Abstract
This review briefly outlines the importance of molecular imaging, particularly imaging of endogenous gene expression for noninvasive genetic analysis of radiographic masses. The concept of antisense imaging agents and the advantages and challenges in the development of hybridization probes for in vivo imaging are described. An overview of the investigations on oncogene expression imaging is given. Finally, the need for further improvement in antisense-based imaging agents and directions to improve oncogene mRNA targeting is stated.
Keywords: Antisense, Imaging, Peptide nucleic acid (PNA), Peptide-PNA conjugates, Positron emission tomography (PET)
1. Introduction
Early diagnosis of cancer remains challenging. Investigational approaches are increasingly focused on development of specific probes that take advantage of the targets that are uniquely expressed or markedly over expressed on tumors. Molecular imaging has emerged as a novel multidisciplinary field that provides potential for earlier detection and characterization of disease, elucidation of mechanisms at the molecular level, and evaluation of therapy by use of specific molecular probes. Advantages of such molecular imaging approaches include noninvasiveness and the ability to measure both spatial and temporal biodistribution of a molecular probe in intact living subjects for real time and serial studies.
Analysis of genomic sequence data generated by the human genome project is expected to give a better understanding of the genes involved in cancer development and chronic diseases [1]. Recent research reveals cancer to be a disease involving dynamic changes in the genome. Mutations that produce oncogenes with dominant gain of function and tumor suppressor genes with recessive loss of function have been identified for elicitation of cancer phenotypes in several cancers [2]. Modulations in the expression of key proteins of the cellular signaling pathways are at the forefront of molecular abnormalities found in cancer. Protein products of proto-oncogenes and tumor suppressor genes are involved in cell growth as growth factors, receptors, intracellular mediators or transcription factors and have been found to be altered through multiple mechanisms of oncogene activation. These include enhanced or ectopic expression, deletions, single point mutations and generation of chimeric proteins [3]. Considerable success has been achieved in the development of molecular imaging agents based on peptide–receptor, antibody-antigen, and substrate-enzyme interactions targeting such overexpressed or mutated proteins in cancers [4, 5, 6]. However, due to tertiary folding and complex structures in each individual protein, it is not feasible to design ligands for any protein based on peptide sequences alone.
Antisense chemotherapy based on the complementary hybridization of an antisense with a target oncogene mRNA sequence is being exploited for treatment of various types of cancers [7]. Based on the same targeting principle, radiolabeled antisense sequences have been explored for imaging applications. Design of antisense sequences for mRNA is theoretically straightforward, based on complementary base pairing rules. Noninvasive, real time imaging of oncogene expression in vivo would provide information on cellular gene expression patterns and might reveal molecular changes in diseased tissues at relatively early stages providing opportunities for gene therapy, especially against overexpressed oncogene mRNAs [8]. In this approach of mRNA targeting, high specificity may be achieved upon binding of radiolabeled oligonucleotides to the target due to sequence complementarity [9]. This approach is not only specific but also sensitive as it has been proposed that mRNA concentrations as low as 1pmol/L in the tissues can probably be imaged with positron emission tomography (PET) using radiolabeled probes of specific activity 1000 to 10000 Ci/mmol [10]. However, there are challenges in imaging endogenous gene expression with radiolabeled oligonucleotides such as in vivo stability, transport to the target, entry into the cell and hybridization with target specific sequences [11]. Development of antisense imaging agents is in its infancy, compared to other approaches of molecular imaging. However, imaging with antisense technology for early, specific and noninvasive detection of oncogene expression is unique and warrants greater attention.
2. Cancer diagnosis via endogenous gene expression
Out of the 20,000 to 25,000 genes in human genome, about 100 have been identified as protooncogenes and tumor suppressor genes [12] CCND1, HER2, MYCC, KRAS and BCL2 oncogenes, as well as the tumor suppressor TP53, are reported to be frequently mutated or overexpressed in cancer cells.
HER2 oncogene, also known as ERBB2/neu, encodes a transmembrane glycoprotein, Her2 (185kDa) with tyrosine kinase activity. Amplification of Her2 expression, consequent to gene amplification, has been demonstrated in a subset of breast cancers corresponding to approximately one third of the patients affected by the disease [13]. Overexpression of Her2 is also reported in other human tumors including ovarian carcinomas, head and neck cancer, lung and gastro-intestinal tumors [14]. Specific down regulation of HER2 mRNA and protein by antisense phosphorothioate oligoucleotide is reported indicating possibility of HER2 mRNA as a specific target for tumor imaging [15].
Activated RAS oncogenes have been identified in precancerous lesions of some common forms of human cancers indicating their role in early stages of carcinogenesis. HRAS mutation is most often associated with bladder and kidney cancers, NRAS mutations with melanoma and hematological malignancies, and KRAS mutations with lung, colorectal, ovarian and pancreatic cancers [16]. The human KRAS proto-oncogene codes for an evolutionary conserved G protein, K-Ras p21, which binds guanine nucleotides with high affinity and is associated with the inner surface of the plasma membrane. K-Ras p21 is involved in transducing signals from growth factors binding to the cell surface receptors [17]. Point mutations in KRAS proto oncogene, mostly confined to codon 12 or 13, are reported to be an early event in pancreatic tumorigenesis indicated both by high gene mutation frequency and by the presence of mutation in low grade tumors [18]. Approximately 95% of ductal pancreatic cancers exhibit activation of KRAS oncogene at the early pancreatic intraepithelial neoplasia (PanIN-1) stage [19].
MYCC oncogene encodes a nuclear DNA binding protein c-Myc (65 kDa) that binds with a small partner protein, Max. The resulting heterodimer binds specifically to the promoter element in the regulatory regions of genes involved in proliferation. MYCC oncogene expression is stimulated by estrogen in hormone responsive breast cancer cells in vitro. Amplification of MYCC is considered to be a powerful prognostic indicator, particularly in node negative and estrogen receptor positive breast cancer [20]. MYCC was the first oncogene targeted by antisense, specifically by MYC6 sequence to treat HL-60 promyelocytic leukemia cells. Gene inhibition and antiproliferative activity were displayed by MYC6 sequence in breast cancer cells [21].
Cyclin D1 protein, encoded by CCND1 (BCL1, PRAD1), is a proto-oncogenic regulator of the G1/S checkpoint in the cell cycle that has been implicated in the pathogenesis of several types of cancers, including breast, prostate, and pancreatic cancer [22]. Cyclin D1 appears to function by binding to cyclin dependent kinases. Overexpression of cyclin D1 in cultured cells leads to a more rapid transition through the G1 phase of the cell cycle and entry into the S phase.
Another oncogene involved in initiation of almost all follicular lymphomas and some diffuse large B-cell lymphomas is BCL2. This gene encodes a cytoplasmic protein that localizes to mitochondria and increases cell survival by inhibiting apoptosis. BCL2 is also important in chronic lymphocytic leukemia and lung cancer. The BCL2 family members BCL-XL and BCL2 inhibit apoptosis and are upregulated in many cancers [23, 24, 25]. Mutations in p53, a tumor suppressor gene has been found to be the most frequent genetic alteration in diverse types of human cancers [26].
Oligonucleotide antisense sequences specific for CCND1 [27], HER2 [15], MYCC [28], KRAS [29], BCL2 [30] and TP53 [31] are reported to downregulate respective gene expression in cancers. Hence, mutated or overexpressed CCND1, HER2, MYCC, KRAS, BCL2, and TP53 mRNAs are expected to serve as potential markers when linked with suitable radionuclides, fluorophores, or contrast agents for nuclear, optical, or magnetic resonance imaging, respectively.
3. In vivo gene imaging agents
Many challenges must be overcome for successful development of imaging agents for gene expression, such as improvement in chemistry to achieve stable oligonucleotides, and conjugation of oligonucleotides to the signaling moieties. Cell specific targeting of antisense sequences and development of biologically compatible probes to facilitate translation of in vitro results to in vivo applications is desired. There is also a need to address mechanisms of cellular uptake of oligonucleotides and estimate hybridization specificity of radiolabeled antisense agents [32].
3.1 Stability
Success has been achieved in design and synthesis of stable oligonucleotides based on modifications in the phosphate backbone, sugar, and heterocyclic base of nucleic acid monomers [33]. Novel oligonucleotide analogs have been synthesized with improved biological stability, solubility, cellular uptake and ease of synthesis. The simplest oligodeoxynucleotide modification involved blocking the 3′ terminus to prevent attack by 3′ exonucleases, the predominant extracellular degradative mechanism for oligodeoxynucleotides [34]. Other modifications focused on protecting the internucleoside linkage by changing the phosphodiester linkages to phosphorothioates, methylphosphonates, or boranophosphates [35]. Although these modifications have led to increased in vivo stability of oligonucleotides, they also have weakened hybridization to the RNA target sites due to the creation of chiral phosphorus diastereomers [36]. The deoxyribose may be modified to 2′-O-alkyl RNAs, such as 2′-O-methyl, strengthening hybridization and resisting nuclease attack [37]. Similar improvements result from preparing 3′-amino phosphoramidates or morpholino phosphorodiamidates [35].
Furthermore, attachment of a base to deoxyribose may be reversed, changing the natural β-anomer to the α-anomer. The α-anomer achieves nuclease resistance without loss of base pairing [38]. Each of these structural changes affect not only the nuclease susceptibility, but also the cellular uptake, the cellular trafficking and recognition as substrate for RNase H [39]. Among the derivatives described, only phosphodiester, phosphorothioates, and boranophosphates DNAs direct RNase H mediated degradation of hybridized RNA. Encouraging results have been obtained suggesting that greater potency and a better specificity might be possible with 2′-O-alkyl RNA, phosphorothioate chimeras, anomeric DNA chimeras, or DNA boranophosphates [35, 40].
The most radical modifications to oligonucleotides are found in peptide nucleic acids (PNA), where both the phosphodiester linkages and sugars are replaced with a peptide-like backbone of (N-2-aminoethyl) glycine units, with the bases directly attached by methylene-carbonyl linkers. Due to their achiral, uncharged and rather flexible peptide backbone, PNAs hybridize more strongly and specifically to RNA [41]. Twelve PNA bases are reported to be sufficient for statistical uniqueness among transcribed mRNAs [42]. Compared with other oligonucleotide derivatives, PNAs display the highest melting temperature (Tm) for duplexes formed with single-stranded DNA or RNA [42]. PNAs are not easily recognized by either nucleases or proteases and are thus resistant to enzymatic degradation and also show stability over a wide range of pH [43]. Hence, PNAs show desired properties for development of antisense imaging agents.
3.2 Antisense mechanism
Antisense oligonucleotides have shown promising results as chemotherapeutic agents. However, phosphorothioate DNAs are the only derivatives that have thus far been administered to humans. Despite their efficacy, phosphorothioate (PS) DNAs exhibit less sequence specificity than phosphodiesters or methylphosphonates due to significant binding to a wide spectrum of plasma and cellular proteins [44]. Requirements for antisense chemotherapy are different than those for antisense imaging. Pharmacokinetic requirements differ sharply as the antisense cellular transport is limited and mRNAs are produced continuously. For chemotherapy, prolonged blood circulation of antisense is desired to eliminate need for multiple administrations while for antisense imaging, rapid plasma clearance for achieving high target to background ratio is required. Among the chemically modified oligonucleotides such as phosphorothioates, a positive correlation exists between their ability to act as substrates for RNaseH due to formation of a suitable RNA: DNA hybrid and their potency for antisense inhibition. This indicates RNaseH-mediated translational arrest as a major mechanism of antisense chemotherapy [11]. However, for antisense imaging, hybridization of probe to the target sequence is the principal requirement. RNA hybridized to uncharged oligonucleotide derivatives, such as PNA, is not recognized and cleaved by RNase H. Hence, PNAs are more promising for diagnostic applications than the charged derivatives because the PNAs, the analytical reagents, do not destroy the target mRNA, the analyte.
3.3 Cell specific targeting of antisense sequences
A number of delivery systems for oligonucleotide transport have been reported. These are lipid-based agents, polypeptides, polylysines, recombinant histones and biotinylated polyamidoamine dendrimers. Among them biotinylated polyamidoamine dendrimers have emerged as novel cationic gene carriers [45, 46]. However, these agents are not specific for tumor cells.
Cellular uptake and nuclear localization are also reported to be dependent on the type of modification in the oligonucleotide sequence. Cellular accumulation of phosphorothioates is reported to be 3–5 fold more than 3′-alkylamino oligodeoxynucleoside phosphodiesters (PONH2) and 2′-O-methyl oligoribonucleotide phosphodiesters (2 OM), 6–7 fold greater than PNAs and 8–10 fold more than oligodeoxynucleoside methylphosphonates [47]. The relative hybridization efficiency to the target sequence ranged in the order 2′-O-methyl phosphodiester (OMe) > phosphodiester (PO) and phosphorothioate (PS) [48].
To improve the uptake of PNAs specifically in tumor cells, PNAs linked to peptide ligands of receptors overexpressed on tumor cells were explored. Peptides such as IGF-1 analogues and Tyr3-ocreotate have been coupled to antisense sequences to improve cell delivery [49, 50]. Dihydrotesterone attached to anti–gene PNA (Anti–DNA PNA) was reported to be a selective cellular/nuclear localization vector in prostrate cancer cells, correlating with recent suggestions of membrane presentation of androgen receptors [51]. General, nonspecific cell penetrating peptides such as polylysine, oligolysine, and membrane permeating transducing peptide PTD-4 have also been coupled to antisense sequences to improve cell delivery [52, 53].
3.4 Radiolabeling
Methods for radiolabeling antisense agents with beta emitters such as 3H, 35S and 32P are well established. Methods for synthesis of oligonucleotides ready to be labeled with radionuclides for imaging applications are also very well reported.
Because of favorable decay properties and availability, 99mTc remains a radionuclide of choice for SPECT imaging. Radiolabeling with 99mTc has been reported using various chelating groups attached to oligonucleotide sequences. Diethylene triamine pentaacetic acid (DTPA), 6-hydrazinonicotinamide (HNH), and mercaptoacetyl-triglycine (MAG3) conjugated to oligonucleotides have been reported for chelation of 99mTc as well as for 111In labeling [9, 54, 55]. A facile method for labeling PNA ligands with 99mTc was reported using a tetrapeptide, Gly-D-Ala-Gly-Gly, providing an N4 conformation for strong and efficient chelation of 99mTc [56]. Radioiodination of oligonucleotides has been reported using tributylstannyl benzamide and p-methoxyphenyl isothiocyanate (PMPITC) conjugates [57].
With the success achieved in PET imaging due to high resolution and sensitivity, a number of positron emitting radionuclides are being employed as tracers. The macrocyclic chelator, 1, 4,7,10 tetraaza cyclododecane tetracaetic acid (DOTA) conjugated to antisense sequences has been utilized for complexation with positron emitters such as 64Cu and 68Ga [58, 59, 60]. DOTA was also reported for radiolabeling of gamma and beta emitting radionuclides such as 111In and 90Y [53]. Probes with an N2S2 chelator were also reported for 64Cu labeling. Diaminopropionic acid was incorporated at the N terminus of the PNA with an aminoethoxyethoxyacetic acid linker (AEEA) coupled to two S-benzoyl thioglycolic acid residues (SBTG2) to generate an N2S2 chelator [61]. Dendrimeric chelator-PNA-peptide probes have been utilized for MRI using Gd (III) as a contrast agent [62].
Synthons are structural units that can be incorporated as part of the synthesis of a larger molecule. Synthons can be radiolabeled prior to synthesizing a molecular probe. Methods for labeling with radiohalogens, mostly positron and gamma emitters like 18F, 131I, and 76Br, typically involve prelabeling of synthons for high yield and stable incorporation of halogens [63]. Purification of the synthon from other radioactive byproducts is done by HPLC followed by regioselective conjugation of the radiolabeled synthon to an oligonucleotide. Prelabeling provides the possibility to work with any oligonucleotide synthesized in house or commercially without tedious unblocking steps [48]. It was reported that the hybridization efficiency of radiolabeled antisense oligonucleotides was not compromised due to the method of radiolabeling [63].
3.5 Pharmacokinetics of antisense agents
Pharmacokinetics of various antisense agents labeled with 18F and 68Ga using different backbones such as phosphodiester (PO), phosphorothioate (PS) and 2′-O-methyl phosphodiester (OMe) were studied and observed to be dependent on oligonucleotide backbone [63, 64, 65]. Tissue distribution of the radioactivity showed that the phosphodiester was eliminated both through renal and digestive system while phosphothioates and 2′-O-methyl RNAs showed only renal excretion [63]. Pharmacokinetics of uncharged PNAs linked to tumor targeting peptides is expected to be governed by the specificity of the peptide for the receptors expressed on tumor cells.
Like development of any new radiopharmaceutical, antisense imaging agents need to undergo intense, systemic and thorough evaluation before they can be used in clinics. Table 1 summarizes the major steps in the research and development of antisense imaging agents. Perhaps the most important aspect is preferential uptake by the target tissue and ability of the probes to hybridize specifically to the target mRNA sequences.
Table 1.
Key components in development of antisense imaging agents
| Stage and goals | Test system | Approaches and requirements |
|---|---|---|
| I. Design of oligonucleotides |
|
|
| II. Target validation: Specificity and efficiency | Cell free system |
|
| III. Radiolabeling |
|
|
| IV. In vitro biological activity: Specificity and efficiency | Cell culture |
|
| V In vivo pharmacokinetics | Animal models |
|
| VI. In vivo toxicology | Animal |
|
| VII. Large scale synthesis and radiolabeling |
|
|
| VIII. Clinical evaluation and regulatory issues | Humans |
|
4. Pioneer oncogene expression imaging investigations
A number of oncogenes like KRAS, HER2 (ERB-B2, neu), MYCC, BCL2 and CCND1 are overexpressed in various types of cancers. Among them, breast and pancreatic cancers have been targeted using radiolabeled antisense probes [8]. Apart from detection of cancers based on overexpression of genes, this technology was also explored for determination of response to anticancer therapy. This was accomplished by studying the expression of an early response gene for DNA damage, p21WAF-1/CIP-1 [66].
One investigation demonstrated preferential in vitro uptake and retention of radiolabeled antisense in tumor cells. A 18mer phosphorothioate antisense to the initiation codon of the HER2 oncogene mRNA was synthesized. The sense strand was also synthesized and both were labeled with 32P. The uptake and retention of radiolabeled antisense HER2 was observed in MCF-7 cells known to express HER2 mRNA. However, studies to show target-specific uptake and in vivo distribution of the agent were not performed [67]. A similar in vitro study reported hybridization of 99mTc and 32P labeled 15mer phosphodiester oligodeoxynucleotide antisense to the complementary MYCC sequence. Efficient binding to the complementary sequence was achieved in cell-free systems. However, uptake in a cell line expressing MYCC and in a MYCC knockout cell line was not statistically different [68]. A 23mer oligonucleotide phosphodiester sequence, complementary to the translation initiation site of TGF-α mRNA, was radiolabeled with 125I [69]. Radioiodination was achieved via a tyramine group conjugated to the 5′ end. In this study, a FITC labeled 23mer TGF-α antisense was used to demonstrate in vitro stability, intracellular and nuclear localization in NS2T2A1 cells, expressing high levels of TGF-α. In in vivo studies, uptake was followed after intratumoral injection. Tumor uptake ranged from 14% at 1 h to 1.2% at 24 h p.i. No study to indicate uptake and retention via antisense mechanism was performed [69].
Synthesis and bioevaluation of a 15mer phosphodiester and a 15mer phosphothioate DNA complementary to a sequence within the initiation codon site of MYCC oncogene mRNA was reported [10, 55]. Sequences were conjugated to a DTPA analogue and were labeled with 111In. Uptake of the radiolabel in murine monocyte leukemia tumor cells appeared to be significantly higher for antisense DNAs compared to sense DNAs. In nude mice bearing mammary adenocarcinoma xenografts, 10 to 12% uptake of radiolabeled antisense DNAs was observed in the tumors as compared to 1% uptake of the sense sequence at 1 h p.i. However, uptake in blood and muscles were 18–20% and 15–17% respectively. Such high uptake might lead to high background and hinder in achieving good scintigraphic images for tumor detection [55]. However, no subsequent investigations verifying the above results have been reported.
Another study reported 68Ga-labeled 17mer oligonucleotide sequences for targeting mutated KRAS oncogene mRNA in human A549 lung cancer xenografts. The intravenously injected tracer revealed high quality PET images that allowed quantification of biokinetics in major organs and in tumors containing KRAS point mutations versus tumors with wild type KRAS oncogene [65].
In the recent past, peptide nucleic acid based antisense sequences labeled with 99mTc, 111In, 64Cu and 90Y showed promising results for targeting oncogene mRNA. In one study, beads conjugated with a complementary DNA sequence were first injected in the thigh region of the mice. A 99mTc labeled PNA sequence was then administered i.v. The PNA probe showed stability and superior pharmacokinetics along with high uptake in the left thigh where the complementary DNA beads were located [9].
Synthesis and characterization of PNA probes with a peptide analog of insulin-like growth factor 1 (IGF1) for increased tumor cell uptake and chelating groups for 99mTc and 64Cu labeling have been studied extensively [70]. A construct of chelator-PNA-peptide is depicted in Figure 1. Antisense sequences specific for CCND1 [71] and MYCC [72] mRNAs were explored for genetic characterization of breast cancer xenografts. Unique PNA sequences targeting the initiation codon regions were selected because the start codon domain has been reported to be available for PNA hybridization.
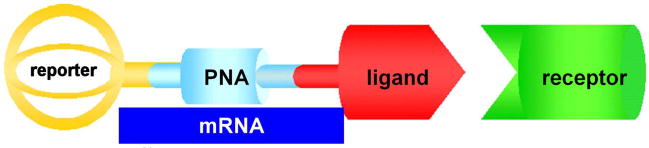
Figure 1.
Schematic representation of reporter-PNA-ligand probe wherein reporter can be radioisotope like 64Cu or 99mTc, or contrast agent like Gd for MRI, bound to macrocyclic chelator DOTA, PNA sequence complementary to oncogene mRNA, and peptide ligand specific for overexpressed receptors on cancer cells.
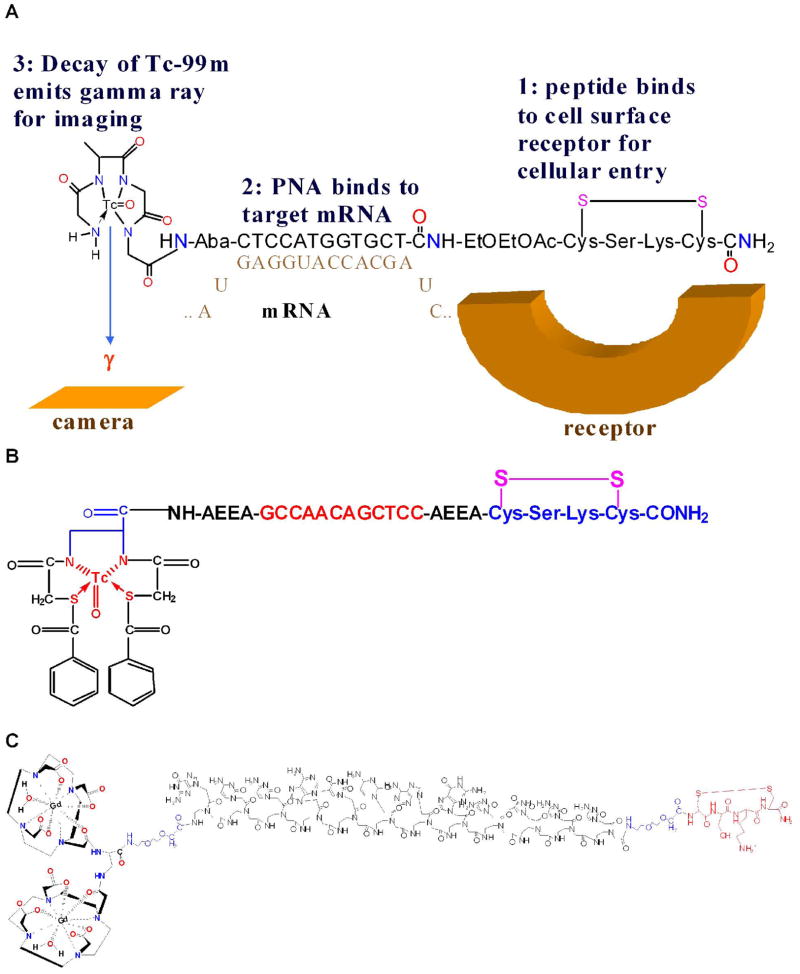
An antisense sequence for the KRAS D12 mutant was selected for targeting KRAS oncogene expression in pancreatic cancer xenografts. In this study, PNA sequences targeting the mutated 12th codon region of KRAS oncogene mRNAs were selected [73]. Probes with 12mer antisense sequence and 12mer mismatch controls for CCND1, KRAS and MYCC were synthesized because PNA oligomers as short as 12 residues are reported to be sufficient for statistical uniqueness [8]. Sequences with one, two, three mismatch served as controls. Hydrophilic aminoethoxyethoxyacetyl (AEEA) and 4-aminobutanoyl (Aba) spacers were introduced at the N terminus and C terminus of PNA hybridization sequences to minimize the steric hindrance of bulky chelator-metal ion complex and the cyclic peptide moieties. Without the spacers, these bulky groups might affect the hybridization efficiency of the PNAs. To achieve tumor cell specificity and improved cellular delivery of PNAs, the IGF1 analog JB9, cyclic D- (Cys-Ser-Lys-Cys), was extended from the solid phase support before coupling of the hydrophilic spacer and the PNA monomers. N4 chelation of 99mTc was achieved by extending a tetrapeptide, Gly-D-Ala-Gly-Gly, from the N terminus of the spacer-PNA-spacer-peptide. Probes with either DO3A or an N2S2 chelator coupled to the N-termini of spacer-PNA-spacer-peptides were synthesized for 64Cu labeling. Their sequences are shown in Table 2. Schematics of chelator-PNA-peptides labeled with 99mTc, 64Cu and Gd are displayed in Figure 2.
Table 2.
Sequence of PNA probes
| Name | Sequence |
|---|---|
| PNA-free (WT990) | Gly-D-Ala-Gly-Gly-Aba- (Gly) 4-D (Cys-Ser-Lys-Cys) |
| MYCC | |
| MYCC PNA antisense (WT4219) | Gly-D-Ala-Gly-Gly-Aba-GCATCGTCGCGG-AEEA-D (Cys-Ser-Lys-Cys) |
| MYCC PNA mismatch (WT4235) | Gly-D-Ala-Gly-Gly-Aba-GCATGTCTGCGG-AEEA-D (Cys-Ser-Lys-Cys) |
| CCND1 | |
| CCND1 PNA antisense (WT4185) | Gly-D-Ala-Gly-Gly-Aba-CTGGTGTTCCAT-AEEA-D (Cys-Ser-Lys-Cys) |
| CCND1 PNA mismatch (WT4172) | Gly-D-Ala-Gly-Gly-Aba-CTGGACAACCAT-AEEA-D (Cys-Ser-Lys-Cys) |
| CCND1 peptide mismatch (WT4113) | Gly-D-Ala-Gly-Gly-Aba-CTGGTGTTCCAT-AEEA-D (Cys-Ala-Ala-Cys) |
| Fl-CCND1 PNA antisense (WT4433) | SFX-AEEA-CTGGTGTTCCAT-AEEA-D (Cys-Ser-Lys-Cys) |
| Fl-CCND1 peptide mismatch (WT4361) | SFX-AEEA-CTGGTGTTCCAT-AEEA-D (Cys-Ala-Ala-Cys) |
| KRAS | |
| KRAS PNA antisense | SBTG2-DAP-AEEA-GCCAACAGCTCC-AEEA-D (Cys-Ser-Lys-Cys) |
| KRAS PNA antisense (WT4286, Asp 12 mutant) | DOTA-AEEA-GCCATCAGCTCC-AEEA-D (Cys-Ser-Lys-Cys) |
| KRAS PNA, 1 mismatch (WT4271, Gly12 wild type) | DOTA-AEEA-GCCACAGCTCC-AEEA-D (Cys-Ser-Lys-Cys) |
| KRAS PNA, 1 mismatch (WT4295, Val12 mutant) | DOTA-AEEA-GCCACAGCTCC-AEEA-D (Cys-Ser-Lys-Cys) |
| KRAS PNA, 2 mismatch (WT4292, Lys 12 mutant) | DOTA-AEEA-GCCTAGCTCC-AEEA-D (Cys-Ser-Lys-Cys) |
| KRAS PNA, 3 mismatch (WT4277, Glu12 mutant) | DOTA-AEEA-GCCTTTGCACC-AEEA-D (Cys-Ser-Lys-Cys) |
| KRAS PNA antisense, peptide mismatch (WT4214, Asp12 mutant) | DOTA-AEEA-GCCATCAGCTCC-AEEA-D (Cys-Ala-Ala-Cys) |
| F1= Fluoresceinyl; SFX= 6-(fluorescein-5-carboxamido) hexanoyl | |
Figure 2.
Radiohybridization and MRI probes
a. 99mTc-PNA-Peptide probe with IGF1 analog specific for IGF1 receptor, PNA sequence complementary to CCND1 oncogene mRNA, and N4 chelating peptide GdAGG for chelating 99mTc for SPECT imaging of breast cancer.
b. SBTG2-KRAS PNA-Peptide probe with IGF1 analog specific for IGF1 receptor, PNA sequence complementary to KRAS D12 mutant, and N2S2 chelator for 99mTc or 64Cu for SPECT or PET imaging of pancreatic cancer.
c. MRI probe with IGF1 analog specific for IGF1 receptor, PNA sequence complementary to KRAS D12 mutant, and DOTA for chelating Gd for MRI of pancreatic cancer.
Although PNAs are internalized poorly into mammalian cells, CCND1 fluoresceinyl-PNA-IGF1 peptide probe was internalized efficiently by cells overexpressing IGF1 receptors, compared to a peptide mismatch probe [8, 49]. Scintigraphic imaging of MCF-7 xenografts in immunocompromised mice revealed 7-fold higher intensity of CCND1 and MYCC 99mTc-chelator-PNA-D(Cys-Ser-Lys-Cys) probes compared to mismatch or contralateral controls [71, 72]. Figure 3 depicts uptake of 99mTc-CCND1 PNA-peptide probe in MCF-7 xenografts in nude mice at 4, 12, 24 h p.i. [71]. A specific 64Cu-DO3A-CCND1 PNA-IGF1 analog radiohybridization probe was injected intravenously into immunocompromised mice bearing breast cancer xenografts. Eight-fold higher PET intensity at the center of breast cancer xenograft was observed compared to intensity in the contralateral tissues at 24 h p.i. PNA mismatch, peptide mismatch probe and IGF1 blocking yielded significantly weaker images [60].
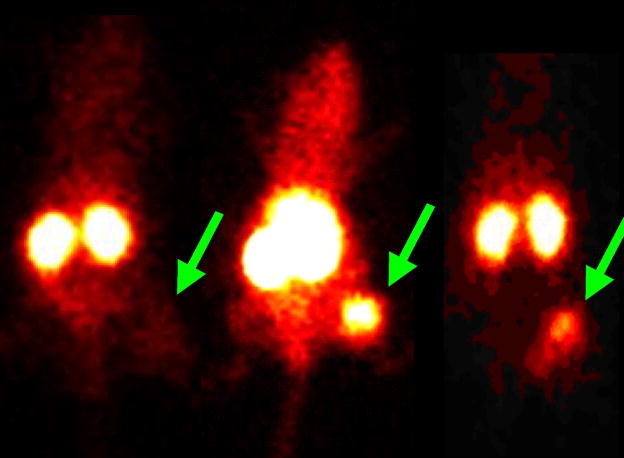
Figure 3.
Scintigraphic images in nude mice bearing human MCF-7 breast tumor xenograft at 4, 12 and 24 h p.i. of 99mTc-CCND1 PNA-peptide probe. (Reprinted, with permission, from reference 67).
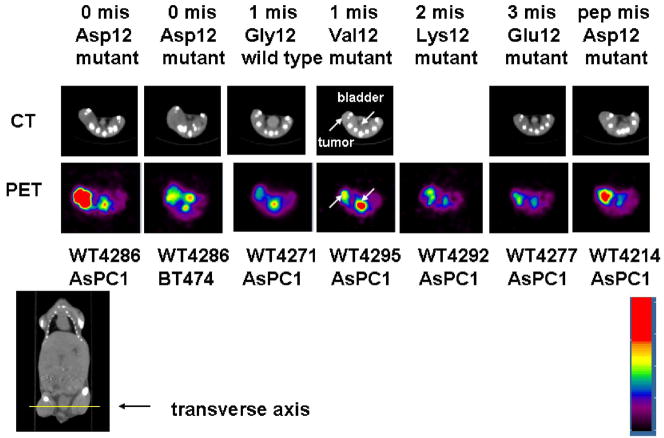
An SBTG2-DAP-PNA-IGF1 analog antisense probe for mutant KRAS mRNA was labeled with 99mTc and also with 64Cu. The radiometal-chelator-PNA-peptide hybridization probes were injected intravenously into immunocompromised mice bearing pancreatic cancer xenografts, followed by scintigraphic and PET imaging [8]. 64Cu-DO3A-KRAS PNA-IGF1 analog (WT4286) radiohybridization probe that was specific for the D12 KRAS mutation in human ASPC1 pancreatic cancer cells gave strong tumor contrast. Eight-fold increase in intensity at the centre of human pancreatic cancer xenografts in PET images was observed compared to the intensity in the contralateral muscle at 4 h p.i as shown in Figure 4 [73]. These results are encouraging and suggest a promising future for early and specific imaging of malignant lesions.
Figure 4.
Transverse CT images (top row) and PET images (second row) of 64Cu KRAS D12PNA peptide probe WT4286 vs.1 mismatch WT4271, 1-mismatch WT4295, 2-mismatch WT4292, 3 mismatch WT4277 and a peptide mismatch WT4214 in human ASPC1 pancreatic xenografts in immunocompromised mice 24 h after probe administration into the tail vein. The mouse second from left bears human BT474 breast cancer xenograft lacking activated KRAS. The yellow line on the coronal CT image shows the level of the transverse images. The color scale of the images was normalized to the max/min of frame to show the dynamic range of tumor uptake. (Reprinted, with permission, from reference 69).
111In and 90Y labeled PNA complementary to the first six codons of BCL2 mRNA were reported. Membrane permeating transducing peptide PTD-4 was coupled to PNA for intracellular delivery. DOTA served as chelating moiety in the probe. 90Y-PTD-4-K (DOTA) – anti BCL2 PNA showed binding similar to 32P labeled analogues when northern analysis was performed using BCL2 mRNA from a cell free system [53]. The same group recently reported an 111In labeled anti BCL2 sequence coupled to Tyr3-octreotate for somatostatin receptor mediated intracellular delivery. Tumors could be imaged by 111In-DOTA-anti BCL2-PNA-Tyr3-ocreotate at 48h p.i. [50].
MicroPET imaging of MCF-7 tumors in mice was reported using PNA targeting murine unr mRNA (upstream of NRAS) sequence. Tetralysines were incorporated at the carboxy termini for cell permeation and PNA was coupled to DOTA for 64Cu labeling. 64Cu-DOTA-Y-PNA50-K4 showed good uptake in tumors with tumor/muscle ratio of 6.6 ± 1.1 at 24 h p.i [52]. Nevertheless, sequence-specific tumor images were not observed. The negative results were ascribed to mouse tissue expression of murine unr mRNA with the same target sequence as human UNR mRNA, and uptake of the PNA universally in all cells due to the nonspecific Lys4 tail.
These studies strengthen the potential of radiolabeled antisense PNAs for utilization as specific molecular probes for early detection of cancer and ultimately for disease specific radiotherapy.
5. Perspectives
Cancer remains the most formidable disease of mankind. Early detection of cancer following effective therapeutic intervention can save lives. Advances in genomics and proteomics are being reported almost every day, shedding new light on the genesis of cancer. Targeting specific biomarkers with radiohybridization probes may play a significant role in early detection of cancer.
Success has been achieved in design and synthesis of novel antisense agents with improved stability and binding affinity. Availability of automated technology for synthesis of antisense agents and recent reports on possibility of single, continuous, solid phase synthesis of various peptide-PNA conjugates indicates the possibility of large scale synthesis of conjugates for commercial applications.
Promising results have been achieved in cell free systems and to some extent in in vitro systems. For in vivo applications, limited success has been achieved. Furthermore, many issues such as in vivo stability, target cell specificity, uptake and retention in the cell and interaction with the target sequences need to be established. Future studies are needed to validate the hypothesis of specificity of the radio hybridization probes to the target sequences in in vitro and in vivo systems. Whether the uptake and retention of antisense agents is proportional to the expression of intracellular message needs to be addressed. Apart from the use of radionuclides for labeling oligonucleotides and PNAs, newer signaling moieties like near infrared (NIR) dyes for optical imaging and supermagnetic iron oxide for magnetic resonance imaging may be useful for in vivo imaging. However, in such approaches, depth and sensitivity are major concerns. Use of dendrimers, conjugated to antisense sequence for incorporation of multiple labels may be helpful in improving the signal for diagnostic applications. These may also be useful for therapeutic applications when labeled with radionuclides of therapeutic importance.
Molecular imaging of oncogene mRNAs with novel and improved hybridization probes for early, specific and noninvasive detection of cancers warrants greater attention.
Acknowledgments
This research was supported by NIH CA109231, EB001809, 1S10RR23709 and PA ME-03-184 grants to M.L.T., and DOE ER63055 and NIH C027175 to E.W.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Archana Mukherjee, Postdoctoral Research Fellow, Department of Radiology, Thomas Jefferson University, 361 JAH, 1020, Locust Street, Philadelphia, PA 19107, USA, Email: Archana.Mukherjee@jefferson.edu, Tel: 215-503-7879, Fax: 215-923-9245.
Eric Wickstrom, Department of Biochemistry and Molecular Biology, Thomas Jefferson University, 233S, 10th street, Suite 219 Philadelphia, PA 19107, USA, Email: eric@tesla.jci.tju.edu, Tel: 215-955-4578, Fax: 215-955-4580.
Mathew L. Thakur, Director, Laboratories of Radiopharmaceutical Research and Molecular Imaging, Department of Radiology, Thomas Jefferson University, Kimmel Cancer Centre. Suite 359 JAH, 1020, Locust Street, Philadelphia, PA 19107, USA, Email: Mathew.Thakur@jefferson.edu, Tel: 215-503-7874, Fax: 215-923-9245.
References
- 1.Weissleder R, Mahmood U. Molecular Imaging. Radiology. 2001;219:316–33. doi: 10.1148/radiology.219.2.r01ma19316. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. The Hallmarks of Cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 3.Weinberg RA. How cancer arises. Sci Am. 1996 sept;:62–70. doi: 10.1038/scientificamerican0996-62. [DOI] [PubMed] [Google Scholar]
- 4.Mankoff DA, Link JM, Linden HM, Sundararajan L, Krohn KA. Tumor receptor imaging. J Nucl Med. 2008;49:149S–63S. doi: 10.2967/jnumed.107.045963. [DOI] [PubMed] [Google Scholar]
- 5.Plathow C, Weber WA. Tumor cell metabolism imaging. J Nucl Med. 2008;49:43S–63S. doi: 10.2967/jnumed.107.045930. [DOI] [PubMed] [Google Scholar]
- 6.Krohn KA, Link JM, Mason RP. Molecular imaging of hypoxia. J Nucl Med. 2008;49:129S–48S. doi: 10.2967/jnumed.107.045914. [DOI] [PubMed] [Google Scholar]
- 7.Gleave ME, Monia BP. Antisense therapy for cancer. Nature Reviews Cancer. 2005;5:468–79. doi: 10.1038/nrc1631. [DOI] [PubMed] [Google Scholar]
- 8.Tian X, Chakrabarti A, Amirkhanov N, et al. External imaging of CCND1, MYC and KRAS oncogene mRNAs with tumor-targeted radionuclide-PNA-peptide chimeras. In: El-Deiry W, editor. Ann N Y Acad Sci. Vol. 1059. 2005. pp. 106–44. [DOI] [PubMed] [Google Scholar]
- 9.Mardirossian G, Lei K, Rusckowski M, Chang F, Qu T, Egholm M, Hnatowich DJ. In vivo hybridization of technetium-99m-labeled peptide nucleic acid (PNA) J Nucl Med. 1997;38:907–13. [PubMed] [Google Scholar]
- 10.Dewanjee MK, Haider N, Narula J. Imaging with antisense oligonucleotides for the detection of intracellular messenger RNA and cardiovascular disease. J Nucl Cardiol. 1999;6:345–56. doi: 10.1016/s1071-3581(99)90047-8. [DOI] [PubMed] [Google Scholar]
- 11.Hnatowich DJ. Antisense and nuclear medicine. J Nucl Med. 1999;40:693–703. [PubMed] [Google Scholar]
- 12.Pickeral OK, Li JZ, Barrow I, Boguski MS, Makalowski W, Zhang J. Classical oncogenes and tumor suppressor genes: A Comparative genomics perspective. Neoplasia. 2000;2(3):280–86. doi: 10.1038/sj.neo.7900090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Revillion F, Bonneterre J, Peyrat JP. ERBB2 oncogene in human breast cancer and its clinical significance. Eur J Cancer. 1998;34(6):791–808. doi: 10.1016/s0959-8049(97)10157-5. [DOI] [PubMed] [Google Scholar]
- 14.D’Incalci M. Correlation of ErbB2 gene status, mRNA and protein expression. Onkologie. 2006;29:246–7. doi: 10.1159/000093385. [DOI] [PubMed] [Google Scholar]
- 15.Vaughn JP, Iglehart JD, Demirdji S, et al. Antisense DNA down regulation of the ERBB2 oncogene measured by a flow cytometric assay. Proc Natl Acad Sci USA. 1995;92:8338– 42. doi: 10.1073/pnas.92.18.8338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bos JL. Ras oncogenes in human cancer: A review [published erratum appears in Cancer Res 1990 Feb 15; 50(4): 1352] Cancer Res. 1989;49:4682–9. [PubMed] [Google Scholar]
- 17.Lowy DR, Willumsen BM. Function and regulation of ras. Annu Rev Biochem. 1993;62:851–91. doi: 10.1146/annurev.bi.62.070193.004223. [DOI] [PubMed] [Google Scholar]
- 18.Pellagata NS, Sessa F, Renault B, et al. K-ras and p53 gene mutations in pancreatic cancer: Ductal and Nonductal tumors progress through different genetic lesions. Cancer Res. 1994;54:1556–60. [PubMed] [Google Scholar]
- 19.Evans DB, Abbruzzese JL, Willett CG. Cancer of pancreas. In: Devita VT, Hellman S, Rosenberg SA, editors. Cancer: Principles and practice of Oncology. Philadelphia: Lippincott, Williams and Wilkins; 2001. pp. 1126–61. [Google Scholar]
- 20.Berns Els MJJ, Klijn JGM, van Staveren IL, Portengen H, Foekens JA. c-myc amplification is a better prognostic factor than Her2/neu amplification in primary breast cancer. Cancer Res. 1992;52:1107–13. [PubMed] [Google Scholar]
- 21.Wickstrom EL, Bacon TA, Gonzalez A, Freeman DL, Lyman GH, Wickstrom E. Human promyelocytic leukemia HL-60 cell proliferation and c-Myc protein expression are inhibited by an antisense pentadecadeoxynucleotide targeted against c-MYC mRNA. Proc Natl Acad Sci USA. 1988;85(4):1028–32. doi: 10.1073/pnas.85.4.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weinstat-Saslow D, Merino MJ, Manrow RE, et al. Overexpression of cyclin D mRNA distinguishes invasive and in situ breast carcinomas from non-malignant lesions. Nat Med. 1995;1:1257–60. doi: 10.1038/nm1295-1257. [DOI] [PubMed] [Google Scholar]
- 23.Hermine O, Haioun C, Lepage E, et al. Prognostic significance of bcl-2 protein expression in aggressive Non-Hodkins lymphoma. Blood. 1996;87:265–72. [PubMed] [Google Scholar]
- 24.Pepper C, Bentley P, Hoy T. Regulation of clinical chemo resistance by bcl-2 and bax oncoproteins in B-cell chronic lymphocytic leukemia. Br J Haematol. 1996;95:513–17. doi: 10.1046/j.1365-2141.1996.d01-1927.x. [DOI] [PubMed] [Google Scholar]
- 25.Croce CM. Oncogenes and cancer. N Engl J Med. 2008;358:502–11. doi: 10.1056/NEJMra072367. [DOI] [PubMed] [Google Scholar]
- 26.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 27.Sauter ER, Herlyn M, Liu SC, Litwin S, Ridge JA. Prolonged response to antisense cyclin D1 in a human squamous cancer xenograft model. Clin Cancer Res. 2000;6:654– 60. [PubMed] [Google Scholar]
- 28.Wickstrom E, Bacon TA, Wickstrom EL. Down regulation of c-MYC antigen expression in lymphocytes of Eμ-c-myc transgenic mice treated with anti-c-myc DNA methylphosphonates. Cancer Res. 1992;52:6741–45. [PubMed] [Google Scholar]
- 29.Wickstrom E. Clinical trials of genetic therapy with antisense DNA and DNA vectors. New York: Marcel Dekker; 1998. [Google Scholar]
- 30.Webb A, Cunningham D, Cotter F, et al. BCL-2 antisense therapy in patients with non-Hodgkin lymphoma. Lancet. 1997;349:1137–41. doi: 10.1016/s0140-6736(96)11103-x. [DOI] [PubMed] [Google Scholar]
- 31.Bayever E, Haines KM, Iversen PL, et al. Selective cytotoxicity to human leukemic myoblasts produced by oligodeoxyribonucleotide phosphorothioates complementary to p53 nucleotide sequences. Leuk Lymphoma. 1994;12:223–26. doi: 10.3109/10428199409059593. [DOI] [PubMed] [Google Scholar]
- 32.Paroo Z, Corey DR. Imaging gene expression using oligonucleotides and peptide nucleic acids. J Cell Biochem. 2003;90:437–42. doi: 10.1002/jcb.10626. [DOI] [PubMed] [Google Scholar]
- 33.Swayze EE, Balkrishen B. The medicinal chemistry of oligonucleotides. In: Crooke ST, editor. Antisense drug technology: Principles, strategies and applications. CRC press; USA: 2008. pp. 144–72. [Google Scholar]
- 34.Zendegui JG, Vazquez KM, Tindley JH, et al. In vivo stability and kinetics of absorption and disposition of 3′ phosphopropyl amine oligonucleotides. Nucleic acids Res. 1992;20(2):307–14. doi: 10.1093/nar/20.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thakur ML, Tian X, Amirkhanov N, et al. The role of radiolabeled peptide nucleic acid chimeras and peptides in imaging oncogene expression. Indian J Nucl Med. 2004;19(3):98–114. [Google Scholar]
- 36.Lebedev AV, Wickstrom E. The chirality problem in P-substituted oligonucleotides. In: trainor G, editor. Perspectives in Drug Discovery and Design. Leiden: ESCOM science publishers; 1996. pp. 17–40. [Google Scholar]
- 37.Monia BP, Lesnik EA, Gonzalez C, et al. Evaluation of 2′ modified oligonucleotides containing 2′-deoxy gaps as antisense inhibitors of gene expression. J Biol Chem. 1993;268:14514–22. [PubMed] [Google Scholar]
- 38.Bacon TA, Morvan F, Rayner B, et al. Alpha-oligodeoxynucleotide stability in serum, subcellular extracts and culture media. J Biochem Biophys Methods. 1988;16(4):311–8. doi: 10.1016/0165-022x(88)90065-6. [DOI] [PubMed] [Google Scholar]
- 39.Ho PT, Ishiguro K, Wickstrom E, et al. Non-sequence–specific inhibition of transferin receptor expression in HL-60 leukemia cells by phosphorothioate oligodeoxynucleotides. Antisense Research & Development. 1991;1(4):329–42. doi: 10.1089/ard.1991.1.329. [DOI] [PubMed] [Google Scholar]
- 40.Cook D. Antisense medicinal chemistry. In: Crooke ST, editor. Antisense research and applications. Berlin-Heidelberg: Springer-Verlag; 1998. pp. 51–92. [Google Scholar]
- 41.Good L, Nielsen PE. Progress in developing PNA as a gene targeted drug. Antisense Nucleic Acid Drug Dev. 1997;7(4):431–37. doi: 10.1089/oli.1.1997.7.431. [DOI] [PubMed] [Google Scholar]
- 42.Tian X, Wickstrom E. Continuous solid phase synthesis and disulfide cyclization of peptide–PNA-peptide chimeras. Org Lett. 2002;4:4013–16. doi: 10.1021/ol026676b. [DOI] [PubMed] [Google Scholar]
- 43.Nielsen PE. Antisense properties of peptide nucleic acids. In: Crooke ST, editor. Antisense research and applications. Berlin-Heidelberg: Springer-Verlag; 1998. pp. 545–57. [Google Scholar]
- 44.Agrawal S. Antisense oligonucleotides: towards clinical trials. Trends in Biotechnology. 1996;14(10):376–87. doi: 10.1016/0167-7799(96)10053-6. [DOI] [PubMed] [Google Scholar]
- 45.Olivier S, Bolard J. Delivery agents for oligonucleotides. In: Sioud M, editor. Methods in Molecular Biology. Vol. 252. Totowa, New Jersey: Humana Press; 2004. pp. 545–68. Ribozymes and siRNA protocols. [DOI] [PubMed] [Google Scholar]
- 46.Sato N, Kobayashi H, Saga T, et al. Tumor targeting and imaging of intraperitoneal tumors by use of antisense oligo-DNA complexed with dendrimers and/or avidin in mice. Clin Can Research. 2001;7:3606–12. [PubMed] [Google Scholar]
- 47.Gray GD, Basu S, Wickstrom E. Transformed and immortalized cellular uptake of oligodeoxynucleoside phosphorothioates, 3′ Alkylamino oligodeoxynucleosides, 2′-O-methyl oligoribonucleotides, Oligodeoxynucleosides methyl phosphonates, and peptide nucleic acids. Biochem Pharmacol. 1997;53:1465–76. doi: 10.1016/s0006-2952(97)82440-9. [DOI] [PubMed] [Google Scholar]
- 48.Tavitian B, Terrazzino S, Kuhnast B, et al. In vivo imaging of oligonucleotides with positron emission tomography. Nat Med. 1998;4(4):467–71. doi: 10.1038/nm0498-467. [DOI] [PubMed] [Google Scholar]
- 49.Basu S, Wickstrom E. Synthesis and characterization of a peptide nucleic acid conjugated to a D-peptide analog of insulin like growth factor1 for increased cellular uptake. Bioconj Chem. 1997;8:481–88. doi: 10.1021/bc9700650. [DOI] [PubMed] [Google Scholar]
- 50.Jia F, Figueroa SD, Gallazzi F, et al. Molecular imaging of bcl-2 expression in small lymphocytic lymphoma using 111In-labeled PNA-peptide conjugates. J Nucl Med. 2008;49:430–38. doi: 10.2967/jnumed.107.045138. [DOI] [PubMed] [Google Scholar]
- 51.Boffa LC, Scarfi S, Mariani MR, et al. Dihydrotestosterone as a selective cellular/nuclear localization vector for anti-Gene peptide nucleic acid in prostratic carcinoma cells. Cancer Res. 2000;60:2258–62. [PubMed] [Google Scholar]
- 52.Sun X, Fang H, Li X, et al. MicroPET imaging of MCF-7 tumors in mice via unr mRNA targeted peptide nucleic acids. Bioconj Chem. 2005;16:294–305. doi: 10.1021/bc049783u. [DOI] [PubMed] [Google Scholar]
- 53.Lewis MR, Jia F, Gallazzi F, et al. Radiometal-labeled peptide–PNA conjugates for targeting bcl-2 expression: Preparation, characterization and in vitro mRNA binding. Bioconj Chem. 2002;13:1176–80. doi: 10.1021/bc025591s. [DOI] [PubMed] [Google Scholar]
- 54.Hnatowich DJ, Winnard P, Virzi F, et al. Technetium-99m labeling of DNA oligonucleotides. J Nucl Med. 1995;36:2306–14. [PubMed] [Google Scholar]
- 55.Dewanjee MK, Ghafouripour AK, Kapadvanjwala M, et al. Noninvasive imaging of c-myc oncogene messenger RNA with indium-111-antisense probes in a mammary tumor bearing mouse model. J Nucl Med. 1994;35:1054–63. [PubMed] [Google Scholar]
- 56.Rao PS, Tian X, Qin W, et al. 99mTc-peptide nucleic acid probes for imaging oncogene mRNAs in tumors. Nucl Med Commun. 2003;24(8):857–63. doi: 10.1097/01.mnm.0000084583.29433.df. [DOI] [PubMed] [Google Scholar]
- 57.Dewanjee MK, Ghafouripour AK, Werner RK, Serafini AN, Sfakianakis GN. Development of sensitive radioiodinated antisense oligonucleotide probes by conjugation technique. Bioconj Chem. 1991;2:195–200. doi: 10.1021/bc00010a001. [DOI] [PubMed] [Google Scholar]
- 58.Smith SV. Molecular imaging with 64Cu. J Inorg Biochem. 2004;98:1874–1901. doi: 10.1016/j.jinorgbio.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 59.Roivainen A, Tolvanen T, Salomäki S, et al. 68Ga-labeled oligonucleotides for in vivo imaging with PET. J Nucl Med. 2004;45:347–55. [PubMed] [Google Scholar]
- 60.Tian X, Aruva MR, Zhang K, Cardi CA, Thakur ML, Wickstrom E. PET imaging of CCND1 mRNA in human MCF7 estrogen receptor-positive breast cancer xenografts with an oncogene-specific [64Cu] DO3A-PNA-IGF1 analog radiohybridization probe. J Nucl Med. 2007;48(10):1699–1707. doi: 10.2967/jnumed.107.042499. [DOI] [PubMed] [Google Scholar]
- 61.Tian X, Chakrabarti A, Amirkhanov N, et al. Receptor mediated internalization of chelator-PNA-peptide hybridization probes for radioimaging or magnetic resonance imaging of oncogene mRNAs in tumors. Biochem Soc Trans. 2007;35:72–6. doi: 10.1042/BST0350072. [DOI] [PubMed] [Google Scholar]
- 62.Amirkhanov NV, Dimitrov I, Opitz AW, Zhang K, Lackey JP, Cardi CA, et al. Design of (Gd-DO3A) n-polydiamidopropanoyl-peptide nucleic acid-D (Cys-Ser-Lys-Cys) magnetic resonance contrast agents. Biopolymers. 2008;89:1061–76. doi: 10.1002/bip.21059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tavitian B. In vivo imaging with oligonucleotides for diagnosis and drug development. Gut. 2003;52:40–7. doi: 10.1136/gut.52.suppl_4.iv40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lendvai G, Velikyan I, Bergström M, et al. Biodistribution of 68Ga-labelled phosphodiester, phosphorothioate and 2′-O-methyl phosphodiester oligonucleotides in normal rats. Eur J Pharm Sci. 2005;26(1):26–38. doi: 10.1016/j.ejps.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 65.Roivaninen A, Tolvanen T, Salomäki S, et al. 68Ga-labelled oligonucleotides for in vivo imaging with PET. J Nucl Med. 2004;45:347–55. [PubMed] [Google Scholar]
- 66.Wang J, Chen P, Mrkobrada M, et al. Antisense imaging of epidermal growth factor –induced p21WAF-1/CIP-1 gene expression in MDA-MB-468 human breast cancer xenografts. Eur J Nucl Med. 2003;30:1273–80. doi: 10.1007/s00259-003-1134-0. [DOI] [PubMed] [Google Scholar]
- 67.Urbain JLC, Shore SK, Vekemans MC, et al. Scintigraphic imaging of oncogenes with antisense probes: does it make sense? Eur J Nucl Med. 1995;22:499–504. doi: 10.1007/BF00817271. [DOI] [PubMed] [Google Scholar]
- 68.Stalteri MA, Mather SJ. Hybridization and cell uptake studies with radiolabeled antisense oligonucleotides. Nucl Med Commun. 2001;22 (11):1171–79. doi: 10.1097/00006231-200111000-00002. [DOI] [PubMed] [Google Scholar]
- 69.Cammilleri S, Sangrajrang S, Perdereau B, et al. Biodistribution of iodine-125 tyramine transforming growth factor a antisense oligonucleotide in athymic mice with a human mammary tumour xenograft following intratumoral injection. Eur J Nucl Med. 1996;23:448–52. doi: 10.1007/BF01247375. [DOI] [PubMed] [Google Scholar]
- 70.Wickstrom E, Thakur ML, Sauter ER. Radiolabeled peptide nucleic acid oncogene probes conjugated to receptor-specific peptide analogs. In: Janson CG, During MJ, editors. Peptide Nucleic Acids, Morpholinos, and Related Antisense Biomolecules. New York: Landes Bioscience/Kluwer Academic/Plenum Publishers; 2006. pp. 59–86. [Google Scholar]
- 71.Tian X, Aruva MR, Qin W, Zhu W, Duffy KT, Sauter ER, Thakur ML, Wickstrom E. External imaging of CCND1 cancer gene activity in experimental human breast cancer xenografts with [99mTc] peptide-PNA-peptide chimeras. J Nucl Med. 2004;45(12):2070–82. [PubMed] [Google Scholar]
- 72.Tian X, Aruva MR, Qin W, et al. Noninvasive molecular imaging of MYC mRNA expression in human breast cancer xenografts with a [99mTc] peptide-peptide nucleic acid-peptide chimera. Bioconj Chem. 2005;16:70–9. doi: 10.1021/bc0497923. [DOI] [PubMed] [Google Scholar]
- 73.Chakrabarti A, Zhang K, Aruva MR, Cardi CA, Opitz AW, Wagner NJ, Thakur ML, Wickstrom E. Radiohybridization PET imaging of KRAS G12D mRNA expression in human pancreas cancer xenografts with [64Cu] DO3A-peptide nucleic acid-peptide nanoparticles. Cancer Biology & Therapy. 2007;6(6):948–56. doi: 10.4161/cbt.6.6.4191. [DOI] [PubMed] [Google Scholar]