Abstract
Mechanisms that couple protein turnover to cell cycle progression are critical for coordinating the events of cell duplication and division. Despite the importance of cell cycle-regulated proteolysis, however, technologies to measure this phenomenon are limited, and typically involve monitoring cells that are released back into the cell cycle after synchronization. We describe here the use of laser scanning cytometry (LSC), a technical merger between fluorescence microscopy and flow cytometry, to determine cell cycle-dependent changes in protein stability in unperturbed, asynchronous, cultures of mammalian cells. In this method, the ability of the LSC to accurately measure whole cell fluorescence is used, together with RNA fluorescence in situ hybridization and immunofluorescence, to relate abundance of a particular RNA and protein in a cell to its point at the cell cycle. Parallel monitoring of RNA and protein levels is used, together with protein synthesis inhibitors, to reveal cell cycle-specific changes in protein turnover. We demonstrate the viability of this method by analyzing the proteolysis of two prominent human oncoproteins, Myc and Cyclin E, and argue that this LSC-based approach offers several practical advantages over traditional cell synchronization methods.
Keywords: Laser scanning cytometry, cell cycle, proteolysis, Myc, Cyclin E
INTRODUCTION
Ordered progression through the eukaryotic cell cycle depends on mechanisms that tie the abundance and activity of critical regulatory molecules to the events of DNA replication and cellular division. One of the most pervasive of these mechanisms is ubiquitin (Ub)-mediated proteolysis, a process in which covalent attachment of Ub to target proteins signals their destruction by the 26S proteasome. By destroying proteins at specific points in the cell cycle, the Ub–proteasome system (UPS) provides directionality to the events of cell duplication, and insures that each phase of the cycle occurs after the previous one is completed [1].
Despite the biological importance of cell cycle-regulated proteolysis, techniques to study this phenomenon are limited. One of the most popular approaches is to arrest cells at specific cell cycle stages, either by chemical or growth-factor blockade, and to then release cells back into the cycle and monitor them as they move in synchrony through subsequent cell cycle transitions. These synchronization methods, however, involve significant disruption to normal cellular physiology, and—because of the perturbations involved—can influence the apparent behavior of molecules with respect to the cell cycle [e.g., [2–4]]. An alternative strategy, centrifugal elutriation [5], separates cells based on their size, which increases linearly during the cell cycle. In this way, relatively pure fractions of cells can be obtained that are in either the G1, S, or G2/M cell cycle phases. Elutriation has the advantage of not requiring disruption to the cell cycle to provide synchronicity, but its practical application is restricted because it is best suited to non-adherent cells and requires relatively large volumes of cell cultures.
We are interested in the mechanisms controlling Ub-mediated proteolysis of the oncoprotein Myc [6]. There are conflicting reports as to whether Myc levels and stability are influenced by the cell cycle. Some studies demonstrated that Myc synthesis and stability are not cell cycle regulated [e.g., [7]], whereas others concluded that Myc RNA and protein levels peak at the G1/S transition [e.g., [8, 9]], or that Myc is stabilized during mitosis [10]. Moreover, phosphorylation events within Myc—at residues threonine 58 (T58) and serine 62 (S62)—that control its ubiquitylation by the SCFFbw7 Ub-ligase [11] have been reported to peak during late G2/M-phase [e.g., [12]], lending support to the idea that Myc destruction is cell cycle-regulated [13]. Because true cell cycle-dependent changes in Myc levels or stability could have a profound impact on the mechanism through which Myc promotes cell growth and proliferation, it is important that the issue of whether or not Myc proteolysis is cell cycle-regulated be resolved.
We reasoned that some of the contradictory findings on the relationship between Myc and the cell cycle may have resulted from the different techniques used in the various studies. In some cases, centrifugal elutriation was employed to monitor Myc levels and stability [e.g., [7]], whereas other studies used either nocodazole or double-thymidine block and release strategies. We sought to develop an additional protocol that would allow us to take a comprehensive look at the influence of the cell cycle on Myc synthesis, location, and stability in unperturbed cultures of cells. The recent development of laser scanning cytometry (LSC) created an opportunity to develop this protocol. The LSC, which is a technical merger between fluorescence microscopy and flow cytometry [14], allows for whole cell quantification of fluorophores targeted to DNA, RNA, or protein. By accurately quantifying total cellular DNA content (using fluorescent dyes such as Hoechst 33342), the LSC can determine the cell cycle state of an individual cell, and then relate this state to some other fluorescent parameter, such as the signal from a fluorescently-labeled antibody. In this way, levels of a particular RNA or protein can be measured in individual cells and expressed relative to the particular cell cycle stage. By compiling data from thousands of cells in this way, highly quantitative cell cycle analysis can be performed without disruption to normal cellular physiology. Importantly, by comparing RNA and protein levels for a particular gene product, and by monitoring protein levels after transient inhibition of protein synthesis, cell cycle dependent changes in protein stability can be inferred. Here we describe how LSC-based assays can be used to monitor cell cycle-dependent changes in protein stability in small numbers of unperturbed cells growing on a coverslip. We demonstrate the utility of this approach by analyzing the cell cycle expression profile of Myc and comparing it with that of another prominent human oncoprotein, Cyclin E.
MATERIALS AND METHODS
Cell culture and immunofluorescence
Human U2OS and HeLa cells were grown in DMEM supplemented with antibiotics and 10% fetal calf serum. For analysis, cells were plated onto glass coverslips, grown at 37°C for 24 hours, and then fixed, either using methanol or paraformaldehyde [15]. Immunofluorescence was performed as described [15] using the following antibodies: (i) α-Myc (N-262, Santa Cruz), (ii) α-Cyclin E (HE12, Santa Cruz), (iii) α-Actin (AC-15, Sigma), and (iv) α-phospho T58 Myc (9401S, Cell Signaling). Immune complexes were detected using FITC-tagged secondary anti- mouse or anti-rabbit antibodies, as appropriate. DNA was stained with Hoechst 33342 (2 μg/ml). For Nocodazole arrest, cells were seeded for 24 hours, and then treated with Nocodazole (100 ng/ml) for 16 hrs. Nocodazole was removed and cells were released from G2/M block for varying timepoints. For double-thymidine (DT) arrest, cells were treated with 2.5 mM thymidine for 14 hrs, washed and released for 12 hrs, and then re-arrested in 2.5 mM thymidine for 14 hrs before FC analysis.
CENTRIFUGAL ELUTRIATION
Centrifugal elutriation was performed as described [16]. Briefly, actively growing U2OS cells were elutriated using the Beckman JE-6B rotor, at a rotor speed of 1500 rpm and rotor temperature of 20°C. Cells were eluted in DMEM+1% FBS, by applying an increasing medium flow rate ranging from 40 ml/min – 150 ml/min. Approximately 10 fractions of 250 ml were collected. Cells were rapidly harvested by centrifugation and either fixed in methanol and analyzed by flow cytometry or lysate prepared for western blotting.
RNA FISH
RNA-FISH analysis for detection of nuclear RNAs was performed on triton-extracted, fixed, U2OS cells as described [15]. Four anti-sense probes were used for Myc: Myc1–TAGTCGAGGTCATAGTTCCTG; Myc2–TCGAGGAGAGCAGAGAATCCG; Myc3–TTCAACTGTTCTCGTCGTTTC; Myc4–TGTTCGCCTCTTGACATTCTC. Two antisense probes were used for actin: Act1–ATAGCACAGCCTGGATAGCAA; Act2– TGGAAGCAGCCGTCGCCATCTCTTGCTCGA. In each case, the corresponding sense probes were used as a control. Pooled probes were end-labeled with Dig-11-ddUTP using the Digoxigenin End-labeling kit (Roche). FISH was performed as described [15].
LSC analysis
All experiments were performed using the LSC-iCys system (Compucyte, MA) attached to an Olympus IX-71 microscope. Samples with multiple fluorophores were compensated by measuring the spectral bleed into other channels using controls containing each individual fluorophore, and subtracting the corresponding non-specific leakage. Thresholds for measuring each fluorophore were set at signal intensities where there was negligible background signal. The fluorescence units obtained thereafter were used to compare the expression patterns within different cell cycle populations. Approximately 2000–3000 cells were scanned in each individual analysis.
Cell cycle profiles were gated based on DNA content into G1, S, and G2/M populations. For sub-cellular visualization experiments, galleries of cells in different cell cycle phases were created; localization of the signal was compared by merging the pictures obtained from the Green, Blue, and Scatter channels. For quantification, the mean signals of the respective proteins were measured within the given cell cycle gates. Fluorescence values were normalized either to mean signal in the G1 phase, or reported as a ratio of green/blue fluorescence to quantify relative levels of protein or RNA in different cell cycle stages as a function of DNA.
RNA interference and protein synthesis inhibition
Myc and Fbw7 knockdown was performed using RNA interference. Pools of siGenome RNA against Myc, Fbw7, and Luciferase (control) were obtained from Dharmacon, and transiently transfected into U2OS cells using the Oligofectamine reagent (Invitrogen). Transfected cells were analyzed by LSC and Western blotting. For experiments involving cyclohexamide, HeLa cells, growing on coverslips, were treated with 50 μg/ml cyclohexamide, fixed at the indicated time points, and Myc and actin levels at each point quantified by immunofluorescence and LSC. The corrected fluorescence value in each of the cell cycle stages at the”0” time point was arbitrarily fixed as 100% and values of corrected fluorescence from every subsequent time point were represented as relative percentage of the”0” (100%) time point. The effect of USP28 was studied by transfecting shRNA pools against USP28 into U2OS cells. Knockdown of USP28 expression was confirmed using quantitative RT-QPCR, and found to be ~50% (not shown).
RESULTS AND DISCUSSION
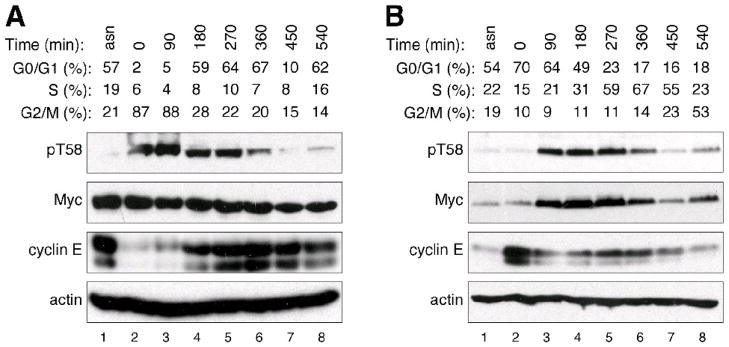
We began our study of the relationship between Myc and the cell cycle by testing the notion that different methods of cell synchronization can lead to different conclusions regarding the behavior of Myc. We compared two common methods of cell synchronization—nocodazole block, which arrests cells in the G2/M stage of the cell cycle—and double-thymidine (DT) block, which produces a G1/S arrest. We used Western blot to monitor steady-state Myc levels during subsequent release of cells back into the cycle; we also monitored phosphorylation of Myc at residue T58, which is required for its destruction via the SCFFbw7 Ub-ligase, and which has been reported to be cell cycle regulated [12]. As hypothesized, these two approaches gave different results (Figure 1 and Supplemental Figure 1). Nocodazole arrest gave the impression that, although Myc levels are constant throughout the cell cycle, T58 phosphorylation peaks in mitosis (Figure 1A). DT-block, on the other hand, gave the impression that total Myc levels peak during release from G1/S, and that T58 phosphorylation does not appreciably differ from total Myc levels (Figure 1B). These differences were not observed when we examined another SCFFbw7 substrate, Cyclin E—both nocodazole and DT block showed that Cyclin E levels were high at G1/S (Figure 1A, lanes 4–6; Figure 1B, lane 2), and low at G2/M (Figure 1A, lanes 2–3; Figure 1B, lanes 6–8). We conclude that apparent cell cycle-related changes in Myc levels and phosphorylation can be profoundly influenced by chemical synchronization methods. Importantly, because cyclin E behaved consistently after both nocodazole and DT blockade, we further conclude that synchronization approaches can influence the status of different proteins in different ways.
Figure 1. Myc levels and phosphorylation are influenced by cell-synchronization.
(A) U20S cells were synchronized by treatment with nocodazole to arrest at the G2/M transition. Cells were released from arrest, and samples taken at the indicated time-points for analysis of cell cycle by FC, and protein levels by western blot. The numbers for G0/G1, S, and G2/M percentages were derived from FC. ‘pT58’ refers to an antibody that recognizes the phosphorylated form of residue threonine 58 within Myc. (B) As in (A), except that cells were subjected to double-thymidine (G1/S) block and release. FC profiles are presented in Supplemental Figure 1. Detailed methods are presented in Supplemental Information.
Given the discrepancy between the two synchronization approaches, we sought to develop an LSC-based protocol that would allow us to measure the influence of the cell cycle on Myc levels and stability in unperturbed, asynchronous, cultures of cells growing on a glass coverslip. As an initial validation, we found that cell cycle profiles determined by LSC were comparable to those determined by traditional flow cytometry (FC; Supplemental Figure 2A–2B); we also found that we could detect a robust signal for Myc using immunofluorescence (IF), and that Myc was predominantly nuclear through all stages of the cell cycle (Supplemental Figure 2C). We therefore chose to use the LSC determine whether steady-state Myc RNA and protein levels, or stability, are influenced by the cell cycle. By triangulation of all three parameters, we hoped to make an informed conclusion regarding cell cycle control of Myc.
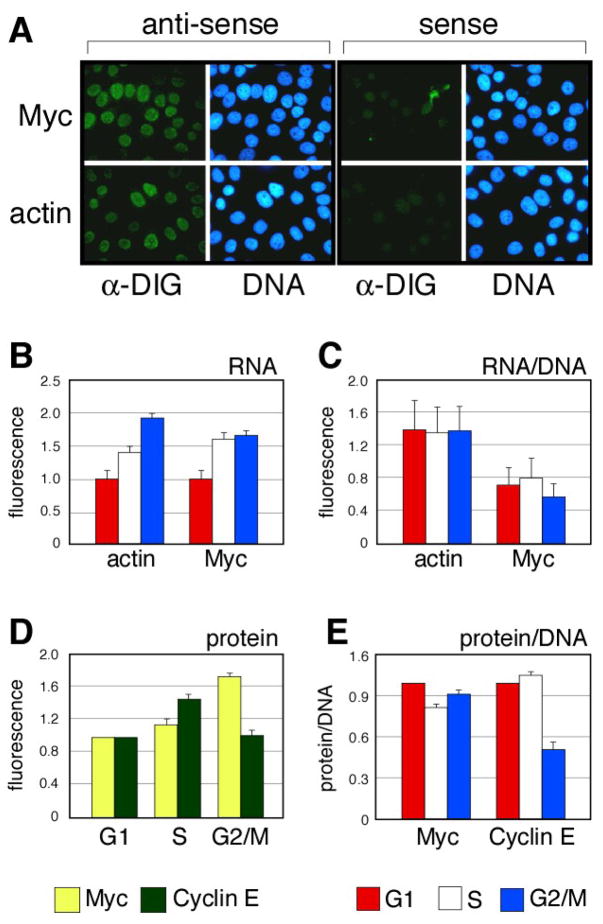
We combined LSC with RNA-fluorescence in situ hybridization (RNA-FISH) to compare the levels of Myc and (as a control) actin RNAs in human U2OS cells (Figure 2). For these studies, we used a detergent pre-extraction protocol to remove most of the cytosolic transcripts [15], allowing us to enrich for the population of newly-synthesized RNA molecules. Results of this analysis, performed in human U2OS cells, are shown in Figure 2. Using LSC/RNA-FISH, we were able to detect robust levels of Myc and actin RNAs using labeled anti-sense, but not the corresponding sense, probes (Figure 2A). Quantification of these hybridizations (Figure 2B) revealed that the absolute levels of both RNAs increase as cells passage through the cell cycle, being lowest in G1 cells, and highest in cells from the G2/M population. This apparent increase, however, appears to be a result of the increase in cell mass that occurs during the cell cycle, because, when normalized for either cell volume (data not shown) or DNA content (Figure 2C), the relative levels of newly-synthesized Myc and actin RNAs are equivalent in each of the cell cycle populations. We conclude, therefore, that the levels of Myc RNA do not fluctuate during the cell cycle. Moreover, because Myc transcripts are extremely unstable, with a half-life of ~25 minutes [17], we infer that the relatively similar levels of nuclear Myc RNA in G1, S, and G2/M phase cells reflects constitutive transcription from the c-Myc gene.
Figure 2. Parallel analysis of Myc RNA and protein levels throughout the cell cycle.
(A) Visualization of nuclear Myc and actin transcripts in U2OS cells by RNA FISH. Anti-sense probe cocktails detect bona-fide transcripts; sense probe cocktails reveal background. (B) Quantification of absolute FISH signals. Fluorescence intensities were quantified by LSC in cells binned into G1, S, or G2/M populations, and expressed relative to the signal from G1 phase cells. (C) Quantification of relative FISH signals. Fluorescence intensities were quantified by LSC in cells binned into G1, S, or G2/M populations, normalized to the signal for DNA content in those cells, and expressed relative to the signal from G1 phase cells (n=2, mean +/− S.D.). Arbitrary units of fluorescence are used. (D) Quantification of absolute levels of Myc and cyclin E protein. Fluorescence intensities were quantified by LSC in cells binned into G1, S, or G2/M populations, and expressed relative to the signal from G1 phase cells. (E) Quantification of relative protein levels. Fluorescence intensities were quantified by LSC in cells binned into G1, S, or G2/M populations, normalized to the signal for DNA content in those cells, and expressed relative to the signal from G1 phase cells (n=4, mean +/− S.D.). Detailed methods are presented in Supplemental Information.
We next examined endogenous Myc protein in the different sub-populations of cells. We compared Myc with cyclin E, because both proteins are targets for the SCFFbw7 Ub-ligase [11], and because cyclin E is tagged for destruction soon after cells enter S-phase [e.g., [18]]. In these assays (Figure 2D–E), Myc protein levels mirrored those of Myc RNA and—after normalization to DNA content—were unaffected by cell cycle status (Figure 2E). Cyclin E, in contrast, displayed pronounced cell cycle dependency, with both its relative (Figure 2D) and absolute (Figure 2E) levels being lowest in G2/M cells, as expected. We validated this pattern of expression independently, by comparing the bivariate distribution of DNA content versus Myc or Cyclin E immunofluorescence as described by Gong et al., [2] and presented in Supplemental Figure 2D. Comparison of these results with those from analysis of parallel U2OS cell cultures by centrifugal elutriation (Supplemental Figure 3) showed remarkable consistency between the LSC and elutriation approaches, with Myc levels relatively fixed throughout the cell cycle, and Cyclin E being lowest in G2/M cells. From these data, we conclude that Myc stability is likely to be unchanged throughout the cell cycle, and that LSC-based methods combining RNA-FISH and IF provide a viable way to reveal cell cycle-dependent changes in protein levels and stability.
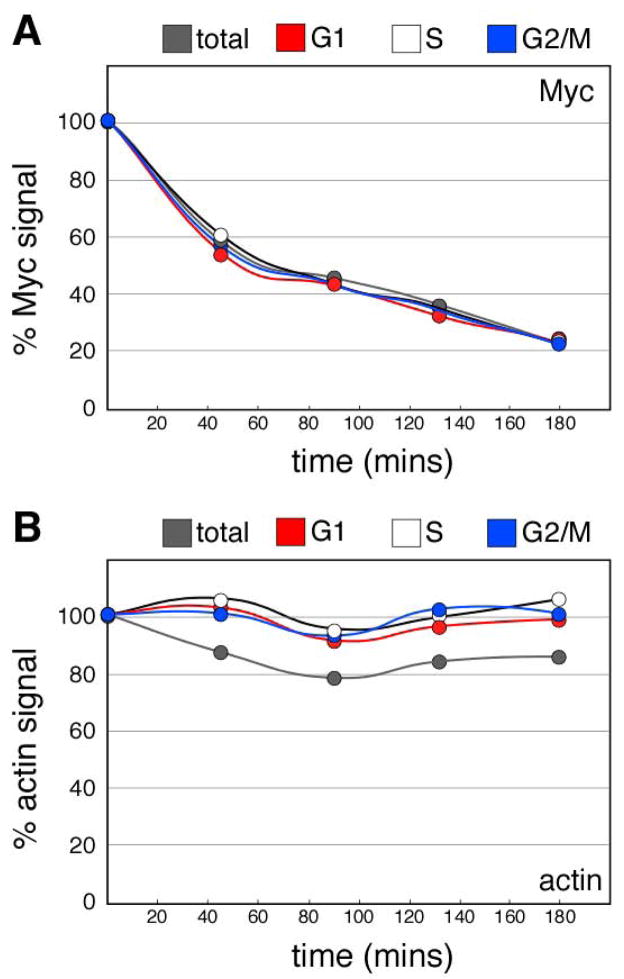
To more directly probe for cell cycle-dependent changes in Myc turnover, we transiently inhibited protein synthesis with cyclohexamide and used the LSC to monitor the levels of Myc protein in each cell subpopulation as a function of time. This approach—which is analogous to the ‘cyclohexamide chase’ protocol that is often used to monitor Myc stability [19–21]—allowed us to compare the rates with which Myc protein disappeared in each cell-cycle subpopulation of cells. Under these conditions, Myc disappeared quickly following addition of cyclohexamide, with an apparent half-life of ~50 minutes (Figure 3A). Actin, in contrast, was relatively stable during the period of the experiment (Figure 3B). Importantly, when we examined the rate of decay of Myc in the distinct cell subpopulations, both the apparent half-life of Myc and its decay kinetics were identical in G1, S, and G2/M phase cells. These results support early studies of Myc proteolysis [7, 17, 22] which showed that the rate of Myc destruction is constant throughout the cell cycle, and are consistent with the conclusions made from our parallel analysis of Myc RNA and protein levels.
Figure 3. The metabolic stability of Myc is constant throughout the cell cycle.
HeLa cells were treated with cyclohexamide for the indicated time points, fixed, and Myc (A) and actin (B) levels for each cell cycle subpopulation measured by immunofluorescence and LSC. In each case, the relative signals are normalized to DNA content and presented as a percentage of the zero-time point samples. Detailed methods are presented in Supplemental Information.
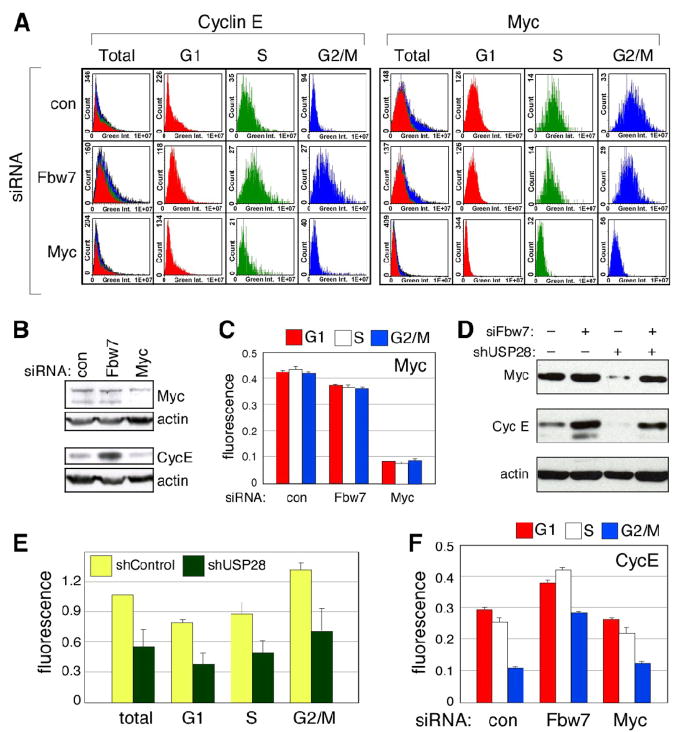
One of the most practical advantages of the LSC-based approach is that it can be performed on a small number of cells (typically less than 5,000), allowing investigators to quickly and economically examine the effects of agents such as drugs or siRNAs on cell cycle-dependent proteolysis; these type of experiments are difficult or prohibitively expensive with the scale needed for centrifugal elutriation. To illustrate this point, we compared the effects of siRNA-mediated knockdown of Fbw7 on two of its substrate proteins, Myc and Cyclin E (Figure 4). For Myc, we also examined the effects of knock-down of USP28, a Ub-specific protease that antagonizes Fbw7-dependent Myc destruction [23]. Knockdown of Fbw7 had little if any effect on levels of Myc protein detected by either LSC (Figure 4A) or western blot (Figure 4B), and no influence on the cell cycle distribution of Myc (Figure 4C). This lack of an effect is likely due to the reported antagonism between Fbw7 and USP28, as knockdown of USP28 reduced steady-state Myc levels (Figure 4D) equally in all stages of the cell cycle (Figure 4E), and this effect was reversed by simultaneous knockdown of Fbw7 (Figure 4D). We thus conclude that the Fbw7–USP28 pathway is unlikely to act in a cell cycle-dependent way on Myc. In contrast, however, Fbw7 did appear to act in a cell cycle-dependent manner upon Cyclin E. Knockdown of Fbw7 increased Cyclin E levels in total cell populations (Figure 4B, E), with the most pronounced effects observed in the S and G2/M populations of cell, as expected. This result demonstrates that the LSC-based approach can be used to address physiologically important questions relating to the role of specific Ub-ligases in cell cycle-dependent proteolysis.
Figure 4. Differential regulation of Myc and Cyclin E by Fbw7 during the cell cycle.
(A) Effects of siRNA-mediated knockdown of Fbw7 on Myc and Cyclin E. U2OS cells were transfected with the indicated siRNAs [control (=luciferase); Fbw7, and Myc], and Myc and Cyclin E levels determined in each cell cycle subpopulation of cells. Protein expression histograms show that Fbw7 knockdown substantially increases Cyclin E levels in S and G2 phase (rightward shift), whereas it has no significant effect on Myc profiles. (B) Analysis of total steady-state levels of Myc and Cyclin E following Fbw7 knockdown. Western blot analysis of cells analyzed in (A). (C) Quantification of relative Myc protein levels. Fluorescence intensities for Myc were quantified by LSC in cells binned into G1, S, or G2/M populations, normalized to the signal for DNA content in those cells, and expressed relative to the signal from G1 phase. (D) Combined effect of USP28 and Fbw7 knockdown. U2OS cells stably expressing an shRNA against USP28 (or control luciferase shRNA) were transfected with siRNA against Fbw7 (or control luciferase siRNA). Cells were harvested and Myc, cyclin E, and actin levels detected by WB. (E) Effect of USP28 on Myc levels during the cell cycle. Immunofluorescence of endogenous Myc was performed in USOS cells expressing either cells stably expressing an shRNA against USP28, or control luciferase shRNA. Cells were counterstained with Hoechst 33342 and analyzed by LSC. (F) Cell cycle-dependent effects of Fbw7 on Cyclin E. Experiment was performed as in (C), except that Cyclin E was detected by IF. Detailed methods are presented in Supplemental Information.
CONCLUSIONS
The LSC-based approach that we describe here is a simple and reliable way to determine how turnover of a specific protein is influenced by the cell cycle. Our comparison with chemical synchronization methods, and the different results that we obtained with the different methods, illustrates the importance of being able to probe cell cycle changes in protein levels in asynchronous cultures of cells. The LSC-based method can obtain highly quantitative data from a much smaller number of cells fixed on a coverslip. The reduced scale of this protocol makes it particularly suited to studying the effects of agents such as siRNAs on cell cycle-related protein turnover (as we did with cyclin E); an approach that is not practical with larger cultures. Finally, the LSC-based method also offers the advantage of being able to simultaneously monitor intracellular protein distribution, offering the potential to expose relationships between the cell cycle, protein localization, and stability.
Acknowledgments
We thank L. Carey, B. Futcher. E. Luther, P. Moody, K. Siddiqui, and A.-M. Torres for technical help, and G. Collins, J. Kurland, and S. Salghetti for comments on the manuscript. W.P.T. was a Leukemia and Lymphoma Society of America Scholar. This work was supported by the CSHL Cancer Center Support Grant CA45508, The Irving Hansen Memorial Foundation, and by US Public Health Service grant CA-13106 from the NCI.
ABBREVIATIONS
- DT
double-thymidine
- FC
flow cytometry
- IF
immunofluorescence
- LSC
laser scanning cytometer
- RNA–FISH
RNA-fluorescence in situ hybridization
- Ub
ubiquitin
- UPS
ubiquitin–proteasome system
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tansey WP. How cells use proteolysis to control their growth. Mol Med. 1999;5:773–782. [PMC free article] [PubMed] [Google Scholar]
- 2.Gong J, Traganos F, Darzynkiewicz Z. Growth imbalance and altered expression of cyclins B1, A, E, and D3 in MOLT-4 cells synchronized in the cell cycle by inhibitors of DNA replication. Cell Growth Differ. 1995;6:1485–1493. [PubMed] [Google Scholar]
- 3.Davis PK, Ho A, Dowdy SF. Biological methods for cell-cycle synchronization of mammalian cells. Biotechniques. 2001;30:1322–1326. 1328, 1330–1321. doi: 10.2144/01306rv01. [DOI] [PubMed] [Google Scholar]
- 4.Cooper S. Is whole-culture synchronization biology’s ‘perpetual-motion machine’? Trends Biotechnol. 2004;22:266–269. doi: 10.1016/j.tibtech.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Zickert P, Wejde J, Skog S, Zetterberg A, Larsson O. Growth-regulatory properties of G1 cells synchronized by centrifugal elutriation. Exp Cell Res. 1993;207:115–121. doi: 10.1006/excr.1993.1169. [DOI] [PubMed] [Google Scholar]
- 6.Salghetti SE, Kim SY, Tansey WP. Destruction of Myc by ubiquitin-mediated proteolysis: cancer-associated and transforming mutations stabilize Myc. Embo J. 1999;18:717–726. doi: 10.1093/emboj/18.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hann SR, Thompson CB, Eisenman RN. c-myc oncogene protein synthesis is independent of the cell cycle in human and avian cells. Nature. 1985;314:366–369. doi: 10.1038/314366a0. [DOI] [PubMed] [Google Scholar]
- 8.Caldani C, Far DF, Birtwisle-Peyrottes I, Ettore F, Rostagno P. Cell cycle expression of p53 protein, c-Myc gene product and tyrosine-phosphorylation level determined by image analysis in human breast cancer cells. Anal Quant Cytol Histol. 1996;18:233–240. [PubMed] [Google Scholar]
- 9.Morgan CJ, Pledger WJ. Cell cycle dependent growth factor regulation of gene expression. J Cell Physiol. 1989;141:535–542. doi: 10.1002/jcp.1041410312. [DOI] [PubMed] [Google Scholar]
- 10.Gregory PD, Schmid A, Zavari M, Munsterkotter M, Horz W. Chromatin remodelling at the PHO8 promoter requires SWI-SNF and SAGA at a step subsequent to activator binding. Embo J. 1999;18:6407–6414. doi: 10.1093/emboj/18.22.6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Welcker M, Orian A, Grim JA, Eisenman RN, Clurman BE. A nucleolar isoform of the Fbw7 ubiquitin ligase regulates c-Myc and cell size. Curr Biol. 2004;14:1852–1857. doi: 10.1016/j.cub.2004.09.083. [DOI] [PubMed] [Google Scholar]
- 12.Seth A, Alvarez E, Gupta S, Davis RJ. A phosphorylation site located in the NH2-terminal domain of c-Myc increases transactivation of gene expression. J Biol Chem. 1991;266:23521–23524. [PubMed] [Google Scholar]
- 13.Dominguez-Sola D, Dalla-Favera R. PINning down the c-Myc oncoprotein. Nat Cell Biol. 2004;6:288–289. doi: 10.1038/ncb0404-288. [DOI] [PubMed] [Google Scholar]
- 14.Pozarowski P, Holden E, Darzynkiewicz Z. Laser scanning cytometry: principles and applications. Methods Mol Biol. 2006;319:165–192. doi: 10.1007/978-1-59259-993-6_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spector DL, Goldman RD, Leinwand LA. Cells: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1998. [Google Scholar]
- 16.Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rabbitts PH, Watson JV, Lamond A, Forster A, Stinson MA, Evan G, Fischer W, Atherton E, Sheppard R, Rabbitts TH. Metabolism of c-myc gene products: c-myc mRNA and protein expression in the cell cycle. Embo J. 1985;4:2009–2015. doi: 10.1002/j.1460-2075.1985.tb03885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ekholm-Reed S, Spruck CH, Sangfelt O, van Drogen F, Mueller-Holzner E, Widschwendter M, Zetterberg A, Reed SI. Mutation of hCDC4 leads to cell cycle deregulation of cyclin E in cancer. Cancer Res. 2004;64:795–800. doi: 10.1158/0008-5472.can-03-3417. [DOI] [PubMed] [Google Scholar]
- 19.Alarcon-Vargas D, Tansey WP, Ronai Z. Regulation of c-myc stability by selective stress conditions and by MEKK1 requires aa 127–189 of c-myc. Oncogene. 2002;21:4384–4391. doi: 10.1038/sj.onc.1205543. [DOI] [PubMed] [Google Scholar]
- 20.Tworkowski KA, Salghetti SE, Tansey WP. Stable and unstable pools of Myc protein exist in human cells. Oncogene. 2002;21:8515–8520. doi: 10.1038/sj.onc.1205976. [DOI] [PubMed] [Google Scholar]
- 21.Gregory MA, Hann SR. c-Myc proteolysis by the ubiquitin-proteasome pathway: stabilization of c-Myc in Burkitt’s lymphoma cells. Mol Cell Biol. 2000;20:2423–2435. doi: 10.1128/mcb.20.7.2423-2435.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson CB, Challoner PB, Neiman PE, Groudine M. Levels of c-myc oncogene mRNA are invariant throughout the cell cycle. Nature. 1985;314:363–366. doi: 10.1038/314363a0. [DOI] [PubMed] [Google Scholar]
- 23.Popov N, Wanzel M, Madiredjo M, Zhang D, Beijersbergen R, Bernards R, Moll R, Elledge SJ, Eilers M. The ubiquitin-specific protease USP28 is required for MYC stability. Nat Cell Biol. 2007;9:765–774. doi: 10.1038/ncb1601. [DOI] [PubMed] [Google Scholar]