10.1046/j.1365-2958.2002.02785.x. [DOI] [PubMed] [Google Scholar]
- Chen SL, Hung CS, Xu J, Reigstad CS, Magrini V, Sabo A, et al. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. Proc Natl Acad Sci U S A. 2006;103:5977–5982. doi: 10.1073/pnas.0600938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng KC, Smith GR. Distribution of Chi-stimulated recombinational exchanges and heteroduplex endpoints in phage lambda. Genetics. 1989;123:5–17. doi: 10.1093/genetics/123.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbeck JC, Hansen LM, Fong JM, Solnick JV. Genotypic profile of the outer membrane proteins BabA and BabB in clinical isolates of Helicobacter pylori. Infect Immun. 2006;74:4375–4378. doi: 10.1128/IAI.00485-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler DC, Lehman IR. On the role of ATP in phosphodiester bond hydrolysis catalyzed by the recBC deoxyribonuclease of Escherichia coli. J Biol Chem. 1977;252:499–503. [PubMed] [Google Scholar]
- Falush D, Wirth T, Linz B, Pritchard JK, Stephens M, Kidd M, et al. Traces of human migrations in Helicobacter pylori populations. Science. 2003;299:1582–1585. doi: 10.1126/science.1080857. [DOI] [PubMed] [Google Scholar]
- Fuchs S, Muhldorfer I, Donohue-Rolfe A, Kerenyi M, Emody L, Alexiev R, et al. Influence of RecA on in vivo virulence and Shiga toxin 2 production in Escherichia coli pathogens. Microb Pathog. 1999;27:13–23. doi: 10.1006/mpat.1999.0279. [DOI] [PubMed] [Google Scholar]
- Gressmann H, Linz B, Ghai R, Pleissner KP, Schlapbach R, Yamaoka Y, et al. Gain and loss of multiple genes during the evolution of Helicobacter pylori. PLoS Genet. 2005;1:e43. doi: 10.1371/journal.pgen.0010043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haijema BJ, Noback M, Hesseling A, Kooistra J, Venema G, Meima R. Replacement of the lysine residue in the consensus ATP-binding sequence of the AddA subunit of AddAB drastically affects chromosomal recombination in transformation and transduction of Bacillus subtilis. Mol Microbiol. 1996;21:989–999. doi: 10.1046/j.1365-2958.1996.601424.x. [DOI] [PubMed] [Google Scholar]
- Harris AG, Wilson JE, Danon SJ, Dixon MF, Donegan K, Hazell SL. Catalase (KatA) and KatA-associated protein (KapA) are essential to persistent colonization in the Helicobacter pylori SS1 mouse model. Microbiology. 2003;149:665–672. doi: 10.1099/mic.0.26012-0. [DOI] [PubMed] [Google Scholar]
- Hennig EE, Allen JM, Cover TL. Multiple chromosomal loci for the babA gene in Helicobacter pylori. Infect Immun. 2006;74:3046–3051. doi: 10.1128/IAI.74.5.3046-3051.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill SA, Woodward T, Reger A, Baker R, Dinse T. Role for the RecBCD recombination pathway for pile gene variation in repair-proficient Neisseria gonorrhoeae. J Bacteriol. 2007;189:7983–7990. doi: 10.1128/JB.00980-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer VN, Szybalski W. A Molecular Mechanism of Mitomycin Action: Linking of Complementary DNA Strands. Proc Natl Acad Sci U S A. 1963;50:355–362. doi: 10.1073/pnas.50.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavermann H, Burns BP, Angermuller K, Odenbreit S, Fischer W, Melchers K, Haas R. Identification and characterization of Helicobacter pylori genes essential for gastric colonization. J Exp Med. 2003;197:813–822. doi: 10.1084/jem.20021531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- Kooistra J, Haijema BJ, Hesseling-Meinders A, Venema G. A conserved helicase motif of the AddA subunit of the Bacillus subtilis ATP-dependent nuclease (AddAB) is essential for DNA repair and recombination. Mol Microbiol. 1997;23:137–149. doi: 10.1046/j.1365-2958.1997.1991570.x. [DOI] [PubMed] [Google Scholar]
- Kooistra J, Venema G. Cloning, sequencing, and expression of Bacillus subtilis genes involved in ATP-dependent nuclease synthesis. J Bacteriol. 1991;173:3644–3655. doi: 10.1128/jb.173.12.3644-3655.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooistra J, Vosman B, Venema G. Cloning and characterization of a Bacillus subtilis transcription unit involved in ATP-dependent DNase synthesis. J Bacteriol. 1988;170:4791–4797. doi: 10.1128/jb.170.10.4791-4797.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusters JG, van Vliet AH, Kuipers EJ. Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev. 2006;19:449–490. doi: 10.1128/CMR.00054-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea DE, Coulson CA. The distribution of the numbers of mutants in bacterial populations. J Genetics. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- Loughlin MF, Barnard FM, Jenkins D, Sharples GJ, Jenks PJ. Helicobacter pylori mutants defective in RuvC Holliday junction resolvase display reduced macrophage survival and spontaneous clearance from the murine gastric mucosa. Infect Immun. 2003;71:2022–2031. doi: 10.1128/IAI.71.4.2022-2031.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meddows TR, Savory AP, Grove JI, Moore T, Lloyd RG. RecN protein and transcription factor DksA combine to promote faithful recombinational repair of DNA double-strand breaks. Mol Microbiol. 2005;57:97–110. doi: 10.1111/j.1365-2958.2005.04677.x. [DOI] [PubMed] [Google Scholar]
- Mehr IJ, Seifert HS. Differential roles of homologous recombination pathways in Neisseria gonorrhoeae pilin antigenic variation, DNA transformation and DNA repair. Mol Microbiol. 1998;30:697–710. doi: 10.1046/j.1365-2958.1998.01089.x. [DOI] [PubMed] [Google Scholar]
- Menard R, Sansonetti PJ, Parsot C. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol. 1993;175:5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Necchi V, Candusso ME, Tava F, Luinetti O, Ventura U, Fiocca R, et al. Intracellular, intercellular, and stromal invasion of gastric mucosa, preneoplastic lesions, and cancer by Helicobacter pylori. Gastroenterology. 2007;132:1009–1023. doi: 10.1053/j.gastro.2007.01.049. [DOI] [PubMed] [Google Scholar]
- O’Rourke EJ, Chevalier C, Pinto AV, Thiberge JM, Ielpi L, Labigne A, Radicella JP. Pathogen DNA as target for host-generated oxidative stress: role for repair of bacterial DNA damage in Helicobacter pylori colonization. Proc Natl Acad Sci U S A. 2003;100:2789–2794. doi: 10.1073/pnas.0337641100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver DB, Goldberg EB. Protection of parental T4 DNA from a restriction exonuclease by the product of gene 2. J Mol Biol. 1977;116:877–881. doi: 10.1016/0022-2836(77)90276-5. [DOI] [PubMed] [Google Scholar]
- Ponticelli AS, Schultz DW, Taylor AF, Smith GR. Chi-dependent DNA strand cleavage by RecBC enzyme. Cell. 1985;41:145–151. doi: 10.1016/0092-8674(85)90069-8. [DOI] [PubMed] [Google Scholar]
- Pride DT, Blaser MJ. Concerted evolution between duplicated genetic elements in Helicobacter pylori. J Mol Biol. 2002;316:629–642. doi: 10.1006/jmbi.2001.5311. [DOI] [PubMed] [Google Scholar]
- Reed LJ, Muench H. A simple method of estimating fifty per cent endpoints. The American Journal of Hygiene. 1938;27:493–497. [Google Scholar]
- Robinson K, Loughlin MF, Potter R, Jenks PJ. Host adaptation and immune modulation are mediated by homologous recombination in Helicobacter pylori. J Infect Dis. 2005;191:579–587. doi: 10.1086/427657. [DOI] [PubMed] [Google Scholar]
- Rocha EP, Cornet E, Michel B. Comparative and evolutionary analysis of the bacterial homologous recombination systems. PLoS Genet. 2005;1:e15. doi: 10.1371/journal.pgen.0010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama NR, Shepherd B, Falkow S. Global transposon mutagenesis and essential gene analysis of Helicobacter pylori. J Bacteriol. 2004;186:7926–7935. doi: 10.1128/JB.186.23.7926-7935.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez H, Kidane D, Castillo Cozar M, Graumann PL, Alonso JC. Recruitment of Bacillus subtilis RecN to DNA double-strand breaks in the absence of DNA end processing. J Bacteriol. 2006;188:353–360. doi: 10.1128/JB.188.2.353-360.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schapiro JM, Libby SJ, Fang FC. Inhibition of bacterial DNA replication by zinc mobilization during nitrosative stress. Proc Natl Acad Sci U S A. 2003;100:8496–8501. doi: 10.1073/pnas.1033133100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt W, Odenbreit S, Heuermann D, Haas R. Cloning of the Helicobacter pylori recA gene and functional characterization of its product. Molecular and General Genetics. 1995;248:563–572. doi: 10.1007/BF02423452. [DOI] [PubMed] [Google Scholar]
- Seyler RW, Jr, Olson JW, Maier RJ. Superoxide dismutase-deficient mutants of Helicobacter pylori are hypersensitive to oxidative stress and defective in host colonization. Infect Immun. 2001;69:4034–4040. doi: 10.1128/IAI.69.6.4034-4040.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sioud M, Forterre P. Ciprofloxacin and etoposide (VP16) produce a similar pattern of DNA cleavage in a plasmid of an archaebacterium. Biochemistry. 1989;28:3638–3641. doi: 10.1021/bi00435a002. [DOI] [PubMed] [Google Scholar]
- Smeets LC, Bijlsma JJ, Boomkens SY, Vandenbroucke-Grauls CM, Kusters JG. comH, a novel gene essential for natural transformation of Helicobacter pylori. J Bacteriol. 2000;182:3948–3954. doi: 10.1128/jb.182.14.3948-3954.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GR. Homologous recombination near and far from DNA breaks: alternative roles and contrasting views. Annual review of genetics. 2001;35:243–274. doi: 10.1146/annurev.genet.35.102401.090509. [DOI] [PubMed] [Google Scholar]
- Smith GR, Kunes SM, Schultz DW, Taylor A, Triman KL. Structure of chi hotspots of generalized recombination. Cell. 1981;24:429–436. doi: 10.1016/0092-8674(81)90333-0. [DOI] [PubMed] [Google Scholar]
- Solnick JV, Hansen LM, Salama NR, Boonjakuakul JK, Syvanen M. Modification of Helicobacter pylori outer membrane protein expression during experimental infection of rhesus macaques. Proc Natl Acad Sci U S A. 2004;101:2106–2111. doi: 10.1073/pnas.0308573100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stohl EA, Seifert HS. Neisseria gonorrhoeae DNA recombination and repair enzymes protect against oxidative damage caused by hydrogen peroxide. J Bacteriol. 2006;188:7645–7651. doi: 10.1128/JB.00801-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suerbaum S, Josenhans C. Helicobacter pylori evolution and phenotypic diversification in a changing host. Nat Rev Microbiol. 2007;5:441–452. doi: 10.1038/nrmicro1658. [DOI] [PubMed] [Google Scholar]
- Suerbaum S, Smith JM, Bapumia K, Morelli G, Smith NH, Kunstmann E, et al. Free recombination within Helicobacter pylori. Proc Natl Acad Sci U S A. 1998;95:12619–12624. doi: 10.1073/pnas.95.21.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor A, Smith GR. Unwinding and rewinding of DNA by the RecBC enzyme. Cell. 1980;22:447–457. doi: 10.1016/0092-8674(80)90355-4. [DOI] [PubMed] [Google Scholar]
- Taylor AF, Smith GR. Monomeric RecBCD enzyme binds and unwinds DNA. J Biol Chem. 1995;270:24451–24458. doi: 10.1074/jbc.270.41.24451. [DOI] [PubMed] [Google Scholar]
- Taylor AF, Smith GR. RecBCD enzyme is a DNA helicase with fast and slow motors of opposite polarity. Nature. 2003;423:889–893. doi: 10.1038/nature01674. [DOI] [PubMed] [Google Scholar]
- Teshima S, Tsunawaki S, Rokutan K. Helicobacter pylori lipopolysaccharide enhances the expression of NADPH oxidase components in cultured guinea pig gastric mucosal cells. FEBS Lett. 1999;452:243–246. doi: 10.1016/s0014-5793(99)00636-5. [DOI] [PubMed] [Google Scholar]
- Thompson SA, Blaser MJ. Isolation of the Helicobacter pylori recA gene and involvement of the recA region in resitance to low pH. Infection and Immunity. 1995;63:2185–2193. doi: 10.1128/iai.63.6.2185-2193.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori [see comments] [published erratum appears in Nature 1997 Sep 25;389(6649): 412] Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- Tomizawa J, Ogawa H. Structural genes of ATP-dependent deoxyribonuclease of Escherichia coli. Nature: New biology. 1972;239:14–16. doi: 10.1038/newbio239014a0. [DOI] [PubMed] [Google Scholar]
- Wang G, Alamuri P, Humayun MZ, Taylor DE, Maier RJ. The Helicobacter pylori MutS protein confers protection from oxidative DNA damage. Mol Microbiol. 2005;58:166–176. doi: 10.1111/j.1365-2958.2005.04833.x. [DOI] [PubMed] [Google Scholar]
- Wang G, Maier RJ. Critical role of RecN in recombinational DNA repair and survival of Helicobacter pylori. Infect Immun. 2008;76:153–160. doi: 10.1128/IAI.00791-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Roos KP, Taylor DE. Transformation of Helicobacter pylori by chromosomal metronidazole resistance and by a plasmid with a selectable chloramphenicol resistance marker. Journal of General Microbiology. 1993;139:2485–2493. doi: 10.1099/00221287-139-10-2485. [DOI] [PubMed] [Google Scholar]
- Wolfson JS, Hooper DC. The fluoroquinolones: structures, mechanisms of action and resistance, and spectra of activity in vitro. Antimicrob Agents Chemother. 1985;28:581–586. doi: 10.1128/aac.28.4.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Chaturvedi R, Cheng Y, Bussiere FI, Asim M, Yao MD, et al. Spermine oxidation induced by Helicobacter pylori results in apoptosis and DNA damage: implications for gastric carcinogenesis. Cancer Res. 2004;64:8521–8525. doi: 10.1158/0008-5472.CAN-04-3511. [DOI] [PubMed] [Google Scholar]
- Yeeles JT, Dillingham MS. A dual-nuclease mechanism for DNA break processing by AddAB-type helicase-nucleases. J Mol Biol. 2007;371:66–78. doi: 10.1016/j.jmb.2007.05.053. [DOI] [PubMed] [Google Scholar]
- Yu M, Souaya J, Julin DA. The 30-kDa C-terminal domain of the RecB protein is critical for the nuclease activity, but not the helicase activity, of the RecBCD enzyme from Escherichia coli. Proc Natl Acad Sci U S A. 1998;95:981–986. doi: 10.1073/pnas.95.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga-Castillo J, Romero D, Martinez-Salazar JM. The recombination genes addAB are not restricted to gram-positive bacteria: genetic analysis of the recombination initiation enzymes RecF and AddAB in Rhizobium etli. J Bacteriol. 2004;186:7905–7913. doi: 10.1128/JB.186.23.7905-7913.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]