Abstract
Anuran (frog) metamorphosis has long-served as a model of how thyroid hormones regulate post-embryonic development in vertebrates. However, comparatively little is known about urodele (salamander) metamorphosis. We conducted a detailed time-course study of induced metamorphosis in the Mexican axolotl (Ambystoma mexicanum) that probed metamorphic changes in morphology and gene expression in the skin. Using morphometrics, quantitative PCR, histology, and in situ hybridization we demonstrate that the development of transcriptional markers is fundamental to the resolution of early metamorphic events in axolotls. We then use linear and piecewise linear models to identify a sequence of morphological and transcriptional changes that define larval to adult remodeling events throughout metamorphosis. In addition, we show that transcriptional biomarkers are expressed in specific larval and adult cell populations of the skin and that temporal changes in these biomarkers correlate with tissue remodeling. We compare our results with other studies of natural and induced metamorphosis in urodeles and highlight what appear to be conserved features between urodele and anuran metamorphosis.
Keywords: Ambystoma, Amphibian, Linear regression, Metamorphosis, Mexican axolotl, Morphometrics, Piecewise regression, Thyroid hormone, Transcription
1. Introduction
Amphibian metamorphosis is a well-studied example of a complex developmental process that is regulated by endocrine factors (Wilder, 1925; Nieuwkoop and Faber, 1967; Rosenkilde and Ussing, 1996; Shi, 2000; Brown and Cai, 2007). Thyroid hormone (TH) control of metamorphosis is broadly conserved across amphibians, and radioimmunoassay (RIA) data from anurans (frogs; e.g., Leloup and Buscaglia, 1977) and urodeles (salamanders; e.g., Larras-Regard et al., 1981; Alberch et al., 1986) support the idea that L-thyroxine (T4; relatively inactive form of TH) and 3,5,3′-triiodothyronine (T3; relatively active form of TH) markedly increase at metamorphic climax. TH (T3 or T4) is necessary and sufficient to induce metamorphosis in anurans (reviewed by Shi, 2000; Brown and Cai, 2007) and urodeles (Prahlad and DeLanney, 1965; Prahlad 1968; Rose, 1995b,c; Rosenkilde and Ussing, 1996) and its biological effects are mediated by nuclear receptors (thyroid hormone receptors α and β; TR-α and TR-β) that repress or activate transcription in a TH-dependent manner (Safi et al., 2004; Buchholz et al., 2006). However, while these and other general features of amphibian metamorphosis are broadly conserved (see Denver et al., 2002), there is considerable variation in the timing, duration, and remodeling patterns that occur across different taxa (Duellman and Trueb, 1994). For example, anurans completely resorb their tails during metamorphosis (Shi, 2000) while urodeles remodel and retain their tails as adults (Duellman and Trueb, 1994). As another example, hind limb formation and growth are intimately linked to metamorphosis in anurans (Shi, 2000), but occur months before metamorphic climax in urodeles (Rosenkilde and Ussing, 1996). Finally, some urodeles (but no anurans) are paedomorphic and altogether fail to undergo a conspicuous metamorphosis thus retaining larval morphological traits and completing their life cycles in water as aquatic adults (see Petranka, 1998).
While the endocrinology (e.g., Leloup and Buscaglia, 1977; Buchholz and Hayes, 2005; Larras-Regard et al., 1981; Alberch et al., 1986), histology (e.g., Heady and Kollros, 1964; Nieuwkoop and Faber, 1967; Fahrmann, 1971a,b,c; Alberch et al., 1985; Ohmura and Wakahara, 1998), and morphology (e.g., Taylor and Kollros, 1946; Nieuwkoop and Faber, 1967; Norman, 1985; Cano-Martinez et al., 1994; Rose, 1995a,b,c) of metamorphosis have been examined in anurans and urodeles, the vast majority of our knowledge about metamorphic gene expression comes from studies of anurans and Xenopus in particular (reviewed by Shi, 2000; Buchholz et al., 2006). This has enabled the conceptualization of models of Xenopus, and, to a lesser extent, Rana metamorphosis that integrate changes in morphology, histology, and gene expression (for examples see Ishizuya-Oka and Shi, 2007; Yoshizato, 2007). However, there have been few attempts to incorporate gene expression data into integrative models of salamander metamorphosis.
Ambystomatid salamanders are a logical urodele family to develop as metamorphic research models because they exhibit interesting life-history variation (Wilbur and Collins, 1973; Shaffer and Voss, 1996; Denver et al., 2002), have a maturing genomic resource infrastructure (Putta et al., 2004; Smith et al., 2005a,b; Monaghan et al., 2009), and are amenable to laboratory culture and conditions (Armstrong et al., 1989; Armstrong and Duhon, 1989; Frost et al., 1989). In particular, the Mexican axolotl (Ambystoma mexicanum) has a long history as a developmental model (Smith, 1989). The axolotl is a paedomorphic salamander that is part of a large species complex (i.e., the tiger salamander species complex) that consists of metamorphic, facultatively paedomorphic, and paedomorphic taxa that have recently diversified from a metamorphic ancestor (Shaffer and Mcknight, 1996; Shaffer and Voss, 1996; Weisrock et al., 2006). Despite rarely spontaneously metamorphosing in nature or the lab, the axolotl can be induced to undergo metamorphosis by adding T3 or T4 to rearing water. We typically induce metamorphosis in the axolotl with T4 because it is generally thought to be the primary iodine containing hormone released by the thyroid and delivered to other tissues where it is locally converted to T3 via deiodinase activity (Denver et al., 2002; Brown, 2005). Fifty nM T4 is an appropriate standard for studying induced metamorphosis in the axolotl because it is well below the threshold of toxicity (80 nM; Rosenkilde and Ussing, 1996), induces metamorphosis at a rate that is not accelerated by higher doses (Rosenkilde and Ussing, 1996), and maximizes rate without altering the sequence of morphological and transcriptional events observed at physiologically relevant concentrations (Page et al., 2008).
During TH-induced metamorphosis, axolotls undergo a series of morphological changes that include reductions in body mass and growth rate, and complete resorption of the tail fins, dorsal ridge, and gills. Comparisons of staging series based on these gross morphological events suggest that the pattern exhibited by TH-induced axolotls (Cano-Martinez, 1994) qualitatively resembles the pattern exhibited by naturally metamorphosing tiger salamanders (Ambystoma tigrinum; Norman, 1985; also see Rosenkilde and Ussing, 1996). Thus the axolotl represents a convenient and informative model system for examining metamorphic gene expression in the tiger salamander complex. In a recent study, we used microarray technology to correlate gene expression in the skin of TH-induced axolotls with metamorphic stage and T4 concentration (Page et al., 2008). These gene expression screens revealed that hundreds of genes are differentially regulated in response to T4 in a way that strongly correlates with morphological stage, but shows little or no correlation with the dose of T4 (5 or 50 nM) used to induce metamorphosis. While potential biomarkers of skin metamorphosis were identified, sampling across developmental time and individuals was too sparse to detail temporal changes in gene expression and morphology. Here, we report on a detailed time-course study of TH-induced metamorphosis of axolotl skin using a group of candidate biomarker genes, quantitative reverse transcriptase real-time PCR (Q-RT-PCR), histology, and in situ hybridization (ISH). We use linear and piecewise linear modeling to identify the onset, offset, and rate of change for morphological and molecular traits during metamorphosis and articulate an integrative model of TH-induced metamorphosis in A. mexicanum. We compare and contrast our results with results obtained from other studies of natural and induced metamorphosis in urodeles and highlight what appear to be conserved features of urodele and anuran metamorphosis.
2. Materials and methods
2.1. Study animals
Sixty Mexican axolotls were obtained from a single inbred cross and reared individually at 20–22 C in 40% Holtfreter’s solution. After hatching, larvae were fed freshly hatched brine shrimp (Artemia) napuli until they were three weeks of age. Beginning at three weeks of age, salamanders were fed California blackworms (Lumbriculus). At 120 days post-fertilization (Day 0), 50 nM T4 (Sigma, St. Louis, MO; T2376) was administered to the animals’ rearing water as described in Page et al. (2007). A ≈ 1 cm2 section of skin was removed from a dorsal region of the head (posterior to the eyes and anterior to the neck) of three individuals beginning on Day 0 (prior to initiating T4 treatment). Skin was then sampled from three individuals every other day for 30 days (i.e., Day 0, Day 2, Day 4, … Day 30). Sampling was also conducted on Day 32; however, only two individuals were sampled at this time point. On Days 0, 12, and 28, additional animals (4, 3, and 3 respectively) were sampled to obtain skin for histological staining and ISH. The remaining 50 tissue samples were used for Q-RT-PCR. All 60 animals were sacrificed at the time of tissue collection. On Day -1, all 60 animals were measured for mass and the seven other morphometric traits described by Fig. 1 (here out referred to as the pre dataset). Just prior to tissue collection, all 60 animals were measured for these same morphometric traits a second time (here out referred to as the post dataset). Morphological staging was conducted according to Cano-Martinez et al. (1994).
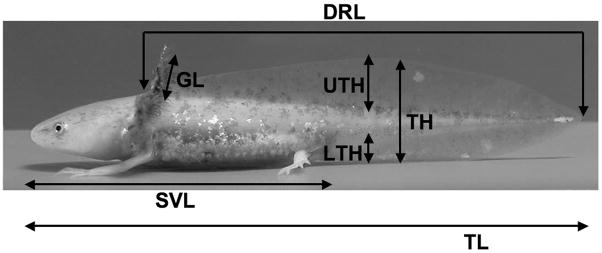
Fig. 1.
The morphometric traits measured throughout the experiment. Dorsal ridge length (DRL) was measured from the most cranial portion of the dorsal ridge to the tip of the tail. Once the dorsal ridge had receded to where it was no longer distinct from the upper tail fin, DRL was scored as zero. Upper tailfin height (UTH) was measured from the most dorsal portion of the tail trunk to the tip of the upper tailfin. Tail height (TH) was measured from the tip of the lower tail fin to the tip of the upper tail fin. Lower tailfin height (LTH) was measured from the ventral tip of the tail trunk to the tip of the lower tailfin. All tail measurements (UTH, TH, and LTH) were taken ≈ 5% of the animal’s total length (TL) posterior to the cloaca. TL was measured from the tip of the nose to the tip of the tail. Snout-vent-length (SVL) was measured from the tip of the nose to the posterior end of the cloaca. Gill length (GL) was measured from where the gills protrude from the cranium to the tip of the epithelium of the gills. Gill filaments were excluded from this measurement. Mass (not shown) was also recorded.
2.2. Modeling changes in morphological traits as a function of time
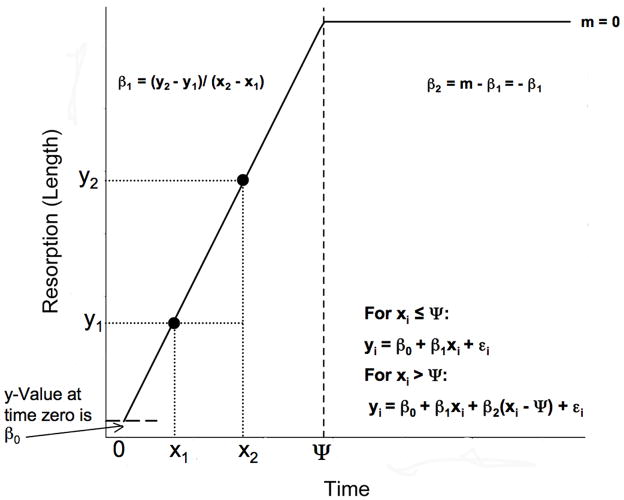
After T4 is administered to an axolotl’s rearing water, there is a latency period before metamorphic changes are observed (Rosenkilde and Ussing, 1996; Page et al., 2008). During this latency period, growth continues at a normal rate and metamorphic changes, such as weight loss and tissue resorption, are not evident. At the end of this latency period, there is an onset and changes in one or more traits become obvious as morphological metamorphosis proceeds. Similarly, because metamorphic change ultimately results in new adult morphology, many metamorphic traits also undergo an offset after which they no longer change because they have taken on their adult form. These onset and offset times can be conceptualized as transitions or breakpoints that connect linear equations with different slopes (rates of change). These slopes describe the direction and rate of change for a trait before the onset/offset and the direction and rate of change after the onset/offset (Fig. 2). Piecewise linear regression is a useful tool because it can enable one to estimate: (1) the time at which a transition (onset/offset) occurs, (2) the direction and rate of change before the transition, and (3) the direction and rate of change after the transition. In its simplest form, a piecewise linear model can be defined as follows: for xi ≤ Ψ, yi = β0 + β1xi + εi and for xi > Ψ, yi = β0 + β1xi + β2(xi − Ψ) + εi where yi is the value observed for a given trait from individual i, β0 is the baseline value for the trait, β1 is the slope or rate of change to the left of the transition, xi is the time at which individual i was sampled, β2 is the slope to the right of the breakpoint minus the slope to the left of the breakpoint (Fig. 2), Ψ is the transition time or breakpoint, and εi is the error associated with individual i (for a detailed discussion of piecewise regression see Toms and Lesperance, 2003). Thus, it follows that the rate of change (slope; m) to the right of the breakpoint can be obtained as follows: m = β1 + β2.
Fig. 2.
A graphical schematic of a four parameter (β0, β1, β2, and Ψ) piecewise linear model for a hypothetical morphometric trait that is resorbed during metamorphosis. Following time Ψ (the breakpoint; vertical dashed line), the slope (m) is zero indicating that the trait is no longer changing due to complete resorption. See the text for further descriptions of the model.
We performed piecewise regression as implemented by the “segmented” software package (Muggeo, 2008) that is freely available for the R statistical computing environment (www.r-project.org). All models in which morphological traits (t) were treated as response variables were conducted using the differences between the pre and post measurements. The following formula: Δt = tpre − tpost makes resorption (for most morphometric traits the phenomenon of interest) positive. Thus, positive values of Δt indicate that a trait is vanishing and negative values of Δt indicate that a trait is growing. For morphological traits where piecewise relationships were not evident, we fit linear regression models of the form yi = β0 + β1xi + εi where β0 is the baseline trait value, β1 is the slope, and ε is the error term (Zar, 1999). Such linear models provide estimates of the direction and rate of change for traits (slopes) that do not exhibit evidence of metamorphic onset or offset within the experimental interval. In all cases, intercept free models (β0 = 0) were fit so as to force lines through the origin as, by definition, all traits have initial values of zero days T4 administration (x-axis) and zero change between the pre and post datasets (y-axis).
2.3. Genes investigated
We previously used microarray technology to identify hundreds of genes that are differentially expressed in response to T4 treatment (Page et al., 2007; Page et al., 2008). This gene list included keratin 6A (KRT6A), keratin 14 (KRT14), uromodulin (UMOD), calmodulin 2 (CALM2), epithelial membrane protein 1 (EMP1), glutamate ammonia-ligase (GLUL), and matrix metalloproteinase 1 (MMP1). To the best of our knowledge, only UMOD and MMP1 are known TH response genes from other amphibian taxa. UMOD-like genes have been identified from Xenopus laevis and are thought to contribute to a larval specific structure on the surface of the skin that has “protective” and “secretory” functions (Furlow et al., 1997). MMP1 is a gene that is known to be directly targeted by TH in Rana catesbeiana (Oofusa and Yoshizato, 1996; Sawada et al., 2001). In R. catesbeiana, MMP1 is involved in collagen turnover that occurs during tail resorption and skin remodeling (Oofusa and Yoshizato, 1991). The functions of the rest of these genes (KRT6A, KRT14, GLUL, EMP1, CALM2) during amphibian metamorphosis are largely unknown. We assign gene names to our assembled and curated expressed sequence tag (EST) based contigs (Putta et al., 2004; Monaghan et al., 2009) in accordance with their presumptive orthologies to human based on BLASTx searches of the human RefSeq protein database (E < 10−7; see Monaghan et al., 2007; Page et al., 2007). Thus, sequence homology leads us to assume that KRT6A and KRT14 encode cytoskeletal proteins that are constituents of intermediate filaments. Similarly, we assume that CALM2 encodes a protein involved in several aspects of calcium regulation and signaling (Friedberg and Rhoads, 2001). We also assume GLUL encodes an enzyme that plays important roles in nitrogen metabolism (Kumada et al., 1993). Finally, we assume EMP1 is associated with cell proliferation (Ben-Porath and Benvenisty, 1996) and epithelial differentiation (Marvin et al., 1995). In addition to genes that were identified by previous expression screens, we have chosen to investigate TR-α and TR-β due to the fundamental roles that these transcription factors play in TH signaling during metamorphosis (see Safi et al., 2004; Buchholz et al., 2006). Table 1 summarizes the genes that we have investigated as well as their presumptive functions.
Table 1.
Primer sequences used for Q-RT-PCR and in situ hybridization
| Gene | Symbol | Presumed Process/Function | Contig ID | Strand | Sequence | Method |
|---|---|---|---|---|---|---|
| Epithelial Membrane Protein 1 | EMP1 | Proliferation/differentiation | TC00532 | F | CTTGGCCATCATCTTTGG | PCR |
| Epithelial Membrane Protein 1 | EMP1 | Proliferation/differentiation | TC00532 | R | AAGAAGCGGCATCCTTTC | PCR |
| Glutamate-ammonia Ligase | GLUL | Nitrogen metabolism | TC00765 | F | ACTTTGGCGTTGTGGCTA | PCR |
| Glutamate-ammonia Ligase | GLUL | Nitrogen metabolism | TC00765 | R | TGGCTTCCGTGCTGTAGT | PCR |
| Keratin 14 | KRT14 | Intermediate filament | MC00279 | F | GCAGCTGACCGATTTGAG | PCR |
| Keratin 14 | KRT14 | Intermediate filament | MC00279 | R | CCAGCCGTGACTTGATGT | PCR |
| Keratin 14 | KRT14 | Intermediate filament | MC00279 | F | AATTAACCCTCACTAAAGGGAGATGCAGTCCCAACTGTCAATG | ISH |
| Keratin 14 | KRT14 | Intermediate filament | MC00279 | R | ATTTAGGTGACACTATAGAAGAGATCCAGCCAGGTGTTGGTAG | ISH |
| Keratin 6A | KRT6A | Intermediate filament | MC02467 | F | CCCTGCAGAAAGCCAAG | PCR |
| Keratin 6A | KRT6A | Intermediate filament | MC02467 | R | TGTCTAGGGCCAGCTTGA | PCR |
| Keratin 6A | KRT6A | Intermediate filament | MC02467 | F | AATTAACCCTCACTAAAGGGAGACGTTTGGATGGTGATCTGAG | ISH |
| Keratin 6A | KRT6A | Intermediate filament | MC02467 | R | ATTTAGGTGACACTATAGAAGAGTCCTGGTAGTCACGCAGTTG | ISH |
| Matrix Metalloproteinase 1 | MMP1 | Collagen degradation | MC01311 | F | CACCCGAAATCTGTGACAA | PCR |
| Matrix Metalloproteinase 1 | MMP1 | Collagen degradation | MC01311 | R | CGGCGCCAGAAGAATC | PCR |
| Matrix Metalloproteinase 1 | MMP1 | Collagen degradation | MC01311 | F | AATTAACCCTCACTAAAGGGAGACAGGAGCTCAGGGAAGAATG | ISH |
| Matrix Metalloproteinase 1 | MMP1 | Collagen degradation | MC01311 | R | ATTTAGGTGACACTATAGAAGAGAAGTGGACGTCTCCACCAAC | ISH |
| Thyroid Hormone Receptor α | TR-α | Transcription factor | N/A | F | GTCGGATGCCATCTTTGA | PCR |
| Thyroid Hormone Receptor α | TR-α | Transcription factor | N/A | R | CAGCACCGCCTGAAGTAG | PCR |
| Thyroid Hormone Receptor β | TR-β | Transcription factor | N/A | F | AACGCTGAATGGGGAAAT | PCR |
| Thyroid Hormone Receptor β | TR-β | Transcription factor | N/A | R | CGACATCCCAAGGTCAAA | PCR |
| Uromodulin | UMOD | Protection/barrier | MC01737 | F | GCGGATTCATTGACCATTC | PCR |
| Uromodulin | UMOD | Protection/barrier | MC01737 | R | TCGGAGTTGGTGATCTTGA | PCR |
| Uromodulin | UMOD | Protection/barrier | MC01737 | F | AATTAACCCTCACTAAAGGGAGATCCTGCTCCTACCCTCTCAA | ISH |
| Uromodulin | UMOD | Protection/barrier | MC01737 | R | ATTTAGGTGACACTATAGAAGAGGGTCACGGTGGACATTCTCT | ISH |
| Calmodulin 2 | CALM2 | Ca regulation/signaling | MC10283 | F | CCCGTATCTGCGTTTATCC | PCR |
| Calmodulin 2 | CALM 2 | Ca regulation/signaling | MC10283 | R | CTTAACGGAAGGTGGTTGG | PCR |
| Calmodulin 2 | CALM2 | Ca regulation/signaling | MC10283 | F | AATTAACCCTCACTAAAGGGAGAAAGGACTGGCTTGCAGTAGG | ISH |
| Calmodulin 2 | CALM2 | Ca regulation/signaling | MC10283 | R | ATTTAGGTGACACTATAGAAGAGAGGCATAACGCCAGCTAAGA | ISH |
| Transcriptional Intermediary Factor 1 | TIF1 | Control gene | MC00782 | F | CCTCTGGTCTCGGATCG | PCR |
| Transcriptional Intermediary Factor 1 | TIF1 | Control gene | MC00782 | R | TCCTGCGGAGGCTCTT | PCR |
PCR refers to Q-RT-PCR. Contig ID is the Sal-Site (www.ambystoma.org) identifier associated with each gene. Strand refers to the strand to which primer design was targeted. F = forward strand primer and R = reverse strand primer. Primers for TR-α and TR-β were designed from sequences obtained from the National Center for Biotechnology Information. For the “Contig ID” column, N/A implies that a gene does not have a Sal-Site contig ID.
2.4. RNA isolation
Total RNA was extracted from 50 tissue samples using TRIzol (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. RNA samples were further purified using Qiagen RNeasy mini columns. All samples were treated with RNase-Free DNase Sets (Qiagen, Valencia, CA) according to the manufacturer’s protocol. UV spectrophotometry and gel electrophoresis were used to quantify and qualify RNA preparations.
2.5. Q-RT-PCR
Relative transcript abundances for the genes listed in Table 1 were estimated via Q-RT-PCR according to the protocol described in Page et al. (2008). Briefly, primers (Table 1) were designed using Primer3 (Rozen and Skaletsky, 2000). All PCRs were 25 μL reactions that contained cDNA corresponding to 10 ng total RNA, 41 ng forward and reverse primers, and iQ SYBR-Green real-time PCR mix (Bio-Rad, Hercules, CA). Reaction parameters were 10 minutes at 50 C, five minutes at 95 C, 45 cycles of 10 seconds at 95 C followed by 30 seconds at 55 C, one minute at 95 C, and one minute at 55 C. For all reactions, melting curve analysis was performed to ensure that only a single product was amplified. PCRs were performed on a Bio-Rad iCycler iQ Multi-Color Real Time PCR Detection System. All plates contained template free controls (Bustin and Nolan, 2004). Primer efficiencies were estimated via linear regression and relative expression ratios (R) were calculated according to Pfaffl (2001). All expression ratios are relative to the average of Day 0 animals, and normalized to the expression of transcriptional intermediary factor 1 (Table 1; Page et al., 2008).
2.6. Modeling gene expression changes in the skin as a function of time
Just as many of the morphological changes that occur during induced metamorphosis can be conceptualized as offset times, so can many of the gene expression changes that occur during metamorphic skin remodeling. For example, larval specific and adult specific genes are often down and up-regulated respectively during metamorphosis to new adult expression levels. Thus, as is the case with morphological traits, there are transitions or breakpoints that connect the rate of change observed during the metamorphic period with the rate of change observed after metamorphic gene regulation has ended (i.e., metamorphic offset). Hence, the simple piecewise model described in Fig. 2 allows one to estimate several useful parameters including: β1 the rate of change in transcript abundance during the metamorphic period, Ψ the offset time of metamorphic gene regulation, and m the rate of change in transcript abundance after the offset of metamorphic gene regulation. This same piecewise model (Fig. 2) also enables the estimation of meaningful parameters for genes associated with metamorphic remodeling processes. Such remodeling genes are frequently transiently up or down-regulated before returning to baseline expression levels (Page et al., 2008). Thus, β1 provides an estimate of the rate of change in transcript abundance prior to reaching maximum/minimum expression levels, Ψ provides an estimate of the time at which maximum/minimum expression levels are reached, and m provides an estimate of the rate of return to baseline expression levels. In keeping with these ideas, we applied piecewise and linear regression, as described above for morphological traits, to our gene expression data. In order to decide between piecewise and linear models for each gene, we used Akaike’s information criterion (AIC; Venables and Ripley, 2002) which attaches a numerical value (smaller AIC is better) to each model that accounts for the tradeoff between model fit and model complexity (i.e., the number of parameters in a model). All models of gene expression treat days of T4 treatment as the predictor variable and log2 transformed R-values as the response variable. As with the morphology data, we used models that lack intercepts (β0 = 0) to estimate β1, Ψ, and m (m is estimated indirectly via knowledge of β1 and β2; see Fig. 2) as larval expression levels on a log2 scale (y-axis) are by definition zero and sampling began following zero days of T4 exposure (x-axis). By assuming that baseline expression for each gene is equal to the mean of the three individuals observed at Day 0 (i.e. that β0 = 0), we are able to obtain consistent estimates of β1, Ψ, and m that put the expression of all genes on a common scale. However, because our estimation of R for each gene at Day 0 rests on a small sample size, we use models that estimate β0 (baseline expression) from all 50 data points via least squares to conduct hypothesis testing (i.e., to make statements about differential regulation in a statistical sense). When we were unable to estimate piecewise models due to convergence problems in the model fitting routine, we used quadratic regression to test for the presence of curvilinear relationships that are similar in shape to the piecewise model described in Fig. 2. Quadratic models take on the following form: log2(R)i = β0 + γ1xi + γ2x2i + εi where β0 and ε are the baseline expression value and error term respectively and γ1 and γ2 are the linear and quadratic regression coefficients respectively. Quadratic models provide a well-established framework for testing for the presence of curvilinear relationships (Zar, 1999), but do not provide parameter estimates with interpretations as straight forward as the model described in Fig. 2.
2.7. Histology
In order to show that TH induces tissue remodeling events in the skin, we conducted hematoxylin and eosin staining at three time points: Days 0, 12, and 28. Skin samples were fixed at 4 C in 1x phosphate buffer saline (PBS), 4% paraformaldehyde (PFA) overnight. Following cryoprotection in 30% sucrose, tissues were sectioned at 20 μm using a Microm 500 HM cryostat (Richard Allen Scientific, Kalamazoo, MI) and stained with Gill’s hematoxylin #2 (Sigma, St. Louis, MO) and eosin Y (Amresco, Solon, OH). Microscopy was performed using an Olympus AX80 microscope and images were acquired with an Olympus DP70 camera (Olympus, Center Valley, CA).
2.8. In situ hybridization
We used ISH to verify Q-RT-PCR results and spatially localize gene expression at the cellular level. Tissues were collected and prepared as described above for histological staining. Day 0 and Day 28 sections were collected on the same SuperFrost Plus microscope slides (VWR, West Chester, PA) and analyzed simultaneously. ISHs were performed as described in Monaghan et al. (2007), with minor modifications. Tissues were not decalcified and sectioning was conducted at 20 μm rather than 16 μm. Images were captured using an AX-80 Olympus microscope (Olympus, Center Valley, CA). Hybridizations and washes were performed overnight at 63.5 C, except for CALM2 for which hybridizations and washes were performed at 58 C. Probes for each gene are shown in Table 1.
3. Results
3.1. Morphological and histological changes
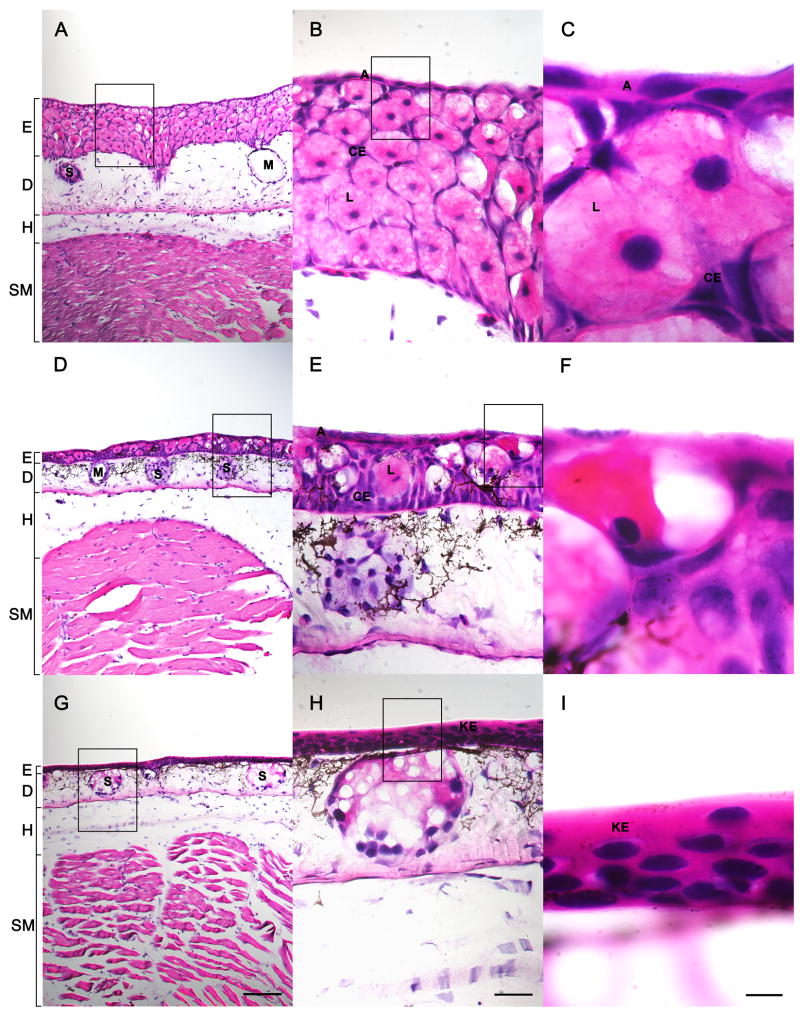
We observed morphological and histological changes that are consistent with TH-induced metamorphosis. The first Stage 1 (Cano-Martinez et al., 1994) animal was observed at Day 8 and individuals completed metamorphosis (Stage 4) between Day 28 and Day 32 (Fig. 3). We note that the time required for a given animal to reach a particular Cano-Martinez stage was quite variable (Fig. 3), suggesting that even among full siblings raised individually under identical conditions, there is variation in response to T4. Histological changes were consistent with Fahrmann’s (1971a,b,c) descriptions of metamorphic skin remodeling (Fig. 4). Briefly, larval cell types (Fig. 4A–C) such as apical cells and Leydig cells were removed while adult cell types (Fig. 4G–I) increased in abundance. By Day 12 (Fig. 4D–F), the skin was chimeric and consisted of larval and adult cell types. By Day 28, the epidermis consisted of a heavily keratinized stratified squamous epithelium similar to that observed in metamorphosed anurans and adult mammals (Fig. 4G–I).
Fig. 3.
A sunflower plot of the metamorphic stage of each of 60 individuals just prior to being sacrificed for skin tissue.
Fig. 4.
Histological staining of metamorphosing head skin following 0 days of T4 treatment (AC), 12 days of T4 treatment (D–F), and 28 days of T4 treatment (G–I). E = epidermis, D = dermis, H = hypodermis, SM = skeletal muscle, S = serous gland, M = mucous gland, L = Leydig cell, A = apical cell, CE = common epithelial cell, and KE = keratinized stratified squamous epithelium. The scale bar is 200 μm in A, D, and G. The scale bar is 50 μm in B, E, and H and 10 μm in C, F, and I.
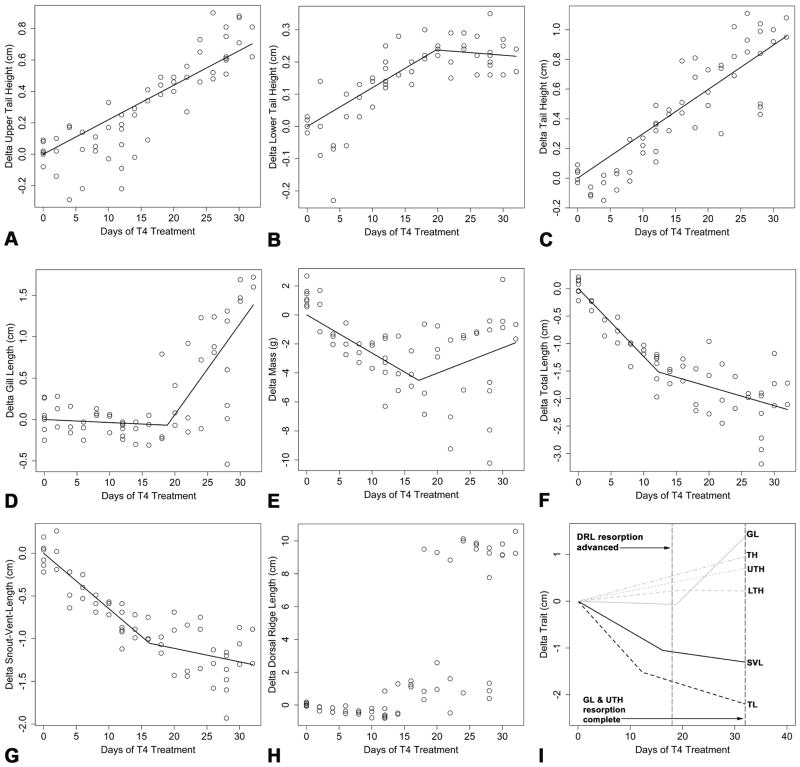
Of the eight morphological traits investigated (Fig. 1), five (LTH, GL, mass, TL, and SVL) exhibited piecewise relationships and two (UTH and TH) exhibited linear relationships (Fig. 5A–G). As measured, DRL is not a continuous variable (Fig. 1) and therefore could not be modeled in a regression framework. However, graphical inspection indicates that the dorsal ridge is completely resorbed between Day 18 and Day 32 (Fig. 5H). Inspection of the slope of the linear regression on ΔUTH (Fig. 5A; β1 = 0.022 cm/day T4 treatment) and the left line segment of the piecewise model fit to ΔLTH (Fig. 5B; β1 = 0.012 cm/day of T4 treatment) suggest similar resorption rates for the upper and lower tail fins. Thus, it is not surprising that the rate of reduction in total tail height was also of a similar magnitude (Fig. 5C; β1 = 0.030 cm/day of T4 treatment). The offset time corresponding to complete lower tail fin resorption was estimated to occur following 19.73 days of T4 treatment; however, there is considerable uncertainty associated with this estimate (lower 95% confidence interval (LCI) = 14.69 days, upper 95% confidence interval (UCI) = 24.76 days). As expected, following complete resorption, the slope of the right line segment for ΔLTH was not statistically different from zero (m = −0.002, LCI = −0.009, UCI = 0.006).
Fig. 5.
Scatter plots and prediction lines for the models fit to the morphometric data. (A) ΔUTH, (B) ΔLTH, (C) ΔTH, (D) ΔGL, (E) ΔMass, (F) ΔTL, (G) ΔSVL, (H) ΔDRL, and (I) a summary of the equations estimated for all the morphometric traits except mass. Note that no model has been fit to ΔDRL because as measured it is not a continuous trait.
The pattern observed for gill resorption (Fig. 5D) was markedly different than the patterns observed for upper and lower tail fin resorption. The slope of the left line segment of the piecewise model fit to ΔGL was not statistically different from zero (β1 = −0.004, t = −0.596, P = 0.554) suggesting that gill resorption does not begin immediately. The onset of gill resorption was estimated to occur following 18.82 days of T4 treatment (LCI = 15.26 days, upper UCI = 22.39 days). Upon commencement, gill resorption was rapid and occurred at a rate that was nearly an order of magnitude faster than lower tail fin resorption (m = 0.110 cm/day of T4 treatment).
Analyses of the traits associated with body size (mass, TL, SVL) showed general concordance. For example, piecewise regression models for all three traits had negative β1 estimates indicating that animals continued to grow in length and mass during the first half of the induction interval (Fig. 5E–G). Similarly, the breakpoints for Δmass (Fig. 5E; Ψ = 17.14 days of T4 treatment, LCI = 11.87 days, UCI = 22.42 days), ΔTL (Fig. 5F; Ψ = 12.29 days of T4 treatment, LCI = 8.23 days, UCI = 16.34 days), and ΔSVL (Fig. 5G; Ψ = 16.24 days of T4 treatment, LCI = 11.29 days, UCI = 21.20 days) suggest that a reduction in growth rate occurred following ≈ 12–17 days of exposure to 50 nM T4. The relationship between Δmass and days of T4 administration deteriorated following metamorphic onset (Fig. 5E). However, as expected, TL (Fig. 5F) and SVL (Fig. 5G) showed similar patterns throughout metamorphosis. Before metamorphic onset the slope (β1) for ΔTL was estimated to be −0.124 cm/day of T4 treatment whereas after metamorphic onset, the slope for ΔTL (m) was reduced to −0.035 cm/day of T4 treatment. For ΔSVL β1 = −0.065 cm/day of T4 treatment and m = −0.016 cm/day of T4 treatment. These results suggest that, after a relatively long latency period, growth rates are reduced by 70–75% during T4 induced metamorphosis of A. mexicanum. Fig. 5I summarizes the equations that we estimated for each morphometric trait using linear and piecewise regression.
3.2. Estimating transitions and rates of change in gene expression with intercept free models
We recovered four basic temporal gene expression profiles. First, we observed piecewise patterns in which m ≈ zero (e.g., UMOD). Second, we observed piecewise patterns in which m < β1 but m ≠ zero (e.g., CALM2). Third, we observed piecewise patterns that are consistent with the notion of a transient response to T4 that is followed by a return to baseline expression levels (e.g., MMP1). Fourth, we observed linear expression profiles (e.g., TR-α). The shapes (piecewise/quadratic versus linear) of the intercept free models (β0 = 0) that were selected for each gene and their parameter estimates are shown in Table 2. Attempts to fit intercept free piecewise models to TR-α and GLUL were thwarted by convergence issues that are due to the difficulties associated with estimating Ψ for these genes (i.e., several possible breakpoints are essentially equally satisfactory). However, the intercept free quadratic models (β0 = 0) fit to TR-α and GLUL both had larger AIC values than the linear models fit to these genes that were selected. These results support the notion that linear models are sufficient to describe TR-α and GLUL expression.
Table 2.
Results from linear and piecewise model selection when β0 = 0.
| Gene | n | df | β1 ± SE (log2R/day) | Ψ ± SE (days) | m ± SE (log2R/day) | AIC | Model |
|---|---|---|---|---|---|---|---|
| UMOD | 50 | 49 | −0.556 ± 0.025 | N/A | N/A | 119.5 | Linear |
| UMOD* | 50 | 47 | −0.891 ± 0.041 | 14.340 ± 1.039 | −0.006 ± 0.066 | 55.6 | Piecewise |
| KRT6A | 50 | 49 | −0.235 ± 0.017 | N/A | N/A | 82.6 | Linear |
| KRT6A* | 50 | 47 | −0.527 ± 0.050 | 11.540 ± 1.218 | 0.071 ± 0.037 | 28.6 | Piecewise |
| CALM2 | 50 | 49 | −0.157 ± 0.009 | N/A | N/A | 11.5 | Linear |
| CALM2* | 50 | 47 | −0.554 ± 0.075 | 4.676 ± 0.787 | −0.048 ± 0.012 | −50.8 | Piecewise |
| KRT14 | 50 | 49 | 0.391 ± 0.018 | N/A | N/A | 88.3 | Linear |
| KRT14* | 50 | 47 | 0.770 ± 0.042 | 10.380 ± 0.826 | 0.063 ± 0.031 | 9.4 | Piecewise |
| MMP1 | 50 | 49 | 0.004 ± 0.011 | N/A | N/A | 36.7 | Linear |
| MMP1* | 50 | 47 | 0.044 ± 0.020 | 20.890 ± 3.227 | −0.196 ± 0.098 | 31.6 | Piecewise |
| EMP1 | 48 | 47 | 0.147 ± 0.012 | N/A | N/A | 45.0 | Linear |
| EMP1* | 48 | 45 | 0.254 ± 0.024 | 16.330 ± 1.878 | −0.093 ± 0.055 | 18.3 | Piecewise |
| TR-β* | 50 | 49 | −0.055 ± 0.005 | N/A | N/A | −34.9 | Linear |
| TR-β | 50 | 47 | −0.054 ± 0.006 | 29.05 ± 10.85 | −0.117 ± 0.325 | −31.0 | Piecewise |
| TR-α* | 50 | 49 | −0.074 ± 0.007 | N/A | N/A | −11.7 | Linear |
| GLUL* | 50 | 49 | −0.036 ± 0.009 | N/A | N/A | 13.3 | Linear |
Piecewise models are not listed for TR-α and GLUL because attempts to fit such models failed to converge.
indicates models that were selected using Akaike’s information criterion (AIC). N/A indicates that a value (Ψ or m) is not obtained via linear regression. df is the sample size (n) minus the number of parameters estimated for a given model (for piecewise models β1, β2, and Ψ = three parameters and for linear models β1 = one parameter).
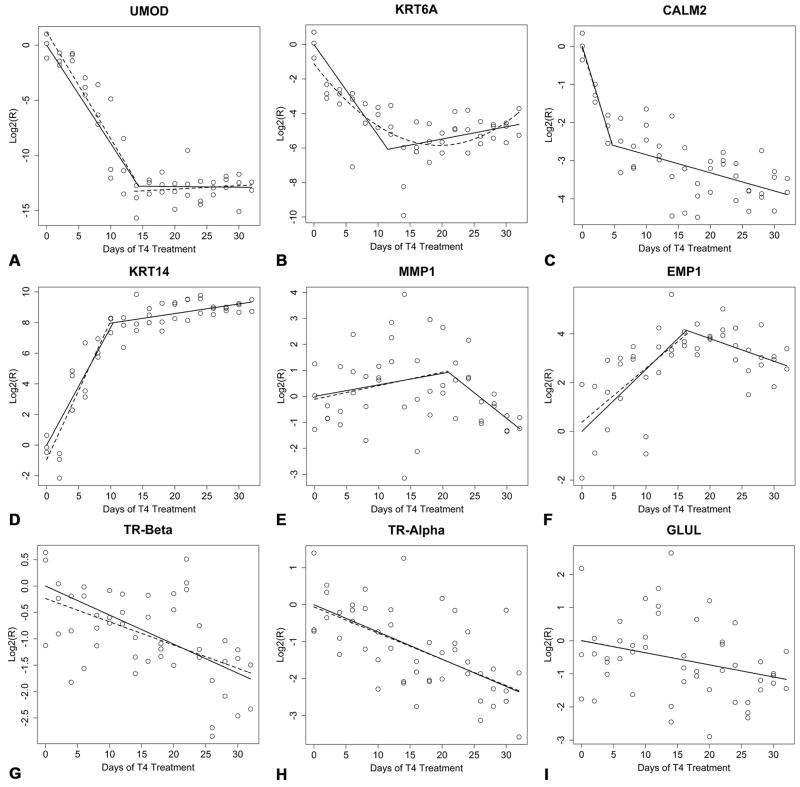
Three down-regulated genes (UMOD, KRT6A, and CALM2) exhibited piecewise relationships. UMOD was drastically down-regulated in response to T4 (Fig. 6A). Taking the absolute value of the log2(R) predicted for UMOD at time Ψ (≈ 12.78) and back transforming to the raw scale (212.78) results in an estimate of ≈ 7000 fold down-regulation in UMOD transcripts prior to metamorphic offset (Ψ = 14.340 days). This suggests UMOD transcripts are down-regulated at an average rate of approximately (7000/14 = 500) 500 fold per day of T4 treatment prior to metamorphic offset. Analogous calculations for KRT6A (Fig. 6B) suggest that KRT6A is down-regulated by ≈ 65 fold by its time of metamorphic offset (Ψ = 11.540 days). This gives an average rate of a (65/11.540 = 5.6) ≈ 5.6 fold decrease in KRT6A transcript abundance per day of T4 treatment prior to metamorphic offset. The piecewise model fit to CALM2 (Fig. 6C) requires a different interpretation than the models fit to UMOD and KRT6A. The breakpoint in the CALM2 model was estimated to occur following 4.68 days of T4 treatment and corresponds to a transition in the rate of CALM2 down-regulation. The estimate of β1 for CALM2 is over an order of magnitude larger than the estimate of m for CALM2 (Table 2). By time Ψ, CALM2 was down-regulated by approximately 5 fold and by the end of the experimental period (Day32) CALM2 was down-regulated by approximately 15 fold.
Fig. 6.
Scatter plots and prediction lines for the intercept free models (β0 = 0; solid lines) and the models for which β0 was estimated from the Q-RT-PCR data (dashed lines). (A) UMOD, (B) KRT6A, (C) CALM2, (D) KRT14, (E) MMP1, (F) EMP1, (G) TR-β, (H) TR-α, and (I) GLUL.
We observed three up-regulated genes (KRT14, MMP1, and EMP1) that exhibited piecewise profiles. KRT14, was strongly up-regulated in response to TH (Fig. 6D). Calculations analogous to those computed previously suggest that KRT14 is up-regulated by ≈ 250 fold by time Ψ resulting in an average rate of a (250/10.380 = 24.1) ≈ 24 fold increase in transcript abundance per day of T4 treatment prior to Ψ. By the end of morphological metamorphosis (Day 32) KRT14 transcripts were up-regulated by approximately 650 fold relative to Day 0 expression levels. MMP1 expression was variable throughout metamorphosis and our temporal expression data for MMP1 are equivocal (Fig. 6E). However, model selection (AIC) resulted in a model whose general interpretation (transient expression) is consistent with the presumptive role of MMP1 during amphibian metamorphosis (collagen remodeling). The estimate for Ψ suggests that MMP1 expression levels were maximal following roughly 21 days of T4 treatment. However, there is considerable uncertainty associated with the estimate of Ψ for MMP1 (Table 2). EMP1 was up-regulated (Fig. 6F) by approximately 17 fold by time Ψ resulting in an average rate of a (17/16.330 ≈ 1.0) ≈ one fold increase in EMP1 transcript abundance per day of T4 treatment prior to Ψ. However, in the case of EMP1, the interpretation of Ψ is unclear. Because the estimate of m is negative for EMP1 (Table 2), it is not clear whether EMP1 levels remain elevated outside of our experimental interval (i.e., in animals that have completed morphological metamorphosis).
Finally, we observed three genes (TR-β, TR-β, and GLUL) with linear expression profiles. The linear model selected for TR-β (Fig. 6G) suggests that TR-α is down-regulated by approximately 3.4 fold by the end of morphological metamorphosis (Day 32). This implies that TR-α transcripts decrease in abundance at an average rate of roughly (3.4/32 = 0.11) 0.11 fold per day of T4 treatment. Similarly, the linear model selected for TR-α (Fig. 6H) suggests that TR-α is down-regulated by approximately 5.1 fold by Day 32. Thus TR-α transcripts decreased in abundance at an average rate of approximately (5.1/32 = 0.16) 0.16 fold per day of T4 treatment throughout the experimental period. GLUL expression was extremely variable and its magnitude of change was modest (Fig 6I; back transformation of the predicted value at Day 32 suggests roughly a two fold decrease relative to Day 0). Overall, our Q-RT-PCR data highlight the dramatic changes in gene expression that occur in the skin during TH-induced metamorphosis of A. mexicanum. Fig. 7 summarizes the intercept free equations that we estimated via linear and piecewise regression and shows the timing of these gene expression patterns relative to key morphological events.
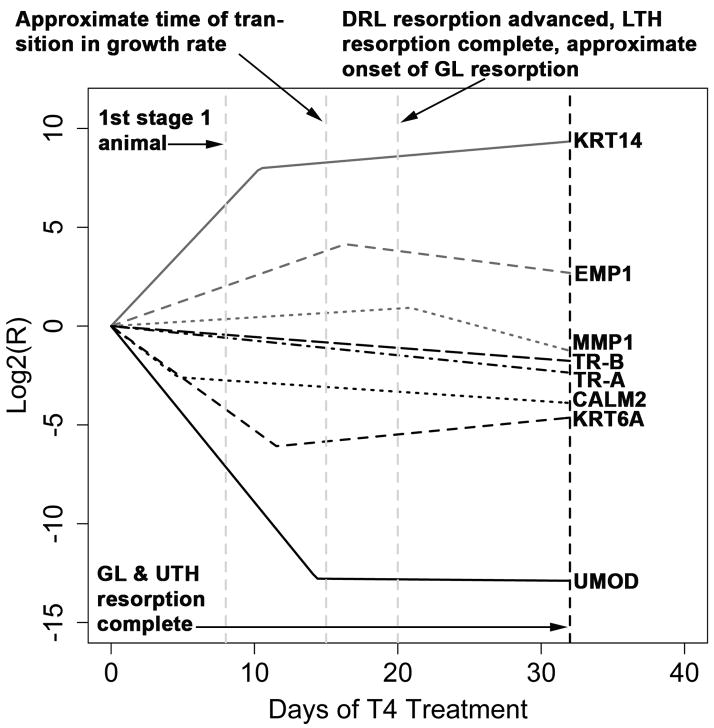
Fig. 7.
The intercept free (β0 = 0) equations estimated from the Q-RT-PCR data. Major morphological events are shown as dashed vertical lines.
3.3. Differential expression and hypothesis testing
Estimating β0 via least squares generally supported the notion that assuming β0 = 0 (i.e., fitting intercept free models) is valid (Fig 6; Table 3). Comparing AIC values from linear models for which β0 was estimated with AIC values from piecewise or quadratic models for which β0 was estimated did not result in the selection of models with different shapes (linear versus piecewise/quadratic) from those presented in Table 2 for any of the genes investigated. The general correspondence between intercept free models and models in which β0 was estimated from the data suggests that our results are robust (Fig 6). We note that seven of the models (UMOD, KRT6A, CALM2, KRT14, EMP1, TRβ, and TR-α) used to estimate β0 account for > 25% of the variation in the data (i.e., R2 > 0.25) with six of these accounting for > 33%, four accounting for > 50%, and two accounting for > 90% (Table 3; see below for KRT6A).
Table 3.
Results from the models selected for hypothesis testing in which β0 was estimated from the data.
| Gene | n | df | β0 | P(β0 = 0) | β1 | P(β1 = 0) | m | LCI | UCI | R2 | Model |
|---|---|---|---|---|---|---|---|---|---|---|---|
| UMOD | 50 | 46 | 1.189 | 0.065 | −0.967 | 1.309 × 10−14 * | 0.031 | −0.073 | 0.135 | 0.909 | PWL |
| CALM2 | 50 | 46 | −0.058 | 0.853 | −0.537 | 5.382 × 10−5 * | −0.048 + | −0.071 | −0.025 | 0.742 | PWL |
| KRT14 | 50 | 46 | −0.956 | 0.043 ◆ | 0.907 | 1.158 × 10−12 * | 0.063 + | 0.012 | 0.113 | 0.908 | PWL |
| MMP1 | 50 | 46 | −0.111 | 0.800 | 0.052 | 0.165 | −0.196 + | −0.391 | −0.001 | 0.153 | PWL |
| EMP1 | 48 | 44 | 0.376 | 0.429 | 0.221 | 3.186 × 10−5 * | −0.093 | −0.202 | 0.016 | 0.441 | PWL |
| TR-β | 50 | 48 | −0.237 | 0.213 | −0.044 | 7.650 × 10−5 * | N/A | N/A | N/A | 0.281 | L |
| TR-α | 50 | 48 | −0.060 | 0.803 | −0.071 | 1.830 × 10−6 * | N/A | N/A | N/A | 0.381 | L |
| GLUL | 50 | 48 | 0.005 | 0.987 | −0.037 | 0.034 * | N/A | N/A | N/A | 0.090 | L |
df is the sample size (n) minus the number of parameters estimated for a given model (for piecewise models β0, β1, Ψ, and β2 = four parameters and for linear models β0 and β1 = two parameters). P(β0 = 0) is the probability that β0 = 0 and P(β1 = 0) is the probability that β1 = 0. LCI is the lower 95% confidence interval for m and UCI is the upper 95% confidence interval for m. Lower and upper 95% confidence intervals for m that do not overlap zero are deemed statistically different from zero (denoted by +). PWL = piecewise linear and L = linear.
indicates β0 terms that statistically differ from zero at the 0.05 level.
indicates β1 terms that statistically differ from zero at the 0.05 level. R2 is the proportion of variation in the data explained by the model. N/A = values that are not obtained via linear models. Results for KRT6A are not shown because attempts to fit a piecewise model that estimated β0 failed. Units for β0 are log2(R). Units for β1, and m are log2(R)/day of T4 treatment.
Inspection of Table 3 reveals that the estimates of β1 for UMOD, CALM2, KRT14, EMP1, TR-β, and TR-α are clearly statistically different from zero. Attempts to fit a piecewise model to KRT6A that estimated β0 failed to converge. However, the quadratic model fit to KRT6A (Fig. 6B) had a lower AIC than the analogous linear model and was highly statistically significant (n = 50, df = 47, β0 = −1.11, t = −2.38, P = 0.022, γ1 = −0.485, t = −7.024, P = 7.52 × 10−9, γ2 = 0.012, t = 5.854, P = 4.47 × 10−7, R2 = 0.567). Although the estimate of β1 for GLUL is statistically different from zero at the 0.05 level (Table 3), expression was too variable to allow for inferences on GLUL’s role during metamorphosis, and we do not attach biological significance to this result. The estimate of β1 for MMP1 was not statistically different from zero (Table 3).
Examination of the estimates for m and their 95% confidence intervals reveal that m is not different from zero at the 0.05 level for UMOD and EMP1 (Table 3). Our interpretation of this with respect to EMP1 was discussed in the previous section. However, for UMOD this result suggests that metamorph expression levels are reached at time Ψ (Fig. 6A). Because the estimates of m for CALM2 and KRT14 statistically differed from zero, it seems likely that expression levels for these genes decrease and increase respectively in a more gradual fashion after their respective transition times (Fig. 6C; Fig. 6D). MMP1 represents an interesting case in the sense that piecewise models consistent with transient regulation were selected in favor of simpler linear models irrespective of whether β0 was estimated (see above). When this result is considered in combination with the observation that a value unique to piecewise models (m) is statistically significant but a parameter with a clear analog in simpler linear models (β1) is null, the idea that MMP1 is transiently up-regulated in response to TH is supported. While acknowledging that MMP1 expression is clearly variable, we note that the ISH data presented below are consistent with the view that MMP1 is up-regulated in response to T4 (see below) as are our results from previous expression screens (Page et al., 2008).
3.4. Spatial changes in gene expression during skin remodeling
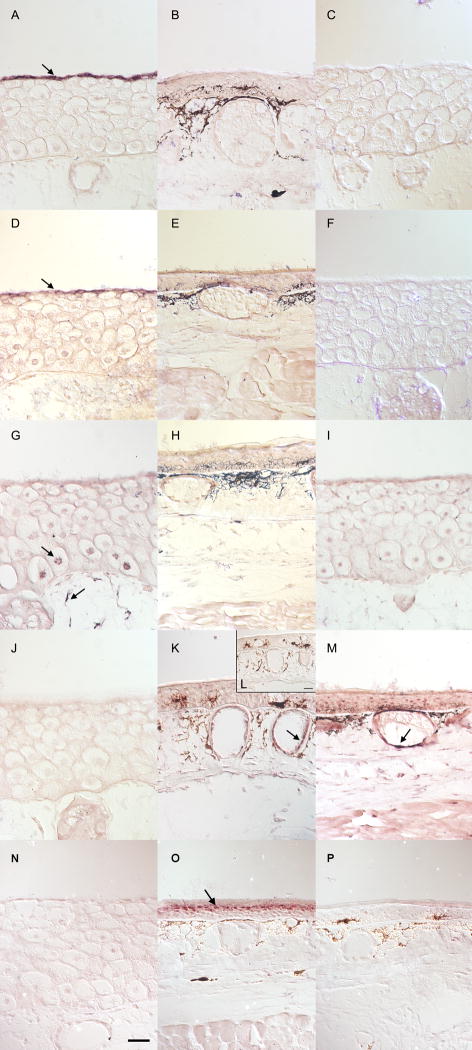
In order to verify the temporal patterns described above and associate candidate biomarkers with specific cell populations, we conducted ISH. The spatial expression data obtained via ISH confirm that UMOD (Fig 8A–C), KRT6A (Fig. 8D–F), and CALM2 (Fig. 8G–I) are larval specific. UMOD and KRT6A were expressed exclusively in the apical cell layer that is present only in the larval epidermis where as CALM2 was expressed by basal Leydig cells and dermal fibroblasts (Fig. 8G–I). MMP1 was not detectable in larval skin (Fig. 8J). However, at Day 12 and Day 28, MMP1 was expressed in dermal glands (Fig. 8K, Fig. 8M). ISH confirmed that KRT14 is metamorph specific (Fig. 8N–P) and revealed that KRT14 is strongly and exclusively expressed in the granular layer of the metamorph epidermis (Fig. 8O).
Fig. 8.
In situ hybridizations of metamorphosing head skin. Control-sense probes were run on slides collected from the same animal as the positive signal. Black deposits in panels B, E, H, K, L, O, and M, and P are melanin deposits and not positive signal. Anti-sense Day 0 samples can be found in panels A, D, G, J, and N. An Anti-sense Day 12 sample can be found in panel K. Anti-sense Day 28 samples can be found in panels B, E, H, M and O. Control-sense Day 0 samples can be found in panels C, F, and I. Control sense Day 12 and Day 28 samples can be found in panels L and P, respectively.. UMOD (A–C) and KRT6A (D–F) are exclusively expressed in the apical cells of larval skin (A and D arrows). CALM2 (G–I), is expressed in basal Leydig cells and highly expressed in dermal fibroblasts (panel G, arrows). MMP1 (J–M) is highly expressed in dermal glands on Day 12 (K, arrow) and Day 28 (M, arrow). KRT14 (N–P) is highly expressed in the granular layer of Day 28 epidermis (O, arrow). The scale bar is 50 μm.
4. Discussion
4.1. Synopsis of developmental sequence
To facilitate further study of metamorphosis in the axolotl, we conducted a detailed time-course study in which we used 50 nM T4 to induce metamorphosis in Mexican axolotls while tracking changes in morphological and transcriptional events. As Fig. 7 makes clear, the earliest events occur at the transcriptional level. In particular, CALM2, KRT6A, KRT14, and UMOD are all substantially differentially regulated between the onset of T4 administration and approximately 14 days of T4 administration. Around the same time that the last of these genes (UMOD) undergoes metamorphic offset, there is a reduction in growth rate, as estimated from SVL and mass, that is accompanied by maximal EMP1 expression levels. This reduction in growth rate is followed by advanced lower tail fin and dorsal ridge resorption, the former of which is completely resorbed following roughly 20 days of T4 treatment. At approximately the same time that the lower tail fin is completely resorbed, MMP1 expression levels are maximal and the gills begin to undergo rapid resorption. Between Day 28 and Day 32, the gills and upper tail fin are completely resorbed, resulting in what has traditionally been considered the end of metamorphosis (Prahlad and DeLanney, 1965; Voss and Smith, 2005). Collectively, these results demonstrate the necessity of transcriptional biomarkers for indexing early metamorphic events and highlight the dynamic changes that occur during induced metamorphosis of the axolotl. Below, we summarize how our gene expression data will enable future studies to index metamorphic progress in the axolotl prior to morphological changes. We then compare and contrast natural and induced metamorphosis in urodeles and discuss the remodeling patterns observed in the epidermis and dermis. Finally, we briefly highlight what appear to be conserved features of urodele and anuran metamorphosis.
4.2. Gene expression changes in the skin: identification of transcriptional indices
Of the genes that we investigated, UMOD, KRT6A, and CALM2 are larval specific. Thus, the responses of these genes to TH are useful for indexing loss of the larval gene expression program in the skin. UMOD and KRT6A are exclusively expressed in the apical cell layer, a larval specific structure that is completely removed during metamorphosis. Hence, UMOD and KRT6A specifically mark the loss of apical cells from the skin during metamorphosis. In addition to these larval specific genes, we have also demonstrated that KRT14 is metamorph specific and expressed exclusively in the granular layer of the metamorph epidermis. For these reasons, KRT14 has considerable utility as a biomarker of differentiation and proliferation of the metamorph epidermis. Of the remaining genes investigated (TR-α, TR-β, EMP1, MMP1, and GLUL), the TRs and EMP1 showed the most potential as transcriptional indices of metamorphosis. Because TH sensitivity is dependent on the presence of TRs (Yoshizato and Frieden, 1975), TR-α and TR-β are useful biomarkers of TH sensitivity. Thus the linear down-regulation that we observed for TR-α and TR-β probably reflects a decrease in TH sensitivity in the skin between metamorphic climax and the end of morphological metamorphosis (see below). With respect to EMP1, in mammals this gene is associated with cell proliferation (Ben-Porath and Benvenisty, 1996) and epithelial differentiation (Marvin et al., 1995). The expression pattern that we recovered for EMP1 is consistent with the axolotl ortholog playing similar roles during TH-induced metamorphosis. Finally, MMP1 and GLUL showed the least potential as metamorphic indices. As mentioned above, MMP1 and GLUL expression were variable; however, MMP1 may have utility as a marker of extra cellular matrix (ECM) remodeling in the dermis (see above; see below). Overall, the genes investigated in this study increase the size of the molecular toolkit for studying TH-induced metamorphosis in the axolotl. These markers identify larval cells (CALM2, KRT6A, UMOD), adult cells (KRT14), factors that influence TH sensitivity (TR-α and TR-β), and putatively, epidermal proliferation and differentiation (EMP1).
4.3. Comparison between natural and induced metamorphosis
In naturally metamorphosing amphibians, serum T3 and T4 gradually rise prior to metamorphic climax (originally proposed by Etkin, 1968 and best documented in anurans; see Shi, 2000 and Denver et al., 2002 for reviews), and the TH competence of cells and tissues increases during the course of larval development (e.g., Rose, 1995b). In contrast, an induced metamorphosis is typically achieved by suddenly exposing larvae to a high dose of one form of TH (T3 or T4); a practice whose effect on serum T3 and T4 levels is not fully understood. Galton (1992) found that juvenile axolotls injected with radiolabeled T4 had undetectable serum T3 at 20 hours post-injection suggesting that T4 is not immediately converted to T3 in large quantities. In agreement with Galton (1992), Alberch et al. (1985) were unable to detect T3 in Eurycea bislineata larvae immersed in 50 nM T4 at 24 hours post-immersion. Moreover, Alberch et al., (1985) observed serum T4 levels as high as 41 nM during the first 48 hours of immersion followed by a leveling off to physiologically relevant T4 concentrations (≈ 1–11 nM; see Alberch et al., 1986). Thus, in addition to discrepancies that may arise between natural and induced metamorphosis due to gradual versus sudden TH exposure, available studies suggest that the serum TH profiles of induced and naturally climaxing urodeles may be quite different, especially early during their respective TH surges.
The dose of T4 that we used to induce metamorphosis is roughly twice the maximum serum T4 level observed in a naturally metamorphosing tiger salamander (28 nM; Larras-Regard, 1981). In a previous study (Page et al., 2008), we found that this relatively high dose of T4 (50 nM) reduces the latency period associated with induced metamorphosis in the axolotl, but does not induce morphological and transcriptional changes that differ from the patterns observed when metamorphosis is induced with a physiologically relevant concentration of T4 (5 nM). Studies that we have conducted on eastern tiger salamanders (Ambystoma tigrinum tigrinum) and tiger salamander-axolotl hybrids show that 120 days post-fertilization is within the range of ages that metamorphic onset occurs spontaneously under laboratory conditions (Voss and Smith, 2005; unpublished data; also see Ducibella, 1974). Thus, the current study presumably reflects a level of TH competence in tissues and cells that is similar to those of naturally metamorphosing members of the tiger salamander complex. While our experimental paradigm is clearly contrived, we have attempted to minimize the disparities between our approach and natural metamorphosis. Despite imperfections, we think that the ability to uniformly administer T4 to animals that are phylogentically close to naturally metamorphosing taxa, but not developing toward metamorphic endpoints, represents a powerful model system that provides information relevant to natural metamorphosis.
4.4. Metamorphic remodeling pattern in the skin: differences between the epidermis and dermis
The remodeling patterns that we observed show that axolotls rapidly remove the larval epidermis while simultaneously constructing the metamorph epidermis. Thus, while gross morphological changes, such as tissue resorption and weight loss are delayed, gene expression changes for some genes in the epidermis are substantial quite early in the induction period. The fact that UMOD, KRT6A, and KRT14 were drastically down, down, and up-regulated respectively shows that axolotls rapidly down-regulate genes that function as larval barriers while rapidly up-regulating genes with protective function in the metamorph epidermis. The mixture of larval and metamorph cell types that we observed at Day 12 corroborates this account of simultaneous removal of the larval epidermis and construction of the metamorph epidermis. In contrast, the remodeling observed in the dermis was far less dramatic, and a well-formed glandular dermis was present in Day 0 axolotls prior to T4 treatment. Holder and Glade (1984) showed that axolotls naturally develop a well-formed glandular dermis during larval development and that dermal glands form as clusters of basal epidermal cells that pass beneath the basement membrane. Ohmura and Wakahara (1998) observed similar patterns for dermis and gland development, but found that these events were associated with epidermal remodeling in Hynobius retardatus. Interestingly, when Ohmura and Wakahara (1998) inhibited thyroid function in H. retardatus via hyphophysectomy, thyroidectomy, or treatment with sodium perchlorate, they found that dermal thickening and gland formation occurred in the absence of epidermal transformation. Thus, either dermis formation is independent of TH in at least some urodeles, or very low concentrations of TH are required for dermis development. While studying the metamorphic cranial and hyobranchial remodeling that occur in E. bislineata, Rose (1995c) found substantial variation in the TH sensitivities of the tissues that comprise these structures. By immersing Eurycea larvae in a series of T4 concentrations ranging from 0.05 nM to 50 nM, Rose (1995c) was able to show that doses as low as 0.05 nM T4 (well below the sensitivity of RIAs used in studies of urodele metamorphosis) are capable of stimulating metamorphic remodeling in some tissues. Thus, it is possible that, as previously suggested by Ducibella (1974), Rosenkilde and Ussing (1996) and Brown (1997), TH plays important roles in the natural development of axolotls despite the low serum T4 estimates from RIAs (Darras and Kuhn, 1983, 1984; Galton, 1992). Irrespective of whether dermis development is TH dependent in the axolotl, our ISH results for MMP1 suggest that T4 induces remodeling in the ECM of the dermis. In addition, our ISH results suggest that dermal fibroblasts down-regulate CALM2 expression following exposure to T4. Thus, cells within the dermis did undergo some differential transcription and/or mRNA metabolism in response to 50 nM T4 despite modest histological remodeling.
4.5. Comparison to gene expression changes during anuran metamorphosis
Most of the genes that we have investigated in this study are not known TH response genes in other amphibian taxa. However, TR-α, TR-β, UMOD, and MMP1 expression have been examined in anurans. In X. laevis, TR-α is expressed early in larval development whereas TR-β abundance increases with TH levels (Yaoita and Brown, 1990; Kawahara et al., 1991). Thus, in Xenopus, TR-α is thought to be a repressor of metamorphic gene expression in the absence of TH (i.e., during pre-metamorphosis) whereas TR-β expression is associated with activation of the metamorphic gene expression program (reviewed by Buchholz et al., 2006). In X. laevis, it is known that TR-β is directly targeted by TH (i.e., contains a thyroid hormone response element; Ranjan et al., 1994; Wong and Shi, 1995) and that exogenous TH can induce Xenopus TR-β expression precocially (e.g., Yaoita and Brown, 1990; Kanamori and Brown, 1992). While axolotl TRs have been shown to function in mammalian cells (Safi et al., 2004), very little is known about their expression patterns during natural development. Galton (1992) found that putative T3 receptors were more abundant in juvenile axolotls than in paedomorphic adults suggesting that beyond a certain age TH sensitivity may decrease in the axolotl naturally. Because frogs and salamanders are separated by over 200 million years of independent evolution (Kardong, 1998; Anderson et al., 2008), and the axolotl (Shaffer, 1993) and Xenopus (Bolker, 1995) represent derived modes of development within their respective orders, it is not clear how to interpret our results on TR expression within the broader context of amphibian metamorphosis. While we did not observe any evidence of an increase in the abundance of TR-β, this could be explained by a number of possible scenarios. For example, TR-β may have been up-regulated in response to low concentrations of endogenous TH, or by a TH independent mechanism, during the larval period prior to the beginning of our experiment. If this is the case, TR-β may have already been at maximal levels by 120 days post-fertilization. Irrespective of these considerations, our results from axolotl are consistent with the observation from Xenopus that TRs decrease in abundance subsequent to metamorphic climax (Yaoita and Brown, 1990; Kawahara et al., 1991).
Despite the evolutionary considerations cited above, the expression of UMOD-like genes is strikingly similar between A. mexicanum and X. laevis. In X. laevis, two genes with clear homology to human UMOD are expressed exclusively in apical cells and are dramatically down-regulated in response to TH (Furlow et al., 1997). Furlow et al. (1997) hypothesized that Xenopus UMOD genes are part of a larval-specific complex that has “protective” and “secretory” functions and is homologous to the mammalian periderm. Our current results are extraordinarily similar to those described for X. laevis demonstrating that the expression of UMOD-like genes is strongly conserved between Xenopus and axolotl with respect to spatial location and TH responsiveness.
Similarly, there appear to be parallels between axolotl MMP1 expression and MMP1 expression in Rana catesbeiana (bullfrog). Using immunohistochemistry, Oofusa and Yoshizato (1991) found that bullfrog MMP1 protein increased in back skin in response to T3 treatment. These results are interesting because skin from the tadpole trunk undergoes remodeling that is histologically similar to the remodeling observed in the current study (reviewed by Yoshizato, 2007). Of particular interest is that Oofusa and Yoshizato (1991) reported that bullfrog MMP1 is found in collagen fibers and fibroblasts located in the dermis. Thus, it seems that MMP1 is expressed in the dermis of metamorphosing axolotls and bullfrogs. Collectively, these findings suggest that several aspects of amphibian metamorphosis appear to be conserved between urodeles and anurans at the level of gene expression.
4.6. Conclusion
Studying metamorphic remodeling in urodeles at multiple levels of biological organization provides comparative perspective and insights into amphibian metamorphosis. In the axolotl, additional temporal and spatial profiling of TH responsive genes from the skin and other tissues is needed to identify additional biomarkers. Within the skin, transcriptional markers of Leydig cells, basal cells, and various adult cell types are needed so that the fate of different epidermal components at metamorphosis can be better understood. In addition, it will be important to characterize TR expression early during axolotl development, so that the roles of these transcription factors during natural development and induced metamorphosis can be better understood. In the current study, we have identified a sequence of transcriptional and morphological events that will provide a useful benchmark for future studies of induced metamorphosis in the Mexican axolotl. In addition, we have identified biomarkers of larval skin loss and adult skin development. This information will enable future studies to assess metamorphic changes in the skin well before any signs of morphological metamorphosis are evident.
Acknowledgments
This project was supported by grants R24-RR016344 and P20-RR016741 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH. This project was also supported by the National Science Foundation funded Ambystoma Genetic Stock Center (DBI-0443496). Phil Crowley provided helpful comments on an earlier draft of this manuscript. Arnold Stromberg provided statistical advice. The content of this manuscript was improved by comments from two anonymous reviewers.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alberch P, Lewbart GA, Gale EA. The fate of larval chondrocytes during the metamorphosis of the epibranchial in the salamander, Eurycea bislineata. J Embryol Exp Morphol. 1985;88:71–3. [PubMed] [Google Scholar]
- Alberch P, Gale EA, Larsen PR. Plasma T4 and T3 in naturally metamorphosing Eurycea bislineata (amphibia; plethodontidae) Gen Comp Endocrinol. 1986;61:153–63. doi: 10.1016/0016-6480(86)90261-3. [DOI] [PubMed] [Google Scholar]
- Anderson JS, Reisz RR, Scott D, Frobisch NB, Sumida SS. A stem batrachian from the early Permian of Texas and the origin of frogs and salamanders. Nature. 2008;453:515–518. doi: 10.1038/nature06865. [DOI] [PubMed] [Google Scholar]
- Armstrong JB, Duhon ST, Malacinski GM. Raising the axolotl in captivity. In: Armstrong JB, Malacinski GM, editors. Developmental Biology of the Axolotl. Oxford University Press; New York: 1989. pp. 220–27. [Google Scholar]
- Armstrong JB, Duhon ST. Induced spawnings, artificial insemination, and other genetic manipulations. In: Armstrong JB, Malacinski GM, editors. Developmental Biology of the Axolotl. Oxford University Press; New York: 1989. pp. 228–235. [Google Scholar]
- Ben-Porath I, Benvenisty N. Characterization of a tumor-associated gene, a member of a novel family of genes encoding membrane glycoproteins. Gene. 1996;183:69–75. doi: 10.1016/s0378-1119(96)00475-1. [DOI] [PubMed] [Google Scholar]
- Bolker JA. Model systems in developmental biology. Bioessays. 1995;17:451–455. doi: 10.1002/bies.950170513. [DOI] [PubMed] [Google Scholar]
- Brown DD. The role of thyroid hormone in zebrafish and axolotl development. Proc Natl Acad Sci USA. 1997;94:13011–13016. doi: 10.1073/pnas.94.24.13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DD. The role of deiodinases in amphibian metamorphosis. Thyroid. 2005;15:815–821. doi: 10.1089/thy.2005.15.815. [DOI] [PubMed] [Google Scholar]
- Brown DD, Cai L. Amphibian metamorphosis. Dev Biol. 2007;306:20–33. doi: 10.1016/j.ydbio.2007.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz DR, Hayes TB. Variation in thyroid hormone action and tissue content underlies species differences in the timing of metamorphosis in desert frogs. Evol Dev. 2005;7:458–467. doi: 10.1111/j.1525-142X.2005.05049.x. [DOI] [PubMed] [Google Scholar]
- Buchholz DR, Paul BD, Fu L, Shi YB. Molecular and developmental analyses of thyroid hormone function in Xenopus laevis, the African clawed frog. Gen Comp Endocrinol. 2006;145:1–19. doi: 10.1016/j.ygcen.2005.07.009. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Nolan T. Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J Biomol Tech. 2004;15:155–166. [PMC free article] [PubMed] [Google Scholar]
- Cano-Martinez A, Vargas-Gonzalez A, Asai M. Metamorphic stages in Ambystoma mexicanum. Axolotl Newsletter. 1994;23:64–71. [Google Scholar]
- Darras VM, Kuhn ER. Effects of TRH, bovine TSH, and pituitary extracts on thyroidal T4 release in Ambystoma mexicanum. Gen Comp Endocrinol. 1983;51:286–291. doi: 10.1016/0016-6480(83)90083-7. [DOI] [PubMed] [Google Scholar]
- Darras VM, Kuhn ER. Difference of the in vivo responsiveness to thyrotropin stimulation between the neotenic and metamorphosed axolotl, Ambystoma mexicanum: failure of prolactin to block the thyrotropin-induced thyroxine release. Gen Comp Endocrinol. 1984;56:321–325. doi: 10.1016/0016-6480(84)90047-9. [DOI] [PubMed] [Google Scholar]
- Denver RJ, Glennemeier KA, Boorse GC. Endocrinology of complex life cycles: amphibians. In: Pfaff D, Arnold A, Etgen A, Fahrbach S, Rubin R, editors. Hormones, Brains, and Behavior. Academic Press; San Diego: 2002. pp. 469–513. [Google Scholar]
- Ducibella T. The occurrence of biochemical metamorphic events without anatomical metamorphosis in the axolotl. Dev Biol. 1974;38:175–186. doi: 10.1016/0012-1606(74)90268-1. [DOI] [PubMed] [Google Scholar]
- Duellman WE, Trueb L. Biology of Amphibians. Johns Hopkins University Press; Baltimore: 1994. [Google Scholar]
- Etkin W. Hormonal control of amphibian metamorphosis. In: Etkin W, Gilbert LI, editors. Metamorphosis: a Problem in Developmental Biology. Appleton-Century-Crofts; New York: 1968. pp. 313–348. [Google Scholar]
- Fahrmann W. Morphodynamics of the epidermis of the axolotl (Siredon mexicanum Shaw) under the influence of exogenously administered thyroxine I. the epidermis of neotenic axolotls. Z Mikrosk Anat Forsch. 1971a;83:472–506. [PubMed] [Google Scholar]
- Fahrmann W. Morphodynamics of the epidermis of the axolotl (Siredon mexicanum Shaw) under the influence of exogenously administered thyroxine II. the epidermis during metamorphosis Z Mikrosk. Anat Forsch. 1971b;83:535–568. [PubMed] [Google Scholar]
- Fahrmann W. Morphodynamics of the epidermis of the axolotl (Siredon mexicanum Shaw) under the influence of exogenously administered thyroxine III the epidermis of the metamorphosed axolotl Z Mikrosk. Anat Forsch. 1971c;84:1–25. [PubMed] [Google Scholar]
- Friedberg F, Rhoads AR. Evolutionary aspects of calmodulin. IUBMB Life. 2001;51:215–221. doi: 10.1080/152165401753311753. [DOI] [PubMed] [Google Scholar]
- Frost SK, Neff AW, Malacinski GM. Primary and long-term culture of axolotl cells. In: Armstrong JB, Malacinski GM, editors. Developmental Biology of the Axolotl. Oxford University Press; New York: 1989. pp. 244–254. [Google Scholar]
- Furlow JD, Berry DL, Wang Z, Brown DD. A set of novel tadpole specific genes expressed only in the epidermis are down-regulated by thyroid hormone during Xenopus laevis metamorphosis. Dev Biol. 1997;182:284–298. doi: 10.1006/dbio.1996.8478. [DOI] [PubMed] [Google Scholar]
- Galton VA. Thyroid hormone receptors and iodothyronine deiodinases in the developing Mexican axolotl, Ambystoma mexicanum. Gen Comp Endocrinol. 1992;85:62–70. doi: 10.1016/0016-6480(92)90172-g. [DOI] [PubMed] [Google Scholar]
- Heady JE, Kollros JJ. Hormonal modification of the development of plical skin glands. Gen Comp Endocrinol. 1964;4:124–131. doi: 10.1016/0016-6480(64)90045-0. [DOI] [PubMed] [Google Scholar]
- Holder N, Glade R. Skin glands in the axolotl: the creation and maintenance of a spacing pattern. J Embyol Exp Morhol. 1984;79:97–112. [PubMed] [Google Scholar]
- Ishizuya-Oka A, Shi YB. Regulation of adult intestinal epithelial stem cell development by thyroid hormone during Xenopus laevis metamorphosis. Dev Dyn. 2007;236:3358–3368. doi: 10.1002/dvdy.21291. [DOI] [PubMed] [Google Scholar]
- Kanamori A, Brown DD. The regulation of thyroid hormone receptor β genes by thyroid hormone in Xenopus laevis. J Biol Chem. 1992;267:739–745. [PubMed] [Google Scholar]
- Kardong KV. Vertebrates: Comparative Anatomy, Function, Evolution. 2. McGraw-Hill; New York: 1998. [Google Scholar]
- Kawahara A, Baker BS, Tata JR. Developmental and regional expression of thyroid hormone receptor genes during Xenopus metamorphosis. Development. 1991;112:933–943. doi: 10.1242/dev.112.4.933. [DOI] [PubMed] [Google Scholar]
- Kumada Y, Benson DR, Hillemann D, Hosted TJ, Rochefort DA, Thompson CJ, Wohlleben W, Tateno Y. Evolution of the glutamine synthetase gene, one of the oldest existing and functioning genes. Proc Natl Acad Sci USA. 1993;90:3009–3013. doi: 10.1073/pnas.90.7.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larras-Regard E, Taurog A, Dorris M. Plasma T4 and T3 levels in Ambystoma tigrinum at various stages of metamorphosis. Gen Comp Endocrinol. 1981;43:443–450. doi: 10.1016/0016-6480(81)90228-8. [DOI] [PubMed] [Google Scholar]
- Leloup J, Buscaglia M. La triiodothyronine: hormone de la metamorphose des amphibiens. C R Acad Sci. 1977;284:2261–2263. [Google Scholar]
- Marvin KW, Fujimoto W, Jetten AM. Identification and characterization of a novel squamous cell-associated gene related to PMP22. J Biol Chem. 1995;270:28910–28916. doi: 10.1074/jbc.270.48.28910. [DOI] [PubMed] [Google Scholar]
- Monaghan JR, Walker JA, Page RB, Putta S, Beachy CK, Voss SR. Early gene expression during natural spinal cord regeneration in the salamander Ambystoma mexicanum. J Neurochem. 2007;101:27–40. doi: 10.1111/j.1471-4159.2006.04344.x. [DOI] [PubMed] [Google Scholar]
- Monaghan JR, Epp LG, Putta S, Page RB, Walker JA, Beachy CK, Zhu W, Pao GM, Verma IM, Hunter T, Bryant SV, Gardiner DM, Harkins TT, Voss SR. Microarray and cDNA sequence analysis of transcription during nerve-dependent limb regeneration. BMC Biology. 2009;7:1. doi: 10.1186/1741-7007-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muggeo VMR. Segmented: An R package to fit regression models with broken-line relationships. R News. 2008;8:20–25. [Google Scholar]
- Nieuwkoop PD, Faber J. Normal Table of Xenopus laevis (Daudin): a Systematical and Chronological Survey of the Development From the Fertilized Egg till the End of Metamorphosis. 2. North-Holland; Amsterdam: 1967. [Google Scholar]
- Norman MF. A practical method for staging metamorphosis in the tiger salamander Ambystoma tigrinum. Anat Rec. 1985;211:102–109. doi: 10.1002/ar.1092110115. [DOI] [PubMed] [Google Scholar]
- Ohmura H, Wakahara M. Transformation of skin from larval to adult types in normally metamorphosing and metamorphosis-arrested salamander, Hynobius retardatus. Differentiation. 1998;62:237–246. [PubMed] [Google Scholar]
- Oofusa K, Yoshizato K. Biochemical and immunological characterization of collagenase in tissues of metamorphosing bullfrog tadpoles. Dev Growth Differ. 1991;33:329–339. doi: 10.1111/j.1440-169X.1991.00329.x. [DOI] [PubMed] [Google Scholar]
- Oofusa K, Yoshizato K. Thyroid hormone-dependent expression of bullfrog tadpole collagenase gene. Roux’s Arch Dev Biol. 1996;205:243–251. doi: 10.1007/BF00365802. [DOI] [PubMed] [Google Scholar]
- Page RB, Monaghan JR, Samuels AK, Smith JJ, Beachy CK, Voss SR. Microarray analysis identifies keratin loci as sensitive biomarkers for thyroid hormone disruption in the salamander Ambystoma mexicanum. Comp Biochem Physiol C, Comp Pharmacol Toxicol. 2007;145:15–27. doi: 10.1016/j.cbpc.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Page RB, Voss SR, Samuels AK, Smith JJ, Putta S, Beachy CK. Effect of thyroid hormone concentration on the transcriptional response underlying induced metamorphosis in the Mexican axolotl (Ambystoma) BMC Genomics. 2008;9:78. doi: 10.1186/1471-2164-9-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petranka JW. Salamanders of the United States and Canada. Smithsonian Institution Press; Washington: 1998. [Google Scholar]
- Pfaffl M. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahlad KV, DeLanney LE. A study of induced metamorphosis in the axolotl. J Exp Zool. 1965;160:137–146. doi: 10.1002/jez.1401600112. [DOI] [PubMed] [Google Scholar]
- Prahlad KV. Induced metamorphosis: rectification of a genetic disability by thyroid hormone in the Mexican axolotl Siredon mexicanum. Gen Comp Endocrinol. 1968;11:21–30. doi: 10.1016/0016-6480(68)90103-2. [DOI] [PubMed] [Google Scholar]
- Putta S, Smith JJ, Walker JA, Rondet M, Weisrock DW, Monaghan JR, Samuels AK, Kump K, King DC, Maness NJ, Habermann B, Tanaka E, Bryant SV, Gardiner DM, Parichy DM, Voss SR. From biomedicine to natural history research: EST resources for ambystomatid salamanders. BMC Genomics. 2004;5:54. doi: 10.1186/1471-2164-5-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan M, Wong J, Shi YB. Transcriptional repression of Xenopus TRβ gene is mediated by a thyroid hormone response element located near the start site. J Biol Chem. 1994;269:24699–24705. [PubMed] [Google Scholar]
- Rose CS. Skeletal morphogenesis in the urodele skull: I. postembryonic development in the hemidactyliini (amphibia: plethodontidae) J Morphol. 1995a;223:125–148. doi: 10.1002/jmor.1052230203. [DOI] [PubMed] [Google Scholar]
- Rose CS. Skeletal morphogenesis in the urodele skull: II effect of developmental stage in thyroid hormone-induced remodeling. J Morphol. 1995b;223:149–166. doi: 10.1002/jmor.1052230204. [DOI] [PubMed] [Google Scholar]
- Rose CS. Skeletal morphogenesis in the urodele skull: III effect of hormone dosage in TH-induced remodeling. J Morphol. 1995c;223:243–261. doi: 10.1002/jmor.1052230303. [DOI] [PubMed] [Google Scholar]
- Rosenkilde P, Ussing AP. What mechanisms control neoteny and regulate induced metamorphosis in urodeles. Int J Dev Biol. 1996;40:665–673. [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the www for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Safi R, Bertrand S, Marchand O, Duffraisse M, de Luze A, Vanacker JM, Maraninchi M, Margotat A, Demeneix B, Laudet V. The axolotl (Ambystoma mexicanum), a neotenic amphibian, expresses functional thyroid hormone receptors. Endocrinology. 2004;145:760–772. doi: 10.1210/en.2003-0913. [DOI] [PubMed] [Google Scholar]
- Sawada T, Oofusa K, Yoshizato K. Thyroid hormone response element-like sequence in anuran metalloproteinase 1 gene is responsive to in vivo thyroid hormone administration. J Endocrinol. 2001;169:477–486. doi: 10.1677/joe.0.1690477. [DOI] [PubMed] [Google Scholar]
- Shaffer HB. Phylogenetics of model organisms: the laboratory axolotl, Ambystoma mexicanum. Syst Biol. 1993;42:508–522. [Google Scholar]
- Shaffer HB, McKnight ML. The polytypic species revisited: genetic differentiation and molecular phylogenetics of the tiger salamander Ambystoma tigrinum (amphibia: caudata) complex. Evolution. 1996;50:417–433. doi: 10.1111/j.1558-5646.1996.tb04503.x. [DOI] [PubMed] [Google Scholar]
- Shaffer HB, Voss SR. Phylogenetic and mechanistic analysis of a developmentally integrated character complex: alternate life history modes in ambystomatid salamanders. Am Zool. 1996;36:24–35. [Google Scholar]
- Shi YB. Amphibian Metamorphosis: From Morphology to Molecular Biology. Wiley-Liss; New York: 2000. [Google Scholar]
- Smith HM. Discovery of the axolotl and its early history in biological research. In: Armstrong JB, Malacinski GM, editors. Developmental Biology of the Axolotl. Oxford University Press; New York: 1989. pp. 3–12. [Google Scholar]
- Smith JJ, Kump DK, Walker JA, Parichy DM, Voss SR. A comprehensive expressed sequence tag linkage map for tiger salamander and Mexican axolotl: enabling gene mapping and comparative genomics in Ambystoma. Genetics. 2005a;171:1161–1171. doi: 10.1534/genetics.105.046433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Putta S, Walker JA, Kump DK, Samuels AK, Monaghan JR, Weisrock DW, Staben C, Voss SR. Sal-site: integrating new and existing ambystomatid research and informational resources. BMC Genomics. 2005b;6:181. doi: 10.1186/1471-2164-6-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CA, Kollros JJ. Stages in the normal development of Rana pipiens larvae. Anat Rec. 1946;94:7–23. doi: 10.1002/ar.1090940103. [DOI] [PubMed] [Google Scholar]
- Toms JD, Lesperance ML. Piecewise regression: a tool for identifying ecological thresholds. Ecology. 2003;84:2034–2041. [Google Scholar]
- Venables WN, Ripley BD. Modern Applied Statistics with S. 4. Springer; New York: 2002. [Google Scholar]
- Voss SR, Smith JJ. Evolution of salamander life cycles: a major-effect quantitative trait locus contributes to discrete and continuous variation for metamorphic timing. Genetics. 2005;170:275–281. doi: 10.1534/genetics.104.038273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisrock DW, Shaffer HB, Storz BL, Storz SR, Voss SR. Multiple nuclear gene sequences identify phylogenetic species boundaries in the rapidly radiating clade of Mexican ambystomatid salamanders. Mol Ecol. 2006;15:2489–2503. doi: 10.1111/j.1365-294X.2006.02961.x. [DOI] [PubMed] [Google Scholar]
- Wilbur HM, Collins JP. Ecological aspects of amphibian metamorphosis. Science. 1973;182:1305–1314. doi: 10.1126/science.182.4119.1305. [DOI] [PubMed] [Google Scholar]
- Wilder IW. The Morphology of Amphibian Metamorphosis. Smith College, Northampton; Massachusetts: 1925. [Google Scholar]
- Wong J, Shi YB. Coordinated regulation of and transcriptional activation by Xenopus thyroid hormone and retinoid X receptors. J Biol Chem. 1995;270:18479–18483. doi: 10.1074/jbc.270.31.18479. [DOI] [PubMed] [Google Scholar]
- Yaoita Y, Brown DD. A correlation of thyroid hormone receptor gene expression with amphibian metamorphosis. Genes Dev. 1990;4:1917–1924. doi: 10.1101/gad.4.11.1917. [DOI] [PubMed] [Google Scholar]
- Yoshizato K, Frieden E. Increase in binding capacity for triiodothyronine in tadpole tail nuclei during metamorphosis. Nature. 1975;254:705–707. doi: 10.1038/254705a0. [DOI] [PubMed] [Google Scholar]
- Yoshizato K. Molecular mechanism and evolutional significance of epithelial-mesenchymal interactions in the body and tail-dependent metamorphic transformation of anuran larval skin. Int Rev Cytol. 2007;260:213–260. doi: 10.1016/S0074-7696(06)60005-3. [DOI] [PubMed] [Google Scholar]
- Zar JH. Biostatistical Analysis. 4. Prentice Hall, Upper Saddle River; New Jersey: 1999. [Google Scholar]