Abstract
We previously reported that the fibronectin (FN) type III domains of FN may unfold to interact with anastellin and form FN aggregates. In the present study, we have focused on the interaction between anastellin and the third FN type III domain (III3) which is a key anastellin binding site on FN. Anastellin binding to III3 was monitored by 8-anilino-1-naphthalene sulfonate (ANS) fluorescence. ANS binding to anastellin dramatically increased its emission intensity, but this was reduced to half by the addition of III3, suggesting that ANS and III3 share a common hydrophobic binding site on anastellin. An engineered mutant of III3 that was stabilized by an intrachain disulfide bond did not interact with anastellin, as seen by its failure to interfere with ANS binding to anastellin. We also mutated hydrophobic core residues to destabilize III3 and found that these mutants were still capable of interacting with anastellin. Anastellin binding to III3 was also monitored using an intramolecular GFP-based FRET construct, in which III3 was flanked by two GFP variants (III3-FRET). Anastellin bound to III3-FRET and caused an increase in the FRET signal. The dissociation constant was estimated to be ~210 nM. The binding kinetics of anastellin to III3-FRET fit a first order reaction with a half time of ~30 s; the kinetics with destabilized III3 mutants were even faster. MALDI-TOF mass spectrometry suggested that the middle part of III3 became destabilized and protease sensitive upon anastellin binding. Thus, the stability of III3 seems to be a key factor in anastellin binding.
Fibronectin (FN) is assembled into matrix fibrils, which are the primary components of the extracellular matrix in embryonic tissues and wound healing (1). Cells in tissue culture can also assemble an FN matrix. The FN molecules must be connected to each other by protein-protein bonds to make the matrix fibrils, but almost nothing is known about the nature of these bonds or their location on the FN molecules. Unlike most protein assemblies, which have been reconstructed in vitro, FN matrix fibrils have so far required cell surface interactions for their assembly. An important step toward FN matrix assembly in vitro was the discovery by Morla et al. (2) that a small fragment of FN, now called anastellin, could cause FN to form a precipitate with a fibrillar substructure. They called this aggregate superFN. Anastellin and superFN, although artificial constructs, are interesting for their potent anti-angiogenic activity (3, 4). In addition, the interactions that form superFN may be related to those that assemble FN in natural matrix fibrils.
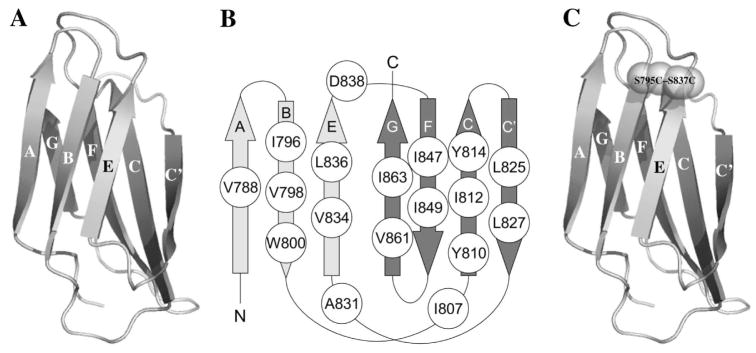
FN is a modular protein containing 15–17 FN type III (FNIII) domains along with 12 FN type I domains and two FN type II domains. FNIII domains are abundant in animal proteins and are one of the most commonly occurring domains (5). Currently, 2,573 domains in 488 proteins in the human genome have been classified as members of the FNIII superfamily in the SUPERFAMILY database (http://supfam.org/SUPERFAMILY) (6, 7). As shown in Fig. 1, FNIII domains consist of a beta-sandwich structure with three strands (A, B and E) on one side and four strands (C, C′, F and G) on the other.
Fig. 1. Structural models of III3 and III3SS, and the positions of mutations.
(A) III3 consists of a beta-sandwich structure with three strands (A, B and E) on one side and four strands (C, C′, F and G) on the other. (B) The positions of mutated residues are shown. These residues were substituted with aspartate, except that D838 was substituted with glycine. (C) An engineered disulfide bond between residues 795 and 837 locks strands B and E together. Structural models of III3 and III3SS were created previously (9).
Anastellin is an artificial protein domain derived from the first FNIII domain of FN by truncating the A and B strands (2). Briknarova et al. (8) extensively studied the structure and function of anastellin and discovered that hydrophobic residues on the C-, F- and G-strands, which are exposed by the truncation, and also a few residues on the E-strand are crucial for inducing superFN. We studied the interaction of anastellin with FN and found that anastellin bound to FNIII domains 1–2, 3 and 11 (9). FN fragments containing FNIII domains 1–3 formed superFN-like aggregates without the rest of the FN molecule. The third FNIII domain (III3) seemed particularly important. This was the only single FNIII domain that could block superFN formation, and FN fragments containing a disulfide bond in III3, engineered to prevent its unfolding, were inhibited for aggregation. This suggested that opening of the beta sandwich of III3 was an essential step in the binding of anastellin.
In the present study, we generated several III3 mutants to study the III3/anastellin interaction. Fluorescence spectroscopy was used to study the binding of anastellin to III3 in solution and MALDI-TOF (matrix assisted laser desorption ionization-time of flight) mass spectrometry was used to identify protease sensitive sites in III3 that were exposed by anastellin binding. Overall, our results indicate that the instability of III3 plays an important role in the interaction with anastellin.
EXPERIMENTAL PROCEDURES
Bacterially expressed proteins
The cDNA fragment of III3 was amplified from the III1–5 expression vector (9) by PCR and was cloned into pET15b (Novagen). This construct has the sequence: mgsshhhhhhssglvprgshmgtTTAP…(III3)… PRSDgt (the sequence in lowercase is from the expression vector). Previous works have found that a C-terminal extension enhances the stability of some FNIII domains (10, 11), so we included several extra amino acids at both the N and C termini. The PCR amplified III3 fragment was also cloned into a GFP-based FRET construct, which was generated for this experiment. The sequence of the III3-FRET construct is mgsshhhhhhssglvprgshmggrMVSK…(mYPet)…LTAAtsgt-TTAP…(III3)… PRSDgt-MVSK…(mECFP*)… ELYKggr (the mYPet, III3 and mECFP* sequences are underlined; ECFP* differs from ECFP by two mutations (12)). To make the FRET signals more sensitive to conformational changes, the flexible 11 amino acid C-terminal segment of mYPet was deleted in this construct. The expression construct for the disulfide mutant, III3(S795C–S837C), which we call III3SS, and the III3SS-FRET construct were also created in the same way as the wild-type constructs using the III1–5 constructs containing these mutations as templates (9). Note that we use the Kornblihtt amino acid numbering, which does not include a 31 amino acid signal sequence (13). III3 mutants were generated by site-directed mutagenesis with pfu turbo DNA polymerase (Stratagene). Several III3 mutants were also cloned into the FRET constructs. III3, III3SS and anastellin were expressed in Escherichia coli BL21 (DE3) at 37°C and purified with a cobalt column (Talon, Clontech) using standard procedures. Proteins were eluted with imidazole from the column and were dialyzed against 20 mM Tris with 150 mM NaCl (TBS, pH 8.0). In the case of anastellin, denaturation and renaturation were required, as reported previously (9). The FRET constructs were expressed in E. coli C41 (DE3) at 20°C to improve their solubility and were purified as described above. Minor degradation products which contained mYPet co-purified because of the N-terminal his-tag. However, a minor acceptor contamination had a negligible effect on FRET signals as reported previously (12). Protein concentrations were determined from the absorbance at 280 nm using the molar extinction coefficient of each protein as calculated (14, 15) by the Protean computer program (DNAstar Inc.). Disulfide bond formation was confirmed by 5,5′-dithio-bis (2-nitrobenzoic acid) assay under denaturing conditions (16, 17). SDS-PAGE was performed using standard procedures.
Fluorescence spectroscopy
Fluorescence measurements were carried out with a Shimadzu RF-5301-PC spectrofluorometer. For intrinsic tryptophan fluorescence measurements, 5 μM purified III3 or various mutants were excited at 280 nm in the presence of 0 to 6 M urea, and emission spectra were collected at 1-nm intervals from 300 nm to 400 nm using slit widths of 5 nm for both excitation and emission. Some experiments were also carried out in the presence of 1 mM DTT. These measurements were performed with triplicate samples at room temperature.
For 8-anilino-1-naphthalene sulfonate (ANS) fluorescence measurements, reactions contained 50 μM ANS and combinations of 10 μM III3 or its mutants and anastellin. ANS was excited at 360 nm and emission spectra were collected at 1-nm intervals from 370 nm to 620 nm using slit widths of 5 nm for both excitation and emission. Some experiments were also carried out in the presence of 1 mM DTT. The concentration of ANS was estimated from the molar extinction coefficient (4,950 M−1cm−1) at 350 nm in 0.1 M phosphate buffer (pH 6.8) (18). The stoichiometry of ANS binding to anastellin was estimated by a Job plot (19). ANS and anastellin were mixed in various ratios at a total concentration of 50 μM. Samples were excited at 360 nm and emission intensities at 470 nm were plotted to estimate the stoichiometry of ANS to anastellin. The stoichiometry of ANS binding to the anastellin/III3 complex was also estimated.
For FRET measurements, 1 μM III3-FRET or III3mutant-FRET was excited at 433 nm in the presence and absence of anastellin (5 μM), and emission spectra were collected at 1-nm intervals from 440 nm to 620 nm using slit widths of 3 nm for excitation and 5 nm for emission. Some experiments were also carried out in the presence of 1 mM DTT. The relative emission intensity was normalized to the donor (CFP) intensity at 475 nm in the trypsin-digested sample. FRET signals were evaluated by determining the “emission ratio”: the ratio of the acceptor intensity at 528 nm to the donor intensity at 475 nm. FRET efficiency was calculated from the decreased donor emission (12). All fluorescence measurements were performed with triplicate samples at room temperature and the measured error was within 5%.
To determine the dissociation constant (Kd), anastellin binding to 0.05 μM III3-FRET was monitored by measuring the emission ratio (528 nm/475 nm) to create a saturation curve. Based on the saturation curve, a Scatchard (Rosenthal) plot was constructed to estimate the Kd with the following equation (B/F)(Kd) + (B) = (Bmax), where B is the concentration of the III3/anastellin complex, F is the concentration of free anastellin, and Bmax is the maximal concentration of the complex.
The binding kinetics of anastellin to III3-FRET were monitored by measuring acceptor fluorescence at 528 nm at 2-second intervals for 5 min. The negligible amount of photobleaching was corrected with FRET signals for III3-FRET without anastellin. The half time was estimated from the single-exponential equation F∞−F(t)= [F∞ −F0]e−kt, where F∞ is the maximal fluorescence, and F(t)is the fluorescence at time, t and F0 is the initial fluorescence after correction for bleaching (t = 0) Fitting was carried out with Excel using the Solver function and allowing, F∞, F0 and the half time (t1/2 = 1n2/k) to vary as reported previously (20).
MALDI-TOF mass spectrometry
To map thermolysin sensitive sites in III3 upon anastellin binding, MALDI-TOF mass spectrometry was performed with a Voyager DE (Applied Biosystems). A 10 μM mixture of III3 and anastellin was digested with thermolysin (1 μg/ml) in TBS containing 2 mM CaCl2 at room temperature for 1 hr; digestion was quenched with 5 mM EDTA. After thermolysin treatment, some samples were further digested with trypsin (1 μg/ml) at room temperature for 30 min. Protease digested samples were mixed with saturated sinapinic acid (1:5), mounted on a plate and dried for MALDI analysis. Spectra were obtained from triplicate samples in linear mode with the accumulation of 128–160 laser shots. Acquired spectra were smoothed with the Data Explorer program.
RESULTS
The stability of III3 and mutants
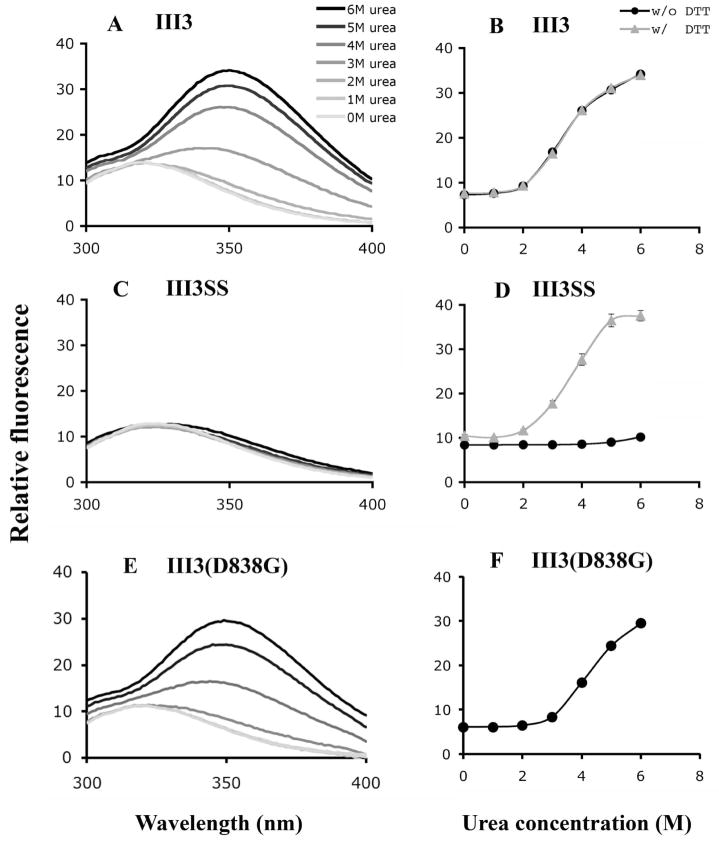
In order to evaluate the stability of wild-type III3, intrinsic tryptophan fluorescence was measured in the presence of urea. For wild-type III3, a broad emission peak centered near 320 nm in the absence of urea was shifted towards 350 nm as the concentration of urea was increased (Fig. 2A). The addition of urea also caused a substantial increase in emission intensity at 350 nm (Fig. 2B). DTT caused no significant change in the emission spectrum for this wild-type domain.
Fig. 2. The stability of III3 and mutants.
III3 and mutants were excited at 280 nm and intrinsic tryptophan fluorescence was measured at wavelengths from 300 nm to 400 nm (A, C and E). Urea was used as a denaturant. Urea denaturation curves (B, D and F) were created by plotting the emission intensity at 350 nm as a function of urea concentration in the presence or absence of DTT.
We have previously generated a disulfide mutant of III1–5 in which an intrachain disulfide bond was engineered in III3 to prevent spontaneous unfolding (9). The disulfide mutant inhibited superFN-like aggregation with anastellin. In the present study, these mutations were introduced into III3 alone, instead of the larger III1–5 construct. The mutant S795C–S837C, here called III3SS, locks the beginning of strand B to the end of strand E (Fig. 1C). The emission spectrum of III3SS was similar to wild-type in the absence of urea, although the broad peak was slightly shifted to around 325 nm (Fig. 2C). Adding urea to 6 M had no effect on the emission spectrum of III3SS. Under reducing conditions, however, this mutant denatured just like wild-type III3 (Fig. 2D). We conclude that the disulfide bond in III3SS was properly formed and greatly enhanced the stability of the domain. In addition to the disulfide mutant, we also mutated D838 on the E–F loop to glycine to enhance its stability (see details in the Discussion). As shown in Figs. 2E and F, this mutant is slightly more stable than wild-type, as indicated by a shift in the denaturation curve to higher urea concentrations.
Anastellin/III3 interaction assayed by ANS fluorescence
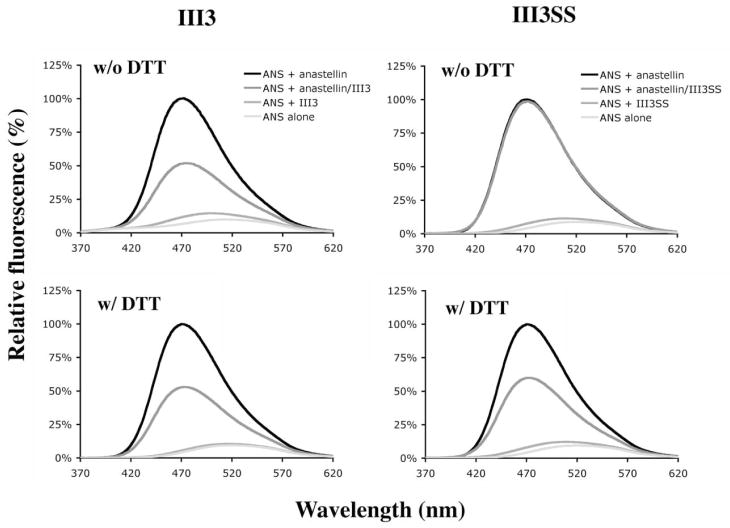
The emission intensity of 8-anilino-1-naphthalene sulfonate (ANS) is enhanced when it binds to hydrophobic regions of proteins, so it can be used to monitor conformational changes and protein-protein interactions. The emission intensity of ANS alone was very low, with a broad emission maximum near 520 nm when it was excited at 360 nm (Fig. 3). When mixed with III3 the spectrum of ANS was only slightly increased, indicating that III3 does not have any large exposed hydrophobic patches. In the presence of anastellin, however, the emission intensity of ANS was greatly increased and the emission maximum was shifted to around 470 nm. This is apparently because anastellin lacks two beta-strands on one side of its beta-sandwich, exposing several hydrophobic residues, as seen in the NMR structure (8). When III3 was included in a mixture of ANS and anastellin, however, the emission intensity was reduced by about half. DTT did not affect the emission spectra of ANS with anastellin, the anastellin/III3 complex, or III3 (Fig. 3).
Fig. 3. Interaction between anastellin and III3 measured by ANS fluorescence.
Anastellin, III3 and a mixture of anastellin and III3 in the presence of ANS were excited at 360 nm and the emission intensities were measured at wavelengths from 370 nm to 620 nm. ANS fluorescence was strongly enhanced in the presence of anastellin but decreased to about half when III3 was also present, indicating that III3 partially blocks ANS binding to anastellin. III3SS had no effect, but under reducing conditions, III3SS interacted with anastellin just like wild-type and led to a reduction in ANS fluorescence.
In contrast to III3, the addition of III3SS to anastellin did not affect the ANS emission spectrum (Fig. 3), indicating that III3SS does not interact with anastellin. Under reducing conditions, however, the reactivity of ANS was decreased just like in the case of the anastellin/III3 complex (Fig. 3), indicating that complex formation was inhibited by the disulfide bond and not by the mutations themselves. ANS binding to anastellin was not inhibited by high salt (1M NaCl) or an ionic detergent (0.1% Chaps), although ANS fluorescence was somewhat reduced. Interestingly, anastellin was able to interact with III3 under high salt conditions, as seen by a reduction of ANS fluorescence. On the other hand, Chaps inhibited the interaction between anastellin and III3 (data not shown). This is consistent with a previous report that Chaps stabilizes the structure of anastellin and eliminates superFN formation (8).
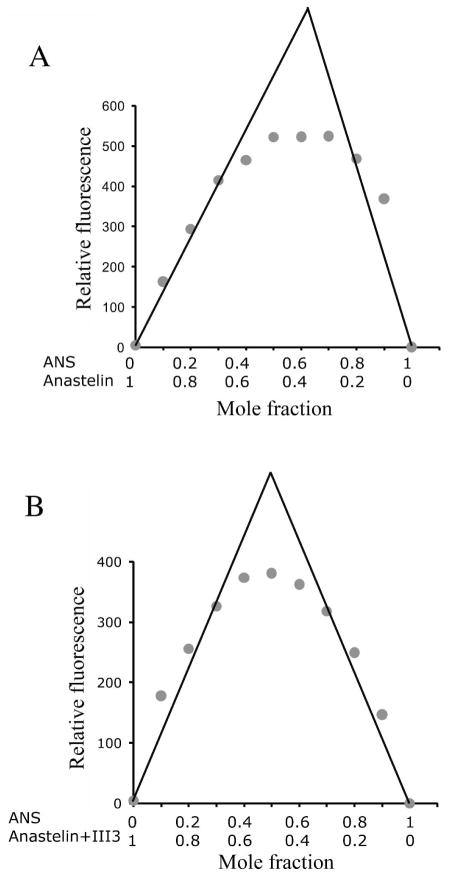
We used a Job plot to estimate the stoichiometry of anastellin to ANS. The plot of ANS binding to anastellin is asymmetric indicating that the stoichiometry cannot be 1:1 but is approximately 2:1 ANS:anastellin (Fig. 4A). In contrast, the plot of ANS binding to the anastellin–III3 complex is symmetric and the stoichiometry appears to be 1:1 (Fig. 4B).
Fig. 4. The stoichiometry of ANS binding to anastellin and the anastellin/III3 complex.
Job plots show an approximately 1:2 stoichiometry of anastellin to ANS (A) and 1:1 stoichiometry of anastellin/III3 complex to ANS (B) at a total concentration of 50 μM.
Mutational analysis of III3/anastellin binding
The structure and stability of FNIII domains have been most extensively studied with the tenth FNIII domain (III10) of FN and the third FNIII domain of tenascin (10,21–26). These studies indicate that hydrophobic core packing is a key factor for stability (21,22,27), although a number of hydrogen bonds and some surface residues seem to contribute to the folding process (10,28,29). Our results so far suggested that anastellin binds to unfolded III3 and that electrostatic interactions do not contribute to complex formation. Therefore, we mutated hydrophobic core residues of III3 to negatively charged aspartate to further explore the stability of III3 and its interaction with anastellin. The positions of mutations are shown in Fig. 1B. Although all mutated domains were soluble, most mutations did affect folding as shown by changes in tryptophan fluorescence and increasing ANS reactivities (see Table S1 of the supporting information). Surprisingly, none of the mutations inhibited anastellin binding as monitored with ANS fluorescence. Because most of the mutants behaved similarly, it was impossible to determine which specific residues contact anastellin. Instead, it seems that anastellin may bind to a non-specific hydrophobic “patch” of some undetermined size when it is exposed.
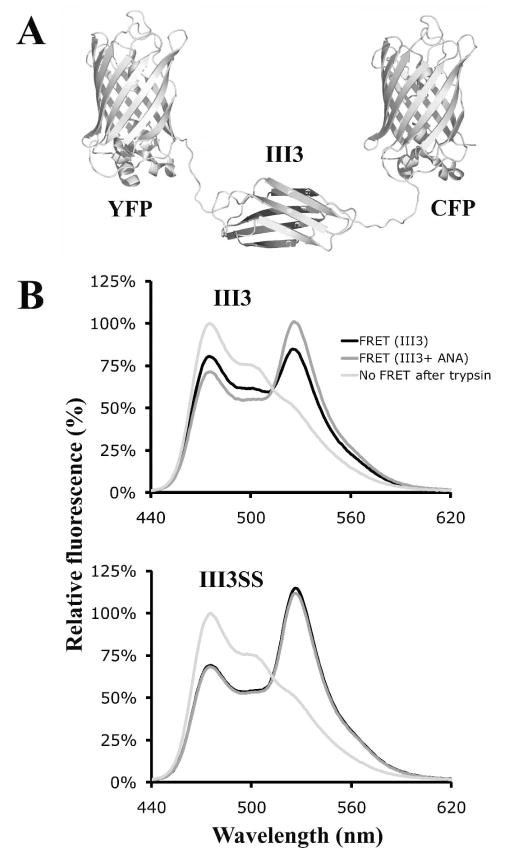
The interaction between III3 and anastellin analyzed by FRET
The III3/anastellin interaction was also monitored by GFP-based FRET, using a construct in which III3 was flanked by YFP and CFP (Fig. 5A). For this construct, we deleted 11 amino acids from the flexible C terminus of YFP. When this construct was digested with trypsin, which does not affect the fluorophores of GFPs (12) but cleaves a sensitive site in the C terminus of III3, the FRET signal decreased, as indicated by the increase in donor fluorescence at 475 nm, and the decrease in acceptor fluorescence at 528 nm (Fig. 5B). Upon binding of anastellin the FRET signal for III3-FRET increased (Fig. 5B and Table 1). DTT had no effect on the FRET signal of wild-type III3-FRET, either before or after anastellin binding (data not shown). Interestingly, the FRET signal for III3SS-FRET was higher than that for III3-FRET, suggesting that the stabilization of III3 makes the conformation more compact, or enhances a weak binding of the GFPs to III3. The FRET signal of III3SS did not change upon addition of anastellin. Under reducing conditions, the emission spectrum of III3SS-FRET was similar to that of wild-type III3-FRET, and the FRET signal was increased by the addition of anastellin just like wild-type III3-FRET (data not shown). This suggests that the cysteine mutations alone did not affect anastellin binding, but the formation of the disulfide bond did. We also tested several III3 mutants in this assay. The FRET signals for III3(V798D), III3(V834D) and III3(D838G) were also significantly increased by anastellin binding (Table 1). The relation between the conformations of the FRET constructs and the FRET signals was also studied by urea treatment (see details in Table S2 and the supplementary results of the supporting information).
Fig. 5. Interaction between anastellin and III3 monitored by GFP-based FRET.
(A) III3 is connected to two GFPs by flexible segments. The diagram does not include the N-terminal his-tag and the unstructured C terminus of CFP. (B) The III3-FRET and III3SS-FRET constructs in the presence or absence of anastellin were excited at 433 nm and the relative emission intensities were measured at wavelengths from 440 nm to 620 nm. The emission spectrum of III3-FRET changed significantly in the presence of anastellin, suggesting that anastellin interacts with III3. On the other hand, anastellin had no effect on the emission spectrum of III3SS-FRET. The emission spectra were normalized to the donor intensity at 475 nm in the trypsin-treated samples.
Table 1.
FRET efficiency with or without anastellin, and the dissociation constant and the binding kinetics of the complex
| FRET efficiencya |
Kd | Half time |
|
|---|---|---|---|
| III3-FRET | 0.19 | - | - |
| III3-FRET + Anastellin | 0.28 | ~210nM | ~30 s |
| III3(V798D)-FRET | 0.21 | - | - |
| III3(V798D)-FRET + Anastellin | 0.28 | ~180nM | < 1 sc |
| III3(V834D)-FRET | 0.21 | - | - |
| III3(V834D)-FRET + Anastellin | 0.27 | <50 nMb | <1 sc |
| III3(D838G)-FRET | 0.18 | - | - |
| III3(D838G)-FRET + Anastellin | 0.27 | ~ 490 nM | ~ 60 s |
| III3SS-FRET | 0.31 | - | - |
| III3SS-FRET + Anastellin | 0.32 | - | - |
FRET efficiency is estimated from the decreased donor fluorescence.
The Kd of anastellin binding to this mutant was too low to be accurately measured by this assay.
The binding kinetics of anastellin to these mutants were too fast to monitor by this assay.
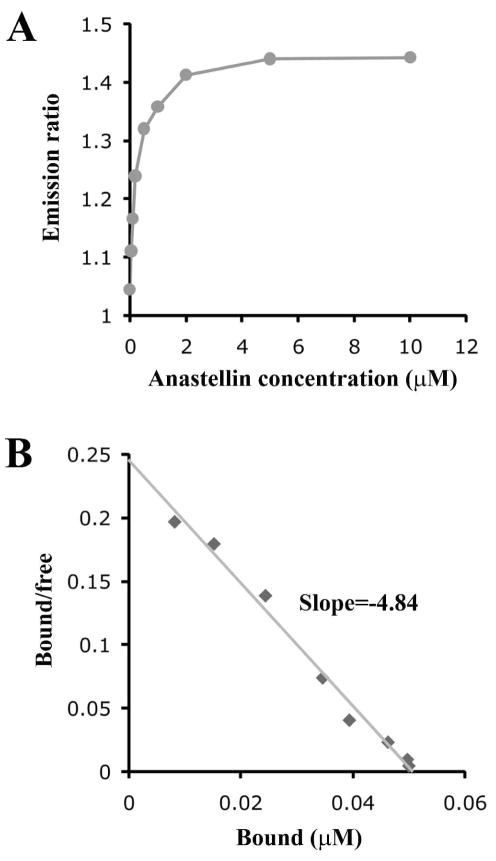
We next used the FRET assay to determine an apparent affinity constant for the binding of anastellin to III3. Fig. 6A shows the emission ratio as the concentration of anastellin was increased from 0.05 to 10 μM. Based on the saturation curve, a Scatchard plot was constructed. The Kd was estimated to be ~210 nM from the slope of the line (−1/Kd) (Fig. 6B). III3(D838G) interacted with anastellin with lower affinity, while III3(V798D) and III3(V834D) appeared to bind anastellin more tightly (Table 1).
Fig. 6. Estimation of the dissociation constant between anastellin and III3 by GFP-based FRET.
(A) Anastellin binding to 0.05 μM III3-FRET was monitored by measuring the emission ratio (528 nm/475 nm) to create a saturation curve. (B) A Scatchard plot was created by transforming the data from the saturation curve to estimate the dissociation constant.
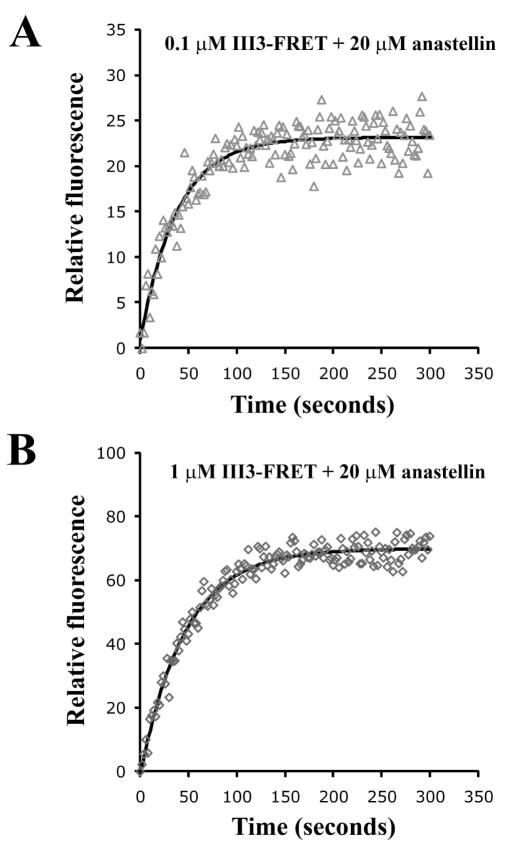
The binding kinetics of anastellin to III3-FRET were monitored by measuring acceptor fluorescence at 528 nm. At 20 μM anastellin the half time was approximately 30 s, at both low (0.1 μM, Fig. 7A) and high (1 μM, Fig. 7B) III3-FRET concentrations. Anastellin concentrations below 20 μM gave slower reactions, while concentrations above 20 μM showed no increase in reaction rates (data not shown), consistent with the saturation seen in Fig. 6A. These results suggested that the kinetics of binding of anastellin to III3-FRET were determined by a first order reaction with a rate constant of ~0.02 s−1. The binding kinetics of anastellin to III3(D838G) were slower than to wild-type. On the other hand, the kinetics of the unstable III3(V798D) and III3(V834D) mutants were too fast to measure in this assay (Table 1). These results are consistent with the measurements of apparent Kd.
Fig. 7. Binding kinetics of anastellin to III3.
0.1 μM (A) and 1 μM (B) III3-FRET were mixed with 20 μM anastellin and excited at 433 nm. Anastellin binding to III3-FRET was measured as the increase in acceptor fluorescence at 528 nm for 5 min. At 20 μM anastellin, the kinetics profiles were not affected by different III3-FRET concentrations.
Thermolysin sensitive sites on III3 exposed by anastellin binding
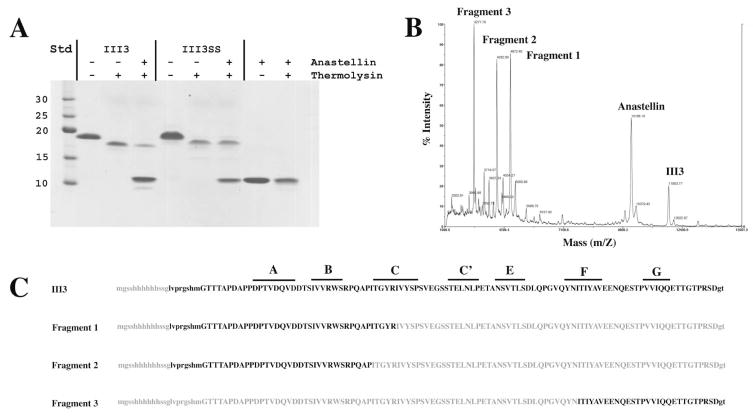
Intact III3 and III3SS are approximately 13 kDa (including the his-tag), but they migrate slowly in SDS-PAGE, as if they are ~19 kDa (Fig. 8A). Mild thermolysin digestion cleaved off the N-terminal his-tags of III3 and III3SS, producing a band that runs at ~17 kDa. The thermolysin cleaved fragments are less sensitive to Coomassie blue staining, perhaps due to the loss of the his-tag. Even with mild digestion, the cleaved fragments continued to degrade gradually under these conditions. In the presence of anastellin, III3 became more sensitive to thermolysin digestion, as reported previously for full-length FN and the III1–5 fragment (9). The primary indication of digestion is the loss of intensity of the ~17 kDa band. In addition, a smaller fragment appears, which can be barely seen just below the anastellin band. On the other hand, anastellin had no effect on the thermolysin sensitivity of III3SS.
Fig. 8. Mapping the thermolysin sensitive sites on III3 upon anastellin binding.
(A) A Coomassie blue stained SDS gel shows that III3 became more thermolysin sensitive in the presence of anastellin. Std, BenchMark protein ladder (Invitrogen). (B) Thermolysin digested fragments derived from III3 in the presence of anastellin were analyzed by MALDI-TOF mass spectrometry. (C) The sequences of the thermolysin digested fragments are shown in bold letters.
We further studied the thermolysin digested III3 fragments by MALDI-TOF mass spectrometry. Mass spectrometry detected three prominent fragments in addition to full-length III3 and anastellin (Fig. 8B). Based on the molecular masses, fragments 1 and 2 were identified as the N-terminal region of III3 (Fig. 8C); they differ by only five amino acids at the C-terminus of fragment 1. Fragment 3 was derived from the C-terminal region of III3. These fragments were further digested with trypsin to verify the locations of the thermolysin sensitive sites (data not shown). These results suggest that the region containing the C-, C′- and E-strands of III3 became thermolysin sensitive upon anastellin binding and are digested into short peptides, which do not show up as discrete fragments.
DISCUSSION
Fluorescence spectroscopy
The hydrophobic probe ANS is often used to study protein structure and function. In the present study, ANS binding to anastellin produced a strong fluorescence enhancement, and the Job plot indicated binding of two ANS per anastellin. III3 binding to anastellin reduced ANS fluorescence to half and the Job plot showed one ANS per III3/anastellin complex. These results suggest that one of the ANS binding sites on anastellin is blocked in the III3/anastellin complex. Another, but less likely, possibility is that III3 binding to anastellin blocks two ANS binding sites and creates a new site on either III3 or anastellin. In any case, III3 and ANS share a common binding site on anastellin, suggesting that anastellin binds to III3 through hydrophobic interactions.
In the present study, we used GFP-based FRET to monitor the interaction between III3 and anastellin. Unlike most FRET applications in which the two interacting proteins are individually tagged with donor and acceptor fluorophores, we fused acceptor (YFP) and donor (CFP) to the N and C termini of III3 to look for a possible conformational change upon anastellin binding. We had expected that if III3 opened up to bind to anastellin, the FRET signal would decrease upon binding. Instead we found that anastellin binding increased the FRET signal. The increased FRET signal could be due to the donor and acceptor being moved closer together, or to their being locked in a more favorable angular orientation. Either case may be explained by a weak interaction between the anastellin/III3 complex and the GFPs. The pseudo-average distance between the two GFP fluorophores changes approximately 0.6 nm by anastellin binding to III3, which is indicative of a weak interaction (for details on distance estimation from FRET efficiency, see our previous work (12)).
The stability of FNIII3
We previously reported that superFN aggregation, at saturating anastellin concentrations, was a first order reaction with a half time of ~600 s (9). Here we found that, at saturating concentrations, anastellin binding to III3 is a first order reaction with a half time of ~30 s. It is reasonable that the binding of anastellin to III3 is faster than the more complex aggregation reaction. We suggest that the rate of ~0.02 s−1 for anastellin binding to III3 is determined by the rate at which III3 spontaneously opens and exposes the anastellin binding site. This rate is much faster than the spontaneous unfolding rate of 0.0002–0.0005 s−1 for III10 and the third FNIII domain from tenascin estimated by chemical denaturation curves (30, 31). On the other hand, a faster unfolding rate of ~0.02 s−1 for III10 was estimated by atomic force microscopy (32). The reason for the difference between unfolding rates deduced from chemical and force measurements is not clear. The rate of opening of the anastellin binding site in III3 seems much faster than the usual spontaneous unfolding rates of other FNIII domains, and this is likely important for its being a key domain for binding anastellin.
NMR studies of FNIII domains have shown that the loops between the beta-strands are highly flexible (23, 24). All mutations in the loops, except the one between strands E and F, had only a minor effect on folding (22, 25, 33, 34). It has also been reported that the E-F loop can interact with the C-terminal residues to stabilize folding (10). The amino acid sequence identity of FNIII domains 1–14 of FN ranges from 14–38% when we aligned them (data not shown). Interestingly, the E–F loop is highly conserved among all FNIII domains, with an average ~65% identity over xLxPGxxY, where Y is the first residue of strand F. The D838G mutant, in which the first residue on the E–F loop was replaced with the more frequently occurring glycine (found in eight out of fourteen FNIII domains) showed slower anastellin binding kinetics. This mutation slightly enhanced the stability of III3 as shown in Figs. 2E and F, and thus probably slows down the unfolding rate, which determines anastellin binding (Table 1). These findings indicate that the E–F loop plays an important role in FNIII domain folding. It is possible that the disulfide bond between strands B and E also locks the E–F loop and stabilizes global folding as shown by the urea denaturation curves in Figs. 2C and D.
Anastellin binding to III3
We previously proposed two possible open configurations of III3: one where strands A and B lifted up with the B–C loop as a hinge and one where strands C′ and E flipped up with the E–F loop as a hinge (9). In the present study, we found that strands C′ and E became thermolysin sensitive upon anastellin binding. These data favor the second model. However, it is possible that the disulfide lock affects global structure rather than local structure, because the disulfide mutation between strands B and E seems to stabilize the entire domain, as suggested by urea denaturation, rather than by locking a particular beta-strand.
Our model involves a mechanism similar to domain swapping (35, 36) but is different in several aspects. Most known domain-swapped oligomers involve association of identical domains. III3 and anastellin have a similar secondary structure but they share only about 20% sequence identity. The III3/anastellin complex must therefore involve interfaces different from those in the folded III3. Since we were unable to map the precise anastellin binding site on III3 by mutagenesis, it is possible that anastellin binds non-specifically to exposed hydrophobic patches when FNIII domains transiently unfold, rather than binding to specific residues.
Our ultimate goal is to discover how FN molecules are bound to each other to make FN matrix fibrils. We suggest that a key to the assembly mechanism may be the unfolding of FNIII domains to interact with FN type I domains and/or unfolded FNIII domains on different molecules. In addition, the folding dynamics that our present study has indicated for III3 may have relevance for the many FNIII domains in other proteins.
Supplementary Material
Acknowledgments
This study was supported by National Institutes of Health Grant CA047056.
We would like to thank Dr. George Dubay (Department of Chemistry, Duke University) for technical advice on MALDI-TOF mass spectrometry.
Abbreviations
- FN
fibronectin
- III3
the third FN type III domain
- GFP
green fluorescent protein
- FRET
fluorescence resonance energy transfer
- ANS
8-anilino-1-naphthalene sulfonate
- MALDI-TOF
matrix assisted laser desorption ionization-time of flight
- CFP
cyan fluorescent protein
- YFP
yellow fluorescent protein
References
- 1.Hynes RO. Fibronectins. Springer-Verlag; New York: 1990. [Google Scholar]
- 2.Morla A, Zhang Z, Ruoslahti E. Superfibronectin is a functionally distinct form of fibronectin. Nature. 1994;367:193–196. doi: 10.1038/367193a0. [DOI] [PubMed] [Google Scholar]
- 3.Yi M, Ruoslahti E. A fibronectin fragment inhibits tumor growth, angiogenesis, and metastasis. Proc Natl Acad Sci U S A. 2001;98:620–624. doi: 10.1073/pnas.98.2.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yi M, Sakai T, Fassler R, Ruoslahti E. Antiangiogenic proteins require plasma fibronectin or vitronectin for in vivo activity. Proc Natl Acad Sci U S A. 2003;100:11435–11438. doi: 10.1073/pnas.1635112100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bork P, Doolittle RF. Proposed acquisition of an animal protein domain by bacteria. Proc Natl Acad Sci U S A. 1992;89:8990–8994. doi: 10.1073/pnas.89.19.8990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gough J, Chothia C. SUPERFAMILY: HMMs representing all proteins of known structure. SCOP sequence searches, alignments and genome assignments. Nucleic Acids Res. 2002;30:268–272. doi: 10.1093/nar/30.1.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson D, Madera M, Vogel C, Chothia C, Gough J. The SUPERFAMILY database in 2007: families and functions. Nucleic Acids Res. 2007;35:D308–313. doi: 10.1093/nar/gkl910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Briknarova K, Akerman ME, Hoyt DW, Ruoslahti E, Ely KR. Anastellin, an FN3 fragment with fibronectin polymerization activity, resembles amyloid fibril precursors. J Mol Biol. 2003;332:205–215. doi: 10.1016/s0022-2836(03)00890-8. [DOI] [PubMed] [Google Scholar]
- 9.Ohashi T, Erickson HP. Domain unfolding plays a role in superfibronectin formation. J Biol Chem. 2005;280:39143–39151. doi: 10.1074/jbc.M509082200. [DOI] [PubMed] [Google Scholar]
- 10.Hamill SJ, Meekhof AE, Clarke J. The effect of boundary selection on the stability and folding of the third fibronectin type III domain from human tenascin. Biochemistry. 1998;37:8071–8079. doi: 10.1021/bi9801659. [DOI] [PubMed] [Google Scholar]
- 11.Niimi T, Osawa M, Yamaji N, Yasunaga K, Sakashita H, Mase T, Tanaka A, Fujita S. NMR structure of human fibronectin EDA. J Biomol NMR. 2001;21:281–284. doi: 10.1023/a:1012947209393. [DOI] [PubMed] [Google Scholar]
- 12.Ohashi T, Galiacy SD, Briscoe G, Erickson HP. An experimental study of GFP-based FRET, with application to intrinsically unstructured proteins. Protein Sci. 2007;16:1429–1438. doi: 10.1110/ps.072845607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kornblihtt AR, Umezawa K, Vibe-Pedersen K, Baralle FE. Primary structure of human fibronectin: differential splicing may generate 10 polypeptides from a single gene. EMBO Journal. 1985;4:1755–1759. doi: 10.1002/j.1460-2075.1985.tb03847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perkins SJ. Protein volumes and hydration effects. The calculations of partial specific volumes, neutron scattering matchpoints and 280-nm absorption coefficients for proteins and glycoproteins from amino acid sequences. Eur J Biochem. 1986;157:169–180. doi: 10.1111/j.1432-1033.1986.tb09653.x. [DOI] [PubMed] [Google Scholar]
- 15.Gill SC, Von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 16.Ellman GL. Tissue sulfhydryl groups. Arch Biochem Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 17.Ohashi T, Erickson HP. The disulfide bonding pattern in ficolin multimers. J Biol Chem. 2004;279:6534–6539. doi: 10.1074/jbc.M310555200. [DOI] [PubMed] [Google Scholar]
- 18.Azzi A. The use of fluorescent probes for the study of membranes. Methods Enzymol. 1974;32:234–246. doi: 10.1016/0076-6879(74)32024-1. [DOI] [PubMed] [Google Scholar]
- 19.Huang CY. Determination of binding stoichiometry by the continuous variation method: the Job plot. Methods Enzymol. 1982;87:509–525. doi: 10.1016/s0076-6879(82)87029-8. [DOI] [PubMed] [Google Scholar]
- 20.Anderson DE, Gueiros-Filho FJ, Erickson HP. Assembly Dynamics of FtsZ Rings in Bacillus subtilis and Escherichia coli and Effects of FtsZ-Regulating Proteins. J Bacteriol. 2004;186:5775–5781. doi: 10.1128/JB.186.17.5775-5781.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cota E, Hamill SJ, Fowler SB, Clarke J. Two proteins with the same structure respond very differently to mutation: the role of plasticity in protein stability. J Mol Biol. 2000;302:713–725. doi: 10.1006/jmbi.2000.4053. [DOI] [PubMed] [Google Scholar]
- 22.Cota E, Steward A, Fowler SB, Clarke J. The Folding Nucleus of a Fibronectin Type III Domain is Composed of Core Residues of the Immunoglobulin-like Fold. J Mol Biol. 2001;305:1185–1194. doi: 10.1006/jmbi.2000.4378. [DOI] [PubMed] [Google Scholar]
- 23.Carr PA, Erickson HP, Palmer AG., III Backbone dynamics of homologous fibronectin type III cell adhesion domains from fibronectin and tenascin. Structure. 1997;5:949–959. doi: 10.1016/S0969-2126(97)00248-7. [DOI] [PubMed] [Google Scholar]
- 24.Akke M, Liu J, Cavanagh J, Erickson HP, Palmer AG., III Pervasive conformational fluctuations on microsecond time scales in a fibronectin type III domain. Nature Struct Biol. 1998;5:55–59. doi: 10.1038/nsb0198-55. [DOI] [PubMed] [Google Scholar]
- 25.Batori V, Koide A, Koide S. Exploring the potential of the monobody scaffold: effects of loop elongation on the stability of a fibronectin type III domain. Protein Eng. 2002;15:1015–1020. doi: 10.1093/protein/15.12.1015. [DOI] [PubMed] [Google Scholar]
- 26.Best RB, Rutherford TJ, Freund SM, Clarke J. Hydrophobic core fluidity of homologous protein domains: relation of side-chain dynamics to core composition and packing. Biochemistry. 2004;43:1145–1155. doi: 10.1021/bi035658e. [DOI] [PubMed] [Google Scholar]
- 27.Plaxco KW, Spitzfaden C, Campbell ID, Dobson CM. Rapid refolding of a proline-rich all-β-sheet fibronectin type III module. Proc Natl Acad Sci U S A. 1996;93:10703–10706. doi: 10.1073/pnas.93.20.10703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koide A, Jordan MR, Horner SR, Batori V, Koide S. Stabilization of a fibronectin type III domain by the removal of unfavorable electrostatic interactions on the protein surface. Biochemistry. 2001;40:10326–10333. doi: 10.1021/bi010916y. [DOI] [PubMed] [Google Scholar]
- 29.Billings KS, Best RB, Rutherford TJ, Clarke J. Crosstalk between the protein surface and hydrophobic core in a core-swapped fibronectin type III domain. J Mol Biol. 2008;375:560–571. doi: 10.1016/j.jmb.2007.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clarke J, Hamill SJ, Johnson CM. Folding and stability of a fibronectin type III domain of human tenascin. J Mol Biol. 1997;270:771–778. doi: 10.1006/jmbi.1997.1147. [DOI] [PubMed] [Google Scholar]
- 31.Cota E, Clarke J. Folding of beta-sandwich proteins: Three-state transition of a fibronectin type III module. Protein Sci. 2000;9:112–120. doi: 10.1110/ps.9.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oberhauser AF, Badilla-Fernandez C, Carrion-Vazquez M, Fernandez JM. The mechanical hierarchies of fibronectin observed with single-molecule AFM. J Mol Biol. 2002;319:433–447. doi: 10.1016/S0022-2836(02)00306-6. [DOI] [PubMed] [Google Scholar]
- 33.Dutta S, Batori V, Koide A, Koide S. High-affinity fragment complementation of a fibronectin type III domain and its application to stability enhancement. Protein Sci. 2005;14:2838–2848. doi: 10.1110/ps.051603005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siggers K, Soto C, Palmer AG., 3rd Conformational dynamics in loop swap mutants of homologous fibronectin type III domains. Biophys J. 2007;93:2447–2456. doi: 10.1529/biophysj.106.100578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, Eisenberg D. 3D domain swapping: as domains continue to swap. Protein Sci. 2002;11:1285–1299. doi: 10.1110/ps.0201402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ding F, Prutzman KC, Campbell SL, Dokholyan NV. Topological determinants of protein domain swapping. Structure. 2006;14:5–14. doi: 10.1016/j.str.2005.09.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.