Abstract
The multi-components of T2 relaxation in cartilage and tendon were investigated by microscopic MRI (μMRI) at 13μm and 26μm transverse resolutions. Two imaging protocols were used to quantify T2 relaxation in the specimens, a 5-point sampling and a 60-point sampling. Both multi-exponential and non-negative-least-square (NNLS) fitting methods were used to analyze the μMRI signal. When the imaging voxel size was 6.76×10−4 mm3 and within the limit of practical signal-to-noise ratio (SNR) in microscopic imaging experiments, we found that (1) canine tendon has multiple T2 components; (2) bovine nasal cartilage has a single T2 component; and (3) canine articular cartilage has a single T2 component. The T2 profiles from both 5-point and 60-point methods were found to be consistent in articular cartilage. In addition, the depletion of the glycosaminoglycan component in cartilage by the trypsin digestion method was found to result in a 9.81% to 20.52% increase in T2 relaxation in articular cartilage, depending upon the angle at which the tissue specimen was oriented in the magnetic field.
Keywords: Multi-component, T2, articular cartilage, bovine nasal cartilage, tendon, MRI, trypsin digestion, magic angle effect
Introduction
The degradation of articular cartilage is a hallmark in musculoskeletal diseases such as osteoarthritis, which affects the majority of the senior population [1]. Due to the lack of specific and sensitive markers to detect subtle changes in the tissue, no routine procedure is currently available to assess nondestructively the functional, structural, and biochemical properties during the early stages of the tissue degradation. One major obstacle that has prevented any successful development of an early detection procedure is the complex molecular and morphological structures of the tissue. On the histological scale, the collagen fibrils (one of the three major molecules in cartilage) changes its spatial orientation significantly across the (thin) depth or thickness of the tissue [2]. (The other two major molecules are water and proteoglycans [3; 4].) Consequently, articular cartilage is often regarded as consisting of three structural zones: the superficial zone (SZ) where the fibrils are parallel to the surface, the transitional zone (TZ) where the fibrils are mostly randomly oriented, and the radial zone (RZ) where the fibrils are oriented perpendicularly to the surface [5; 6; 7; 8; 9]. This depth-dependent organization of the collagen fibrils in articular cartilage defines its depth-dependent anisotropy as viewed by many analytic and imaging instruments, such as the magic angle effect in MRI [8], birefringence in polarized light microscopy [2; 10], and amide anisotropy in Fourier-transform infrared imaging [11; 12; 13].
Among the quantifiable parameters in MRI, T2 relaxation is of particular importance since T2 relaxation has been shown to be sensitive to the fibril structure/orientation and responsible for the MRI visualization of cartilage laminae (the so-called magic angle effect in MRI of cartilage [8]). In recent years, quantitative mapping of T2 relaxation was able to detect the early structural changes in articular cartilage [14; 15; 16; 17; 18; 19; 20; 21]. Furthermore, the anisotropy of T2 relaxation has been used to divide the MRI-visible features of cartilage into MRI-zones nondestructively, which have been proved to be statistically equivalent to the histological zones based on the collagen orientation by quantitative polarized light microscopy [2; 22; 23].
In addition to the anisotropy of T2 relaxation, the study of multi-components of T2 relaxation [24; 25] is also important in biological tissues such as the brain [26; 27; 28], nerve [29; 30], muscle [31; 32], tendon [33; 34; 35; 36; 37], and cartilage [35; 38; 39; 40]. This is because these tissues are known to have several “pools” of water molecules, each with a distinct molecular environment [41; 42]. Therefore, the ability to quantify the signal from each individual pool could enable the examination of sub-tissue populations and provide a better identification of early tissue degradation. Several studies have shown that the bulk T2 in tendon and cartilage are multi-exponential. For example, Fullerton et al [33] considered the FID decays in bulk bovine tendons to be bi-exponential, with the short and long T2 to be about 4 and 22 ms respectively. Peto et al [34] modeled the echo amplitudes from tendon with a four-component equation and found the four T2 peaks at 0° orientation to be at 0.7, 4.7, 14.5, and 68 ms. These four T2 components in tendon were confirmed by the T2 distributions by Henkelman et al [35], who found four peaks at 2, 8, 23, and 56 ms. In bovine articular cartilage, Henkelman et al [35] demonstrated that the distribution of bulk T2 relaxation had at least two peaks, centered around 20 and 55 ms at 0°, and that the 20ms peak largely disappeared when the tissue’s orientation was about 55° to the magnetic field.
The aim of this microscopic MRI (μMRI) project was to investigate if the multiple components of T2 relaxation in articular cartilage could be resolved in imaging at high spatial resolution. In addition to canine articular cartilage (AC), bovine nasal cartilage (BNC) and canine tendon were also studied in this project. Unlike articular cartilage, tendon (bundles of collagen fibers to transfer the force experienced by muscle to the bone) does not have a zonal structure. The same is true in nasal cartilage. Consequently, for tendon and nasal cartilage, a low-resolution imaging or even a bulk experiment should produce the similar result as in high-resolution imaging. Two T2 quantification methods were used in this μMRI project. In one method (5-point method), five T2-weighted intensity images were acquired in μMRI, and a mono-exponential fitting was used to calculate a T2 map at every pixel location in two dimensions. This is the most commonly used method in both clinics and laboratories (including our previous work), and is consistent with the fact that the fastest decay components in the tissue are not visible in MRI because of the long echo time. In the second method (60-point method), 60 T2-weight intensity images were acquired for each specimen, which enables the calculation of multiple components of T2 in the specimen. This method essentially results in a quantitative T2 data cube, with two spatial dimensions and one dimension of T2 distribution. We employed not only the conventional multivariable exponential fitting analysis but also the non-negative least-square (NNLS) method [24; 43] [44] to analyze the T2 distribution. Although this project was carried out using intact tissue blocks at microscopic resolution, it has direct relevance to MRI of cartilage in general. It aimed to answer an important question: does a mono-exponential fitting of T2 in MRI of cartilage result in an accurate T2 value?
Materials and Methods
Eight fresh specimens of articular cartilage were harvested from the central load-bearing area of three canine humeral heads. These dogs were skeletally mature and healthy, sacrificed for an unrelated experimental study. Each specimen consisted of the full thickness of the articular cartilage still attached to the underlying bone and had dimensions of about 1.75×2×10 mm. In addition to articular cartilage, three pieces of Achilles tendons were also harvested from the same canine origin and two pieces of bovine nasal cartilage were harvested from a local slaughter. All NMR/μMRI experiments were performed when the tissues were fresh, except as noted specifically. For imaging experiments, articular cartilage was soaked in physiological saline with 1% protease inhibitor (Sigma, Missouri). The experiments for tendon and BNC were carried out after the tissues were blotted dry.
NMR spectroscopic and microscopic MRI experiments were conducted at room temperature on a Bruker AVANCE II 300 NMR spectrometer equipped with a 7-Tesla/89-mm wide vertical-bore superconducting magnet and micro-imaging accessory (Bruker Instrument, Billerica, MA). A homemade 3mm solenoid coil was used for the cartilage-bone experiments, where the orientation of the collagen fibrils in the radial zone of the cartilage block with respect to the static magnetic field could be adjusted. Bovine nasal cartilage was also imaged using the same homemade coil. A Bruker 5mm birdcage coil with rotation device was used for the tendon experiments, where the long axis of the tendon sample was set at 55° (magic angle) relative to the direction of the magnetic field.
Spectroscopic measurements of bulk T2 relaxation were performed using a standard CPMG pulse sequence,
| (1) |
A τ value of 500 μs was used in the experiments to avoid the spin-locking effect [45]. Only the even echoes were used in the experiments, where the last number of echoes was 800, which corresponds to an echo time of 800ms. The 90° rf excitation pulse had a duration of 8.38 μs and 3.69 μs for the nasal cartilage and tendon, respectively; the repetition time (TR) was 5 s; the number of dummy scans was 8.
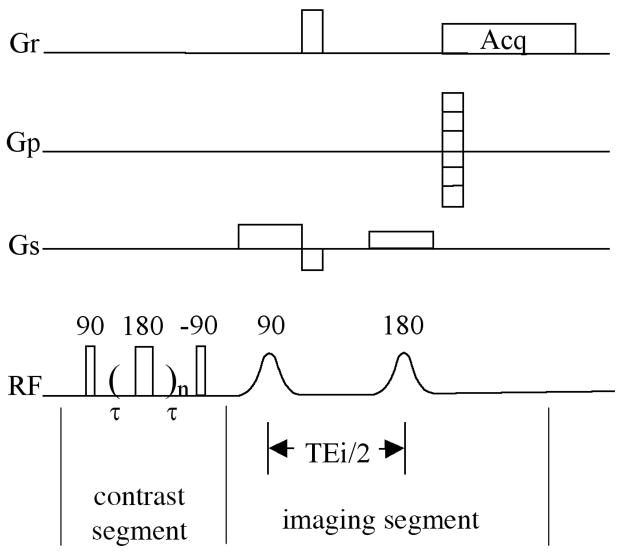
Quantitative T2 imaging experiments were performed using a magnetization-prepared CPMG T2 imaging sequence (Fig 1), which is similar to the magnetization-prepared spin-echo T2 imaging sequence that has been used extensively in our lab [46]. This sequence separates the leading T2-weighting segment that contains no gradient pulse, and a subsequent imaging segment where all timings are kept constant, hence capable of giving an accurate measurement of T2 in the tissue. The echo spacing in the CPMG T2-weighting segment was 1ms, which was the same as that used in the spectroscopy experiments. Two sets of echo times had been used in the leading contrast segment for articular cartilage samples. In the first set (the 5-point method), the number of echoes were 2, 4, 10, 30, 60 for the articular cartilage when its surface was perpendicular to the static magnetic field and 2, 14, 36, 60, 120 when at the magic angle, respectively. In the second set (the 60-point method), the number of echoes had 60 increments; the last number of echoes was 710, which corresponds to an echo time of 710ms. For the first set (the 5-point method) experiment, the 2D imaging parameters were: echo time (TEi) was 8.6 ms; field of view (FOV) was 0.33 cm × 0.33 cm; imaging matrix size was 128 × 256 (256 was in the readout direction); the spectral bandwidth was 50 kHz corresponding to a 20 μs of readout sampling dwell time; 1 ms and 0.75 ms sinc shape pulses were used as excitation and refocusing pulse in imaging segment, respectively. For the second set (the 60-point method) experiment, the following imaging parameters were used to minimize the echo time (TEi) and to achieve higher SNR: Field of view 0.33 cm × 0.33 cm; imaging matrix size 128 × 128; spectral bandwidth 100 kHz corresponding to a 10 μs of readout sampling dwell time; 0.8 ms and 0.507 ms hermite shape pulses were used as excitation and refocusing pulse, respectively. With these imaging parameters, an echo time of 3 ms in the imaging segment was achieved. The TR of the imaging experiment was 2 s. The in-plane pixel size, which was across the depth of the cartilage tissue, was 13 μm for the 5-point method and 26μm for the 60-point method. The 1mm-thick imaging slice was transversely located at the middle of the 10mm-long specimen. For tendon imaging experiments, all imaging parameters are the same except a larger FOV (1.2 cm × 1.2 cm) was used.
Fig 1.
The imaging pulse sequence that was used for the quantification of T2 using CPMG.
From the images acquired using the 5-point method, the T2 relaxation in cartilage was calculated by a single exponential fitting of the data on a pixel-by-pixel basis, which assumes that there is only one T2 component in cartilage. From the intensity images acquired using the 60-point method, the T2 relaxation was calculated based on the same function by two methods, the nonnegative least-square (NNLS) method [24; 43] implemented with the MatLab codes (MathWorks, Natick, MA) and the multi-variable exponential fitting in KaleidaGraph (Synergy Software, Reading, PA). The signal intensities as a function of the TEc (the echo time in the leading contrast segment) can be written as:
| (2) |
where N is the number of echoes, ti is the ith TEc, y(ti) is the signal intensity of the ith echo, M is the number of T2 components, Sj is the intensity of the jth T2 component, and C is a constant accounting for any offset of the signal. While we concentrated on the number of T2 components, no additional constraint was incorporated into the least-squares misfit (Eq(7) in the reference [24]), which was minimized in NNLS algorithm. Therefore only a discrete spectrum of relaxation times was acquired from the data [24]. The bin spacing was 0.2 ms; the range of T2 was from 0.2 ms to 400 ms (M in Eq (2) was 2000). For each T2 NNLS spectrum, the calculated T2 components below a specific threshold (peak intensity less than 2% of the total intensity) were ignored to eliminate the dependence of the fit to the noise [27; 32] [44]. All NNLS results in this report were also verified by the high R-values of the multi-exponential fitting method (in most cases, R > 0.999). The combination of the exponential fitting and NNLS method meant that the results in this report were acquired without a priori assumptions about the number of T2 component and any initial guess of the solution.
Both 2D images and 1D profiles through the depth of the cartilage tissue in the 2D images were used for analysis and presentation. The 2D images enable us to examine any topographical variation in the images, whereas the 1D profiles enable us to examine the depth dependency of the results quantitatively and to compare the profiles from different imaging experiments. All images and profiles have consistent features among all specimens. Since there was no visible variation laterally within the tissue in these images, a rectangular region-of-interest (ROI) with a width of 10 contiguous columns were averaged from the 2D images to enhance the signal-to-noise ratio of the 1D profiles. The width and the location of the ROI were approximately the same in all specimens. There was no manual scaling or adjustment in plotting several cross-sectional profiles, which came from independent experiments, into one figure. Because the averaging occurs perpendicular to the tissue depth, the pixel resolution in the averaged profiles along the tissue depth is still 13μm and 26μm respectively.
Results
Proton Intensity Images
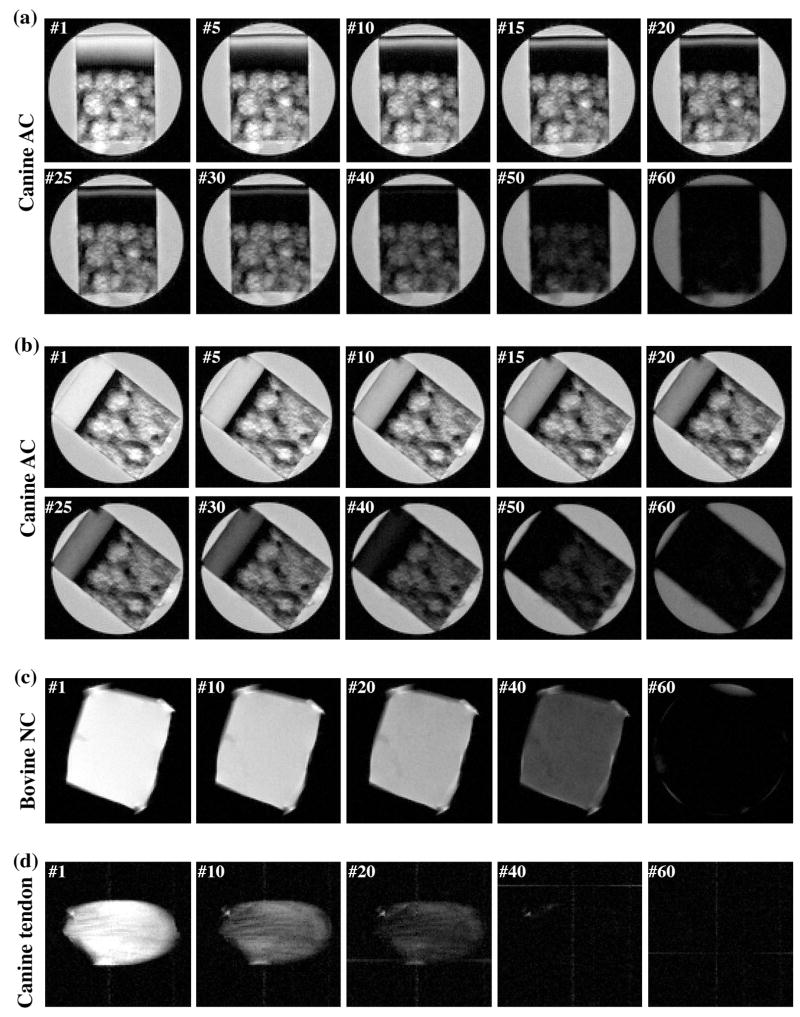
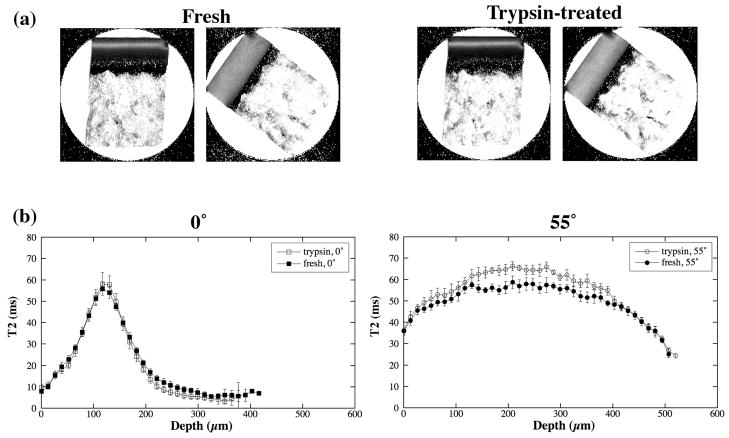
Canine tendon, bovine nasal cartilage and canine articular cartilage were studied using NMR spectroscopy and/or μMRI. Fig 2 shows the selected proton images from the representative T2-imaging experiments where each μMRI experiment had 60 proton intensity images with different T2 weightings. For the articular cartilage (Fig 2a and 2b), it is clear that the tissue has strong depth-dependent laminar structure when the tissue block is oriented with the normal axis of its surface parallel with the external magnetic field, which pointed upwards (Fig 2a, termed as the 0° orientation). This laminar appearance is due to the strong T2 anisotropy in the superficial zone and radial zone of cartilage [47]. When the tissue block is oriented at the magic angle (Fig 2b, the 55° orientation), the minimization of the dipolar interaction results in higher T2 relaxation in these two zones of cartilage, and hence brighter and more uniform images. For both bovine nasal cartilage (Fig 2c) and canine tendon (Fig 2d), there is no μMRI-visible structure in these intensity images.
Fig 2.
Selected proton images from the μMRI experiments where each experiment had 60 proton images with different T2 weightings, (a) a block of canine articular cartilage at 0° orientation, (b) a block of canine articular cartilage at the magic angle (55°), (c) a block of bovine nasal cartilage, and (d) a block of canine tendon. All images were displayed using the same minimum and maximum values. (AC: articular cartilage; NC: nasal cartilage)
T2 Relaxation in Canine Tendon and Bovine Nasal Cartilage
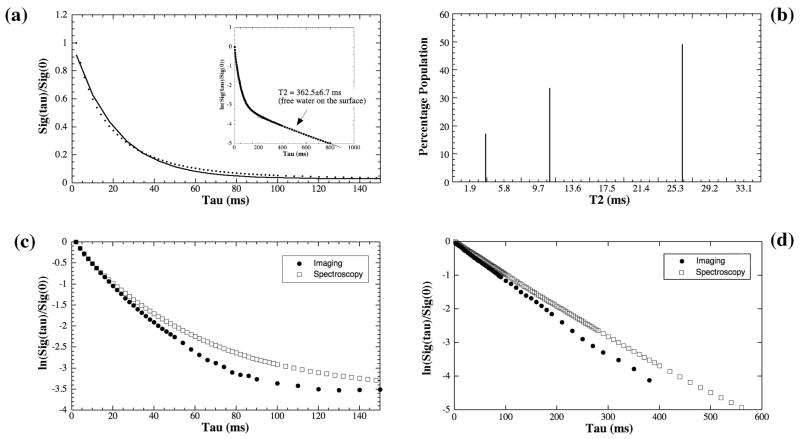
The normalized signal from the NMR spectroscopy experiment of a canine tendon is shown in Fig 3a, where the decay of the signal is clearly not single-exponential. In addition to the long T2 component of 362.5ms that was attributed to the residual free water on the surface of the specimen or NMR tube (see the insert), this set of spectroscopy data was analyzed by the NNLS algorithm to contain three T2 components, shown in Fig 3b. With this a priori knowledge, multi-exponential fitting was also able to resolve three T2 components from the signal, summarized in Table 1. The R-value of this fitting was 1, which means that the R value is larger than 0.999995 because only five significant figures are shown in KaleidaGraph. A visually convenient way of distinguishing whether the data is a single component or not is to plot the data in the natural log scale, shown in Fig 3c for both the spectroscopy and imaging results of canine tendon and in Fig 3d for the results of bovine nasal cartilage, respectively. Since tendon and nasal cartilage have no μMRI-visible structure in their intensity images, one averaged value was extracted from a 2D region-of-interest (10 pixels by 10 pixels) in the middle of each intensity image shown in Fig 2 to improve the signal-to-noise ratio (SNR). (The SNR of NMR spectroscopy and MR imaging (before the averaging) for the first data point were 31250 and 187 for tendon and 21250 and 380 for nasal cartilage respectively) It is clear that canine tendon has multiple T2 components but bovine nasal cartilage has a single T2 component.
Fig 3.
(a) The normalized signal decay in the NMR spectroscopy experiment of a canine tendon, where the solid line in the main figure is an exponential fit with one decay-constant (i.e., one T2 component). (b) The NNLS results using the same data in (a), where three T2 components were found. (c) The natural log plot of the canine tendon data from both NMR spectroscopy and μMRI, where the curvatures of the log plot show visually the multi T2 component of the specimens. (d) The natural log plot of the bovine nasal cartilage data from both NMR spectroscopy and μMRI, where the linearity of the log plot show visually single T2 component of the specimens.
Table 1.
Comparison of multi-component T2 in Tendon and Bovine Nasal Cartilage (BNC)
| Bulk from Spectroscopy | Bulk from Imaging | |||||||
|---|---|---|---|---|---|---|---|---|
| Exp Fit (KG) | NNLS (MatLab) | Exp Fit (KG) | NNLS (Matlab) | |||||
| T2 (ms) | % | T2 (ms) | % | T2 (ms) | % | T2 (ms) | % | |
| tendon | 3.6±0.1 | 30.8 | 3.7±0.2 | 17.3 | 1.7±0.9 | 7.5 | 1.6±0.9 | 7.5 |
| 12.5±0.2 | 29.1 | 11.0±0.4 | 33.6 | 8.1±1.0 | 26.9 | 8.0±0.3 | 26.4 | |
| 29.1±0.2 | 40.1 | 26.1±0.6 | 49.1 | 21.0±0.4 | 65.6 | 20.9±0.8 | 66.1 | |
| BNC | 98.9±0.9 | 100 | 96.0±0.9 | ~100* | 88.3±0.5 | 100 | 85.5±0.9 | ~100* |
There were some minor deviations (~ 2%) in bovine nasal cartilage; but we concluded that they were caused by the noises in the data.
T2 Relaxation in Canine Articular Cartilage
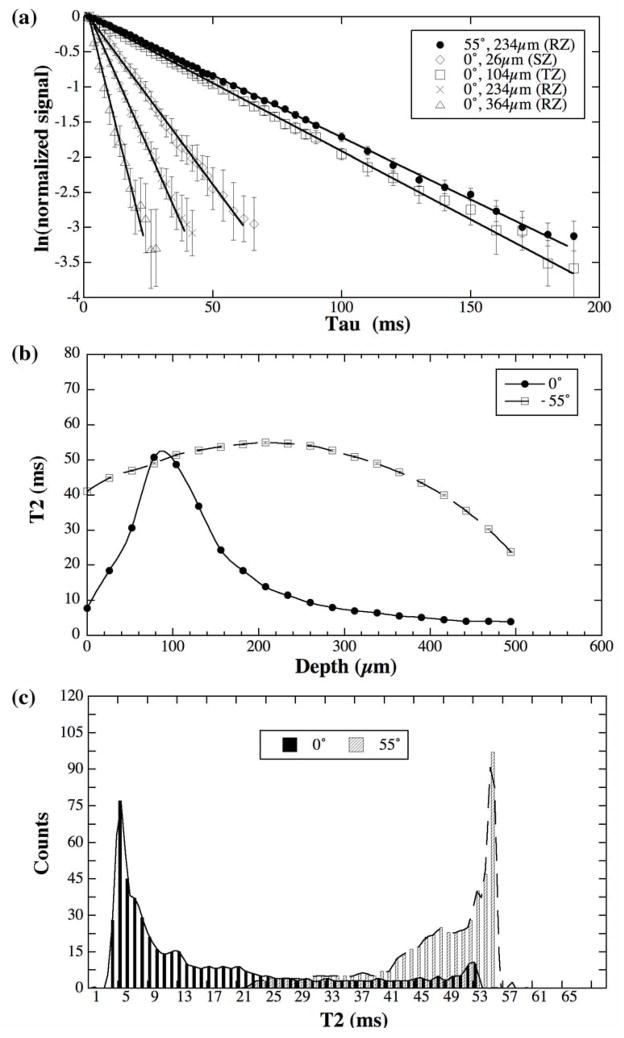
The normalized proton signal from canine articular cartilage in the natural log format is shown in Fig 4a. Since articular cartilage has the depth-dependent morphological structure, every data point in Fig 4a comes from a 1D region-of-interest (1 pixel by 10 pixels) at a fixed tissue depth. The best SNR occurs where the tissue has the longest T2 relaxation, i.e., in (1) all tissue locations when the tissue block is oriented at the magic angle and (2) the transitional zone when the tissue block is oriented at 0°. Within the limit of the imaging SNR (590, 685, 490 and 290 for the first data point at 26 μm, 104 μm, 234 μm and 364 μm location and after averaging) and given our voxel size (26μm×26μm×1mm = 6.76×10−4 mm3), there is no visible multi T2 component in articular cartilage. The NNLS algorithm was also used to verify this visual impression at every pixel along the entire tissue depth of cartilage imaging results; the results were consistently a single-component of T2.
Fig 4.
Fresh canine articular cartilage data from the 60-image μMRI experiments. (a) The natural log plots of the μMRI data of healthy canine articular cartilage, where each plot was from a single pixel location at different zones in articular cartilage. (b) The depth dependent T2 profiles in healthy articular cartilage at 0° and 55°, where each point was calculated individually using the NNLS algorithm. (c) The histograms of the T2 distribution in healthy cartilage based on the interpolated T2 profiles shown in (b). (Note that some error bars, which have been plotted in (a) and (b), are within the symbols.)
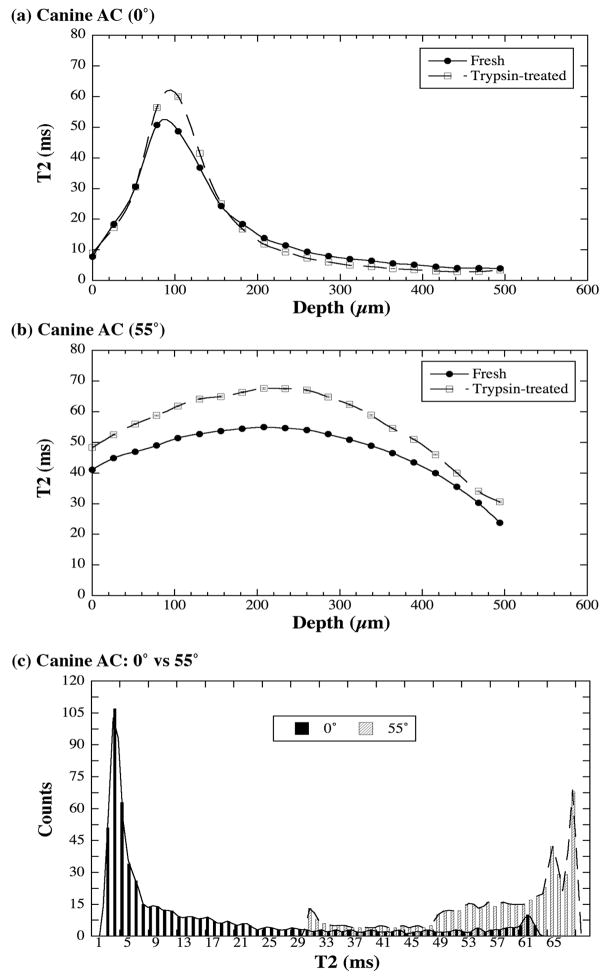
Fig 4b shows the depth-dependent T2 profiles in fresh articular cartilage at 0° and 55°, where each point was calculated individually using the fitting method from all 60 data points from the imaging experiment after NNLS algorithm was performed first, the error bars are the fitting errors. These T2 profiles agree well with the T2 profiles from our previous cartilage imaging work where only 4–5 data points were used [46]. It is clear that the errors in the fitting were extremely small. Fig 4c shows the histograms of the T2 distribution in this fresh cartilage based on the interpolated T2 profiles shown in Fig 4b, where the majority of the T2 relaxation is centered around 54ms when the tissue is oriented at the magic angle and around 4ms when the tissue is oriented at 0°.
Several fresh articular cartilage blocks were treated biochemically using a 10mg/ml trypsin (Sigma-Aldrich, USA) digestion protocol [48], which removes the GAG macromolecules from the tissue. Data analysis of T2 component identical to those used in Fig 4 were applied to this set of images, i.e., along the tissue depth from the articular surface to the bone at each pixel location. No multi-component T2 was found within the limit of SNR at this voxel size. Fig 5a and 5b compare the calculated T2 profiles for the fresh and trypsin-treated cartilage tissues at 0° and 55° respectively. The RMS averages of the entire T2 profiles in Fig 5a and 5b were 23.6±17.8ms for trypsin-treated tissue at 0°, 21.5±14.8ms for fresh tissue at 0°, 56.9±11.2ms for trypsin-treated tissue at 55°, and 47.2±8.7ms for fresh tissue at 55°. Several features can be identified. First, T2 relaxation is sensitive to the GAG concentration in cartilage. After the depletion of GAG, the T2 value increased, presumably due to the increased amount of water and the changed environment of water molecules in the tissue. Second, the sensitivity of T2 to the GAG concentration maximized only when the influence of dipolar interaction was minimized; this happens at all tissue depths when the specimen is oriented at the magic angle (Fig 5b) as well as in the transitional zone when the specimen is oriented at 0° (Fig 5a).
Fig 5.
Trypsin-treated canine articular cartilage data from the 60-image μMRI experiments. The comparisons between the T2 profiles of fresh and trypsin-treated cartilage were shown in (a) and (b). (c) The histograms of the T2 distribution in trypsin-treated cartilage based on their interpolated T2 profiles, which show the increased T2 values in the GAG-depleted cartilage. (The error bars have been plotted in (a) and (b).)
Fig 5c shows the histograms of the T2 distribution in this trypsin-digested cartilage based on the interpolated T2 profiles shown in Fig 5a and 5b. Comparing with Fig 4c, it is clear that the enzymatic digestion shifted the T2 distribution and the most obvious shift of the T2 values occur when the specimen is oriented at 55°, toward higher T2 values.
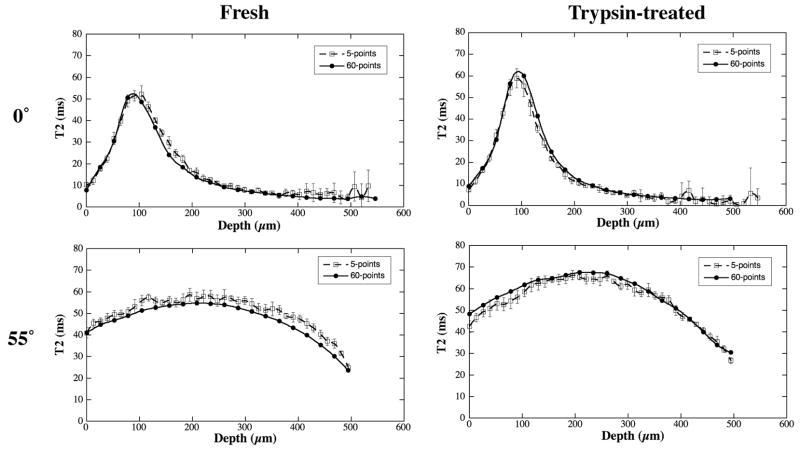
Several tissue blocks were also imaged using our ‘conventional 5-point’ T2-imaging method, shown in Fig 6. Comparing with the T2 images from 60-point method (Fig 5a and 5b), the 5-point method resulted in essentially the similar conclusion, i.e., T2 was sensitive to the trypsin digestion, mostly sensitive when the specimen was oriented at the magic angles. This set of data also shows that the ‘detection sensitivity’, the T2 difference between fresh and digested tissue, is less in the 5-point method. Fig 7 compares the T2 profiles between the 5-image T2 calculation and 60-image T2 calculation. It is clear that both methods produce consistent results.
Fig 6.
Canine articular cartilage data from the 5-image μMRI experiments. The T2 images (a) and profiles (b) of fresh and trypsin-treated articular cartilage.
Fig 7.
Comparison for the T2 profiles between the 5-image T2 calculation and 60-image T2 calculation in μMRI.
Discussions
We have studied the multi-components of T2 relaxation in cartilage and tendon using μMRI and NMR spectroscopy. Since articular cartilage is a thin layer of tissue with a complex depth-dependent structure, any quantification experiment for cartilage needs to be carried out at microscopic resolution, in order to minimize the volume averaging effect among the different molecular and structural environments in articular cartilage.
Single vs. multi-component of T2 in tendon and cartilage
Within the limit of practical SNR in microscopic imaging, we have found in three different tissues that (1) tendon has multiple T2 components; (2) nasal cartilage has single T2 component; and (3) articular cartilage has single T2 components in 26μm resolution along the depth of the tissue. Our tendon results are consistent with the NMR spectroscopy literature, where T2 in tendon has been found to be multi-exponential [34; 35]. Since tendon can be considered as a bundle of ‘uniformly oriented’ collagen fibers, the tendon result does not depend upon the resolution of the MRI/NMR experiments.
It should be point out that the ability to detect multi-components of T2 in NMR/MRI can be influenced by several factors including the regularization algorithms and the inevitable experimental noises. For example, Moody and Xia found that a SNR threshold of about 300 was needed for detecting the presence of the sub-t0 component with a probability of 0.9 or greater when using the second-derivative-squared regularizer in the linear regularization methods [44]. Although the SNR of cartilage imaging experiments in this report was not as good as we would like to have, it was probably better than any other imaging study of articular cartilage at this fine resolution. In fact, we have purposefully conducted an excessively long experiment to see if T2 relaxation in articular cartilage could be measured to be multi-exponential, under these nearly impractical conditions. The answer was no.
The results of the single T2 component in our articular cartilage experiments, however, would appear to be inconsistent with some literature results [35; 38; 40; 49; 50]. For example, Henkelman et al showed that articular cartilage had at least two T2 components, centered around 20 and 55 ms when the tissue block was oriented at 0° [35]. This discrepancy in the number of T2 components in articular cartilage comes from the fact that any full-thickness articular cartilage contains multiple molecular and structural architectures in its organization. If one does NMR spectroscopy or low-resolution MRI experiments, one averages among different architectures of the tissue, each having a unique characteristic. This can result in the observation of multiple T2 components in bulk or large-voxel-imaging experiments. Another critical issue for reliable multi-component T2 measurement is the need of sufficient data points to calculate T2 [24; 35]. In our μMRI experiments at high resolution, when the voxel size was reduced to 6.76×10−4 mm3 and 60 data points were acquired, we found that there was no multi-component T2 in articular cartilage. This conclusion of a single component T2 in articular cartilage is extremely beneficial to MRI T2 mapping in general. Since there is only one T2 component, one does not need to wonder whether a mono-exponential fitting of the high-resolution MRI signal is a faithful assessment of the true T2 characteristics or not.
Our one T2-component result in articular cartilage at all tissue depths, from the superficial zone to the deep zone, however, disagrees with the finding of Keinan-Adamsky et al [50], who showed that there were two T2 components (5ms and 36–57ms) in a deep tissue (~ 20–60%) of porcine articular cartilage of 12 months or older. Since several key experimental details (eg, the thickness of imaging slice, the SNR, the shape and duration of the soft pulses used (which were important to understand the ultra short TE (2ms) used in that paper) were not disclosed in the Keinan-Adamsky report, one couldn’t do a direct comparison between the Keinan-Adamsky experiment and our study. Comparing the actual T2 values, one could wonder if it was possible that we have actually missed the slow (i.e., long) T2 component in our experiments (the 36–57ms component in the Keinan-Adamsky report) in the deep zone (~ 20–60%)? The conclusion is no for the following three reasons. First, in our experiments, the T2 value from the superficial zone to the radial zone changed continuously from 4 ms to about 60ms (by either NNLS or fitting method), which means that all T2 values within this range could be detected in our experiments. Second, if the long T2 component indeed existed in the deep tissue, the long T2 component would be easier to detect than the short T2 component, because the long T2 component decays slower. Finally, the NNLS algorithm used in this report excluded a priori assumption about the number of exponentials.
Several models have been developed to explain the relaxation times and magnetization transfer in articular cartilage [40; 46; 51; 52; 53; 54] and other biological system [55; 56; 57]. It is generally accepted that the proton dipolar interaction is the dominant relaxation mechanism in articular cartilage and that there are at least two proton pools in cartilage: free and bound water with chemical exchanges between them. When the chemical exchanges are faster than the relaxation processes, only one T2 value can be measured in articular cartilage.
The sensitivity of T2 on GAG concentration in articular cartilage
In most cartilage degradation cases in clinics, which portion of the tissue loss GAG first is probably unknown. It would also depend upon what is the trigger of the tissue degradation in each clinical case. For a majority of degradation that is initially diagnosed by a surface roughening, one can assume the GAG loss would be started at the first half of the tissue, which includes the transitional zone. This report demonstrates the depth-dependent ‘sensitivity’ of T2 value towards the GAG depletion in articular cartilage. In the fresh vs. trypsin-treated T2 profiles shown in Fig 5a and 5b, the changes in T2’s RMS averages were 9.8% ((23.6 - 21.5)/21.5) when the specimen was oriented at 0° (Fig 5a) but increased to 20.6% ((56.9 – 47.2)/47.2) when the specimens was oriented at 55° (Fig 5b). This result shows that the sensitivity of T2 relaxation to the GAG depletion in cartilage (one of the early signs of tissue degradation) can be doubled if the cartilage tissue is oriented towards the magic angle (~55°). Furthermore, if one has to measure the tissue degradation when the tissue cannot be oriented at 55° (most likely in human in vivo situations), one can still monitor the tissue regions around the transitional zone. Fig 5a shows that the T2 value in transitional zone was still sensitive to the amount of GAG even though the tissue block was set at 0°, which has the strongest influence of the dipolar interaction.
On a related note, we showed in a previous work [48] that the cartilage GAG loss due to the trypsin incubation was time-dependent, reaching about 40% loss at 40 min of incubation, and about 70% GAG loss at 200 min incubation. Since the tissue blocks used in the previous work and this report are from the same origin (canine humeral cartilage blocks), and since the trypsin protocol in both studies are similar, we can assume that the GAG loss in the current project was also about 70%, which translates into approximately 20% T2 increase in MRI.
Finally, the T2 value itself from any biological experiment is a ‘composite’ measure of several rather complex factors, both at the molecular/structural levels in a biological specimen itself and at the procedural levels in the instrumentation/data-analysis. Consequently, it should be cautionary to attribute any small elevation or depletion in T2 values to some interventional procedure. In this report, it might seem that T2 in the deep part of the fresh cartilage at 0° were somewhat higher than the same in the trypsin-treated cartilage (200–500μm region in Fig 5a). A statistics comparison reveals such visual difference is actually very small: the RMS T2 averages and standard deviations are 5.99±3.19 ms and 5.97±2.86ms for the fresh and trypsin-treated tissues respectively. However, the same experiments clearly confirm that the T2 in the trypsin-treated specimen is significantly higher than the same in the fresh specimen (80–140μm region in Fig 5a). This distinct contrast between the deep tissue and the transitional zone tissue further demonstrates that the measurable T2 difference in the transitional zone in this report was not due to any instrumentation/data-analysis factor, but due to the specimens themselves – one of the two had been trypsin-treated.
In conclusion, the experiments in this report investigated the multi-components of T2 relaxation in three tissues: articular cartilage, nasal cartilage and tendon. While tendon has multi components of T2, articular cartilage was found to have a single component of T2 at microscopic resolution. This study provides an important message to the community of cartilage MRI: As long as one has sufficient resolution, a simple mono-exponential fitting of the T2 data is a valid approach in the quantification of T2 in cartilage. (When one has large voxels, as in clinical MRI, however, one can still have multiple T2 components in MRI, simply because the co-existence of multiple environments.) In addition, the sensitivity of T2 relaxation towards the GAG depletion was investigated. A strong influence of the specimen orientation in the magnetic field was found - One could double the ‘detection sensitivity’ if the specimen could be oriented towards the magic angle. Since a substantial improvement of image resolution in quantitative study of cartilage using the current MRI hardware and methodology is unlikely, the microscopic results in this report can serve as the ‘ultimate’ characteristics of T2 relaxation for future MRI study of articular cartilage.
Acknowledgments
Y Xia is grateful to the National Institutes of Health for the R01 grants (AR 045172 and AR 052353). The authors are indebted to Dr. A Bidthanapally (Dept of Physics, Oakland University) for preparing the solutions used in this study, and Drs. C Les and H Sabbah (Henry Ford Hospital, Detroit) for providing the canine specimens. The authors thank Dr Kenneth P Whittall for his valuable insights in the use of NNLS calculation and Miss Carol Searight (Dept of Physics, Oakland University) for editorial comments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.The Centers for Disease Control and Prevention, Press Release (October 24, 2002). (http://www.cdc.gov/od/oc/media/pressrel/r021024.htm)
- 2.Xia Y, Moody J, Burton-Wurster N, Lust G. Quantitative In Situ Correlation Between Microscopic MRI and Polarized Light Microscopy Studies of Articular Cartilage. Osteoarthritis Cartilage. 2001;9:393–406. doi: 10.1053/joca.2000.0405. [DOI] [PubMed] [Google Scholar]
- 3.Maroudas A. Biophysical chemistry of cartilaginous tissues with special reference to solute and fluid transport. Biorheology. 1975;12:233–248. doi: 10.3233/bir-1975-123-416. [DOI] [PubMed] [Google Scholar]
- 4.Venn M, Maroudas A. Chemical composition and swelling of normal and osteoarthritic femoral head cartilage. Ann Rheum Dis. 1977;36:121–129. doi: 10.1136/ard.36.2.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clarke IC. Articular cartilage: A review and scanning electron microscope study. J Bone Joint Surgery. 1971;53 B:732–750. [PubMed] [Google Scholar]
- 6.Bayliss M, Venn M, Maroudas A, Ali SY. Structure of proteoglycans from different layers of human articular cartialge. Biochem J. 1983;209:387–400. doi: 10.1042/bj2090387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miosge N, Flachsbart K, Goetz W, Schultz W, Kresse H, Herken R. Light and electron microscopical immunohistochemical localization of the small proteoglycan core proteins decorin and biglycan in human knee joint cartilage. Histochem J. 1994;26:939–45. [PubMed] [Google Scholar]
- 8.Xia Y. Magic Angle Effect in MRI of Articular Cartilage - A Review. Invest Radiol. 2000;35:602–621. doi: 10.1097/00004424-200010000-00007. [DOI] [PubMed] [Google Scholar]
- 9.Mow VC, Guo XE. Mechano-electrochemical properties of articular cartilage: their inhomogeneities and anisotropies. Ann Rev Biomed Eng. 2002;4:175–209. doi: 10.1146/annurev.bioeng.4.110701.120309. [DOI] [PubMed] [Google Scholar]
- 10.Nieminen MT, Rieppo J, Toyras J, Hakumaki JM, Silvennoinen J, Hyttinen MM, Helminen HJ, Jurvelin JS. T2 relaxation reveals spatial collagen architecture in articular cartilage: a comparative quantitative MRI and polarized light microscopic study. Magn Reson Med. 2001;46:487–93. doi: 10.1002/mrm.1218. [DOI] [PubMed] [Google Scholar]
- 11.Camacho NP, West P, Torzilli PA, Mendelsohn R. FTIR microscopic imaging of collagen and proteoglycan in bovine cartilage. Biopolymers. 2001;62:1–8. doi: 10.1002/1097-0282(2001)62:1<1::AID-BIP10>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 12.Potter K, Kidder LH, Levin IW, Lewis EN, Spencer RG. Imaging of collagen and proteoglycan in cartilage sections using Fourier transform infrared spectral imaging. Arthritis Rheum. 2001;44:846–55. doi: 10.1002/1529-0131(200104)44:4<846::AID-ANR141>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 13.Xia Y, Ramakrishnan N, Bidthanapally A. The depth-dependent anisotropy of articular cartilage by Fourier-transform infrared imaging (FTIRI) Osteoarthritis Cartilage. 2007;15:780–788. doi: 10.1016/j.joca.2007.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dardzinski BJ, Mosher TJ, Li S, Van Slyke MA, Smith MB. Spatial variation of T2 in human articular cartilage. Radiology. 1997;205:546–50. doi: 10.1148/radiology.205.2.9356643. [DOI] [PubMed] [Google Scholar]
- 15.Gründer W, Wagner M, Werner A. MR-microscopic visualization of anisotropic internal cartilage structures using the magic angle technique. Magn Reson Med. 1998;39:376–82. doi: 10.1002/mrm.1910390307. [DOI] [PubMed] [Google Scholar]
- 16.Mosher TJ, Dardzinski BJ, Smith MB. Human Articular Cartilage: Influence of Aging and Early Symptomatic Degeneration on the Spatial Variation of T2 - Preliminary Findings at 3 T. Radiology. 2000;214:259–266. doi: 10.1148/radiology.214.1.r00ja15259. [DOI] [PubMed] [Google Scholar]
- 17.Nieminen MT, Toyras J, Rieppo J, Hakumaki JM, Silvennoinen J, Helminen HJ, Jurvelin JS. Quantitative MR microscopy of enzymatically degraded articular cartilage. Magn Reson Med. 2000;43:676–81. doi: 10.1002/(sici)1522-2594(200005)43:5<676::aid-mrm9>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 18.Olivier P, Loeuille D, Watrin A, Walter F, Etienne S, Netter P, Gillet P, Blum A. Structural evaluation of articular cartilage: potential contribution of magnetic resonance techniques used in clinical practice. Arthritis Rheum. 2001;44:2285–95. doi: 10.1002/1529-0131(200110)44:10<2285::aid-art391>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 19.Liess C, Lusse S, Karger N, Heller M, Gluer CC. Detection of changes in cartilage water content using MRI T2-mapping in vivo. Osteoarthritis Cartilage. 2002;10:907–13. doi: 10.1053/joca.2002.0847. [DOI] [PubMed] [Google Scholar]
- 20.Gold GE. Dynamic and functional imaging of the musculoskeletal system. Semin Musculoskelet Radiol. 2003;7:245–8. doi: 10.1055/s-2004-815674. [DOI] [PubMed] [Google Scholar]
- 21.Alhadlaq H, Xia Y, Moody JB, Matyas J. Detecting Structural Changes in Early Experimental Osteoarthritis of Tibial Cartilage by Microscopic MRI and Polarized Light Microscopy. Ann Rheum Dis. 2004;63:709–717. doi: 10.1136/ard.2003.011783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xia Y, Moody J, Alhadlaq H, Burton-Wurster N, Lust G. Characteristics of Topographical Heterogeneity of Articular Cartilage over the Joint Surface of a Humeral Head. Osteoarthritis Cartilage. 2002;10:370–380. doi: 10.1053/joca.2002.0523. [DOI] [PubMed] [Google Scholar]
- 23.Xia Y, Moody J, Alhadlaq H, Hu JN. Imaging the Physical and Morphological Properties of a Multi-Zone Young Articular Cartilage at Microscopic Resolution. J Magn Reson Imaging. 2003;17:365–374. doi: 10.1002/jmri.10269. [DOI] [PubMed] [Google Scholar]
- 24.Whittall KP, MacKay AL. Quantitative interpretation of NMR relaxation data. J Magn Reson. 1989;84:134–152. [Google Scholar]
- 25.Bronskill MJ, Santyr GE, Walters B, Henkelman RM. Analysis of discrete T2 components of NMR relaxation for aqueous solutions in hollow fiber capillaries. Magn Reson Med. 1994;31:611–8. doi: 10.1002/mrm.1910310606. [DOI] [PubMed] [Google Scholar]
- 26.Gareau PJ, Rutt BK, Bowen CV, Karlik SJ, Mitchell JR. In vivo measurements of multi-component T2 relaxation behaviour in guinea pig brain. Magn Reson Imaging. 1999;17:1319–25. doi: 10.1016/s0730-725x(99)00064-8. [DOI] [PubMed] [Google Scholar]
- 27.Oh J, Han ET, Pelletier D, Nelson SJ. Measurement of in vivo multi-component T2 relaxation times for brain tissue using multi-slice T2 prep at 1.5 and 3 T. Magn Reson Imaging. 2006;24:33–43. doi: 10.1016/j.mri.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 28.Qin Q, Gore JC, de Graaf RA, Does MD. Quantitative T2 measurement of a single voxel with arbitrary shape using pinwheel excitation and CPMG acquisition. Magma. 2007;20:233–40. doi: 10.1007/s10334-007-0088-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Does MD, Snyder RE. T2 relaxation of peripheral nerve measured in vivo. Magn Reson Imaging. 1995;13:575–80. doi: 10.1016/0730-725x(94)00138-s. [DOI] [PubMed] [Google Scholar]
- 30.Stanisz GJ, Henkelman RM. Diffusional anisotropy of T2 components in bovine optic nerve. Magn Reson Med. 1998;40:405–10. doi: 10.1002/mrm.1910400310. [DOI] [PubMed] [Google Scholar]
- 31.Prior BM, Foley JM, Jayaraman RC, Meyer RA. Pixel T2 distribution in functional magnetic resonance images of muscle. J Appl Physiol. 1999;87:2107–14. doi: 10.1152/jappl.1999.87.6.2107. [DOI] [PubMed] [Google Scholar]
- 32.Saab G, Thompson T, Marsh G. Multicomponent T2 relaxatoin of In Vivo Skeletal Muscle. Magn Reson Med. 1999;42:150–157. doi: 10.1002/(sici)1522-2594(199907)42:1<150::aid-mrm20>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 33.Fullerton GD, Cameron IL, Ord VA. Orientation of tendons in the magnetic field and its effect on T2 relaxation times. Radiology. 1985;155:433–435. doi: 10.1148/radiology.155.2.3983395. [DOI] [PubMed] [Google Scholar]
- 34.Peto S, Gillis P. Fiber-to-field angle dependence of proton nuclear magnetic relaxation in collagen. Magn Reson Imaging. 1990;8:705–12. doi: 10.1016/0730-725x(90)90005-m. [DOI] [PubMed] [Google Scholar]
- 35.Henkelman RM, Stanisz GJ, Kim JK, Bronskill MJ. Anisotropy of NMR properties of tissues. Magn Reson Med. 1994;32:592–601. doi: 10.1002/mrm.1910320508. [DOI] [PubMed] [Google Scholar]
- 36.Gold GE, Pauly JM, Macovski A, Herfkens RJ. MR spectroscopic imaging of collagen: tendons and knee menisci. Magn Reson Med. 1995;34:647–54. doi: 10.1002/mrm.1910340502. [DOI] [PubMed] [Google Scholar]
- 37.Takamiya H, Kusaka Y, Seo Y, Noguchi M, Ikoma K, Morimoto T, Hirasawa Y. Characteristics of proton nmr t(2) relaxation of water in the normal and regenerating tendon. Jpn J Physiol. 2000;50:569–76. doi: 10.2170/jjphysiol.50.569. [DOI] [PubMed] [Google Scholar]
- 38.Lehner KB, Rechl HP, Gmeinwieser JK, Heuck AF, Lukas HP, Kohl HP. Structure, function, and degeneration of bovine hyaline cartilage: Assessment with MR imaging in vitro. Radiology. 1989;170:495–499. doi: 10.1148/radiology.170.2.2911674. [DOI] [PubMed] [Google Scholar]
- 39.Ghiassi-Nejad M, Torzilli PA, Peemoeller H, Pintar MM. Proton spin-spin relaxation study of molecular dynamics and proteoglycan hydration in articular cartilage [In Process Citation] Biomaterials. 2000;21:2089–95. doi: 10.1016/s0142-9612(00)00140-x. [DOI] [PubMed] [Google Scholar]
- 40.Mlynarik V, Szomolanyi P, Toffanin R, Vittur F, Trattnig S. Transverse relaxation mechanisms in articular cartilage. J Magn Reson. 2004;169:300–7. doi: 10.1016/j.jmr.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 41.Maroudas A. Different ways of expressing concentration of cartilage constituents with special reference to the tissue’s organization and functional properties. In: Maroudas A, Kuettner K, editors. Methods in Cartilage Research. Academic Press; London: 1990. pp. 211–219. [Google Scholar]
- 42.Lancaster JL, Andrews T, Hardies LJ, Dodd S, Fox PT. Three-pool model of white matter. J Magn Reson Imaging. 2003;17:1–10. doi: 10.1002/jmri.10230. [DOI] [PubMed] [Google Scholar]
- 43.Lawson CJ, Hanson RJ. Solving Least Squares Problems. Prentice-Hall; Englewood Cliffs, New Jersey: 1974. [Google Scholar]
- 44.Moody JB, Xia Y. Analysis of multi-exponential relaxation data with very short components using linear regularization. J Magn Reson. 2004;167:36–41. doi: 10.1016/j.jmr.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 45.Santyr GE, Henkelman RM, Bronskill MJ. Variation in measured transverse relaxation in tissue resulting from spin locking with the CPMG sequence. J Magn Reson. 1988;79:28–44. [Google Scholar]
- 46.Xia Y. Relaxation Anisotropy in Cartilage by NMR Microscopy (μMRI) at 14 μm Resolution. Magn Reson Med. 1998;39:941–949. doi: 10.1002/mrm.1910390612. [DOI] [PubMed] [Google Scholar]
- 47.Xia Y, Farquhar T, Burton-Wurster N, Lust G. Origin of cartilage laminae in MRI. J Magn Reson Imaging. 1997;7:887–894. doi: 10.1002/jmri.1880070518. [DOI] [PubMed] [Google Scholar]
- 48.Xia Y, Farquhar T, Burton-Wurster N, Vernier-Singer M, Lust G, Jelinski LW. Self-Diffusion Monitors Degraded Cartilage. Arch Biochem Biophys. 1995;323:323–328. doi: 10.1006/abbi.1995.9958. [DOI] [PubMed] [Google Scholar]
- 49.Mlynarik V, Degrassi A, Toffanin R, Vittur F, Cova M, Pozzi-Mucelli RS. Investigation of laminar appearance of articular cartilage by means of magnetic resonance microscopy. Magn Reson Imaging. 1996;14:435–42. doi: 10.1016/0730-725x(96)00025-2. [DOI] [PubMed] [Google Scholar]
- 50.Keinan-Adamsky K, Shinar H, Navon G. Multinuclear NMR and MRI studies of the maturation of pig articular cartilage. Magn Reson Med. 2006;55:532–40. doi: 10.1002/mrm.20775. [DOI] [PubMed] [Google Scholar]
- 51.Adler RS, Swanson SD, Yeung HN. A three-component model for magnetization transfer. Solution by projection-operator technique, and application to cartilage. J Magn Reson B. 1996;110:1–8. doi: 10.1006/jmrb.1996.0001. [DOI] [PubMed] [Google Scholar]
- 52.Lüsse S, Knauss R, Werner A, Gründer W, Arnold K. Action of Compression and Cations on the Proton and Deuterium Relaxation in Cartilage. Magn Reson Med. 1995;33:483–489. doi: 10.1002/mrm.1910330405. [DOI] [PubMed] [Google Scholar]
- 53.Lattanzio PJ, Marshall KW, Damyanovich AZ, Peemoeller H. Macromolecule and water magnetization exchange modeling in articular cartilage. Magn Reson Med. 2000;44:840–51. doi: 10.1002/1522-2594(200012)44:6<840::aid-mrm4>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 54.Xia Y, Moody J, Alhadlaq H. Orientational dependence of T2 relaxation in articular cartilage: A microscopic MRI (μMRI) study. Magn Reson Med. 2002;48:460–469. doi: 10.1002/mrm.10216. [DOI] [PubMed] [Google Scholar]
- 55.Fullerton G, Potter J, Dornbluth N. NMR relaxation of protons in tissues and other macromolecular water solutions. Magnetic resonance imaging. 1982;1:209–228. doi: 10.1016/0730-725x(82)90172-2. [DOI] [PubMed] [Google Scholar]
- 56.Ling GN, Murphy RC. Studies on the physical state of water in living cells and model systems. II. NMR relaxation times of water protons in aqueous solutions of gelatin and oxygen-containing polymers which reduce the solvency of water for NA+, sugars, and free amino acids. Physiol Chem Phys Med NMR. 1983;15:137–54. [PubMed] [Google Scholar]
- 57.Watanabe T, Murase N, Staemmler M, Gersonde K. Multiexponential proton relaxation processes of compartmentalized water in gels. Magn Reson Med. 1992;27:118–34. doi: 10.1002/mrm.1910270112. [DOI] [PubMed] [Google Scholar]