Abstract
A girl with a prenatal 46,XY karyotype was born with a completely normal female phenotype, including uterus and histologically normal ovaries. In mice with a similar phenotype, the ablation of M33, an ortholog of Drosophila Polycomb, causes male-to-female sex reversal. The analysis of the human homolog of M33, Chromobox homolog 2 (CBX2), in this girl revealed loss-of-function mutations that allowed us, by placing CBX2 upstream of SRY, to add an additional component to the still incomplete cascade of human sex development.
Main Text
Factors involved in sexual development were first deduced from animal models and from case studies of people whose genetic or gonadal sex does not match the phenotypic sex. Despite considerable progress in understanding the genetic basis of human sexual development, a specific molecular diagnosis is identified in only 20% of disorders of sex development (DSD)1 cases. Although SRY, the mammalian sex-determining gene on the Y chromosome (MIM 480000), is considered to be a master switch for testicular differentiation, its presence is insufficient to induce testicular development.2 Several other components of the male pathway have been identified, including Sox9,3 Dmrt1,4 Fgf9,5 Dhh,6 Sox8,7 and Dax1.8 WNT4 appears to be essential for sexual differentiation in women.9 An additional player was identified when in mice targeted ablation of M33, an ortholog of Drosophila Polycomb,10,11 caused male-to-female sex reversal.12 Apart from sterility, 50%–75% of M33 knockout Sry-positive mice were phenotypically perfect females (ovaries with follicles, uterus, and normal external genitalia). We identified the human counterpart of this murine model in a girl, born at term (weight: 3.55 kg, length 54 cm) after an uneventful pregnancy with no sign of intrauterine growth retardation, who presented at birth with normal female external genitalia, despite prenatally determined 46,XY karyotype. Abdominal ultrasound showed normal uterus. She is at present the only child of the family.
The 46,XY karyotype, performed prenatally because of the maternal age, was confirmed postnatally. The karyotype in PBL was analyzed twice, as routinely done in a certified medical genetic laboratory in at least 100 mitosis. It is therefore unlikely that a chimerism is present.
Anti-Muellerian hormone was undetectable and basal testosterone was low (<0.03 ng/ml at age 2 years) and failed to increase after hCG stimulation (<0.03 ng/ml at age 2 years). FSH levels were repeatedly elevated (age 1 1/2 years, 60.9 IU/l; age 3 1/2 years, 53.4 IU/l; age 4 10/12 years, 31.1 IU/l), whereas LH levels stayed always normal (age 3 5/12 years, 1.8 IU/l; age 4 10/12 years, 0.9 IU/l). No clinical signs of adrenal insufficiency were present and basal plasma cortisol as well as electrolytes were repeatedly normal (168, 121 ng/ml).
Laparascopy was performed at age 4 1/2 years to evaluate and potentially remove the gonads. Surprisingly, both gonads appeared macroscopically as normal ovaries and were both biopsied. Histology revealed normal ovarian tissues with primordial follicles (Figure 1A). Vaginoscopy showed a normal 7.5 cm vagina and a normally appearing cervix. The study conformed to the guidelines of the institutional review board.
Figure 1.
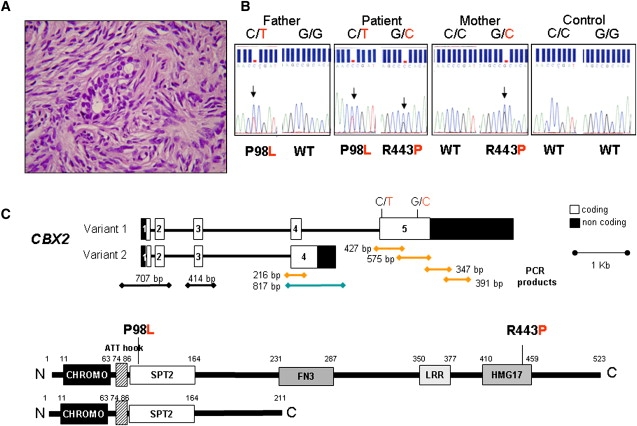
Ovarian Phenotype, Genotype, and Genotyping Strategy
(A) Hystology of bioptic ovarian fragment (0.3 × 0.2 × 0.15 cm) showing ovarian cortex stromal tissue and two primordial follicles.
(B) DNA sequence chromatograms obtained by direct sequencing of PCR products showing the presence of the heterozygote c.C293T and c.G1370C substitution in exon 5, not present in normal individuals (control, representative example out of 320 alleles).
(C) General structure of the CBX2 gene and protein variants and location of the P98L and R443P mutations. Exons are represented by boxes. The PCR products and their lengths are also depicted: black lines, products common to both variants; blue line, product of variant 2 only; yellow lines, products of variant 1 only. The protein domains were identified with the SMART server. Abbreviations: CHROMO, CHRomatin Organization mOdifier; ATT-hook, small DNA binding motif originally described in the high-mobility group (HMG) nonhistone chromosomal protein HMG-I; SPT2, S. cerevisiae chromatin protein involved in transcriptional regulation; FN3, fibronectin 3-repeat region; LRR_R1, leucin-repeat region involved in protein-protein interaction; HMG17, high mobility group nonhistone chromatin component.
After obtaining informed consent, genomic DNA was extracted from peripheral blood leukocytes via the DNA blood and cell culture kit (QIAGEN) and used to perform polymerase chain reaction (PCR) exonic amplifications. SRY amplification was conducted as described.13 The two variants of CBX2 (MIM 602770), the human homolog of M33 (isoforms 1 and 2, NM_005189.1 and NM_032647.2), are located on chromosome 17q25 and share the first 3 exons and were distinguished on the base of size for the amplification products containing exon 4, whereas PCR products generated by using exon 5 primers were specific for isoform 1 (Figure 1B). DNA from 160 unrelated individuals (120 white, 10 of whom were from Turkey; 20 Asian; 20 blacks) was used as controls (320 chromosomes; power > 80% according to Collins & Schwartz14). The coding region of SOX9 (NC_00017.9; MIM 608160; MIM 114290) was also analyzed. SF1/NR5A1 (MIM 184757) was investigated as described.15 The PCR products were sequenced with the Big Dye Terminator Cycle Sequencing Kit and analyzed on an ABI Prism 310 Genetic Analyzer (Applied Biosystems). Primer sequences available in the Supplemental Data available online.
Direct sequencing of the CBX2 gene of the patient revealed the presence of a C to T exchange (c.C293T) and a G to C exchange both in exon 5 (c.G1370C) and both in the heterozygote state, leading to P98L (inherited from the father) and R443P (inherited from the mother) mutations in the CBX2 protein, respectively (Figure 1B). Their absence in 320 alleles of 160 unrelated individuals suggested that they are not common polymorphisms. SRY was normal. We excluded SOX9 and SF1/NR5A1 mutations and DAX1/NR0B1 duplication, the other possible molecular causes of a similar phenotype. The mutated amino acid residues P98 and R463 are conserved in the mouse protein, and P98 lies in a region with similarity to the yeast SPT2 protein, a chromatin binding protein involved in transcriptional regulation (Figure 1C). Proline is an alpha-helix disruptor, so its substitution with a lysine at 98 and its introduction at 443 is likely to change the three-dimensional structure of the protein.
After PCR-mediated addition of an N-terminal myc tag, the CBX2 cDNA, obtained by Origene (Trueclone TC303599, Origene Australia) was inserted into a pCMV6 vector. Mutant cDNAs were constructed with the QuikChange II site-directed mutagenesis kit from Stratagene. Introduction of the mutations was confirmed by sequencing. Wild-type, mutants, and a 1:1 combination of both cDNAs were transfected into 5 × 104 cells of the following cell lines: HepG2 (ATCC HB-8065); NT2-D1 (ICLC HTL97025), and NCI-H295 (ATCC-2128, a generous gift of William Rainey, University of Texas Southwestern, Dallas, TX). These cell lines expressed the main factors essential for a proper sex development with different patterns (details available upon request). The cells were initially seeded in their optimum medium, let grow to 80%–90% confluence, and then transfected with wild-type and/or mutant cDNA with TransFast transfection reagent (Promega; 9 μl transfection reagent/μg DNA/plate). Transfection efficiency was monitored by cotransfection with a β-galactosidase expression plasmid (Promega pSV-β-galactosidase control vector, molar ratio CBX2: β-gal = 2:1). Transfection efficiency was 40%–60%. 48 hr after transfection RNA was extracted with the RNeasy kit (QIAGEN).
Western blot analysis was performed according to standard procedures and with polyclonal goat antihuman-CBX2/M33 antibodies (EB07432, Everest Biotech, Oxford, UK) or an antibody against the myc epitope (9E10, dilution 1:100).
Western blot suggested that there are no significant differences in protein synthesis or stability between WT and mutant CBX2 (not shown).
In the RNA interference experiments, two of the tested siRNAs (designed with the Promega siRNADesigner program) proved, after optimization, to efficiently block the endogenous and the exogenous (transfected) CBX2 expression in OvCar3 cells. These two siRNAs (Ambion) designated siRNA1 (mRNA 145-164) and siRNA2 (411-430) were used in combination at the concentration of 75 pmol each. Scramble siRNAs were used as control. In 24-well plates, 5 × 103 NT2-D1 cells/well were cultured in DMEM medium for 48 hr until they reached 50% confluence. The siRNAs were then transfected with Lipofectamine 2000 (Invitrogen) as suggested by the provider. 48 hr later, RNA was extracted with RNeasy minikit (QIAGEN) and used for qualitative reverse transcriptase PCR and quantitative real-time PCR. The influence of CBX2 on the expression of SRY (acc no. NM_0031401), SF1/NR5A1 (NM_004959), and SOX9 (BC056420) was studied by means of quantitative real-time PCR, performed with an ABI 7000 Sequence Detection System (Applera Europe) and PCR products were quantified fluorometrically with the SYBR Green Core Reagent kit. The reference mRNA cyclophillin was used for normalizing the data (primers sequence available in Supplemental Data). Western blot was performed as above.
Downregulation of CBX2 obtained by siRNA treatment significantly reduced expression of the putative targets (Figure S1) supporting the CBX2-dependent expression of these genes.
Chromatin immunoprecipitation (ChIP) was performed with the protocol described by Katoh-Fukui et al.,16 except for the human SF1/NR5A1 (NG_008176)-specific primers used for PCR (available in Supplemental Data) and for the usage of H295R cells. PCR conditions were validated with genomic DNA as template. Anti-myc antibodies (9E10) that recognize only the transfected CBX2 were used for immunoprecipitation (IP). After optimization, five fragments were found to be the most suitable: fragment 1 (324 bp) from −465 to −85, fragment 2 (651 bp) from −85 to + 524 containing the noncoding first exon, fragment 3 (171 bp) from +239 to + 410, fragment 4 (767 bp) from −1232 to −465, and fragment 5 (920 bp) from 8761 to 9681 encompassing exon 2. Fragment 5 was not amplified after IP enrichment and was used for control (background). Two fragments amplified from genes flanking SF1/NR5A1 on chromosome 9q33, GPR144 and NR6A1, were used as control for unspecific enrichment. Starting DNA amounts were determined by optical density (OD 260 nm). For the quantitative real-time PCR assays, all samples were run in triplicate with the SYBR Green Core Reagent kit with the immunoprecipitate DNA samples (experiment) run in parallel with their INPUT counterparts. Average cycle threshold (Ct) of INPUT was subtracted from the experiment average Ct to give the CtNET. Cts derived from the control fragment (nontarget DNA), named CtCTRL were subtracted from the CtNET to obtain the definitive Ct. The definitive Cts of three independent experiments were plotted as multiple of the value of nontransfected cells set as 1 (mean ± SD ; no DNA = 0) and represent fold enrichment.
Real-time PCR experiments showed that in H295R WT CBX2 increased 6-fold the expression of endogenous SF1/NR5A1. With the two mutated CBX2 proteins, the effect significantly diminished (Figure 2A). To verify a direct influence of CBX2 on SF1 transcription, ChIP experiments were carried out. Whereas WT CBX2 transfected in H295R cells caused enrichment of three fragments corresponding to sequences in the SF1/NR5A1 promoter, mutant CBX2s failed to bind to one of the fragments, which contains the noncoding first exon (fragment 2). These results are confirmed in the quantitative ChIP experiments (Figures 2B and 2C).
Figure 2.
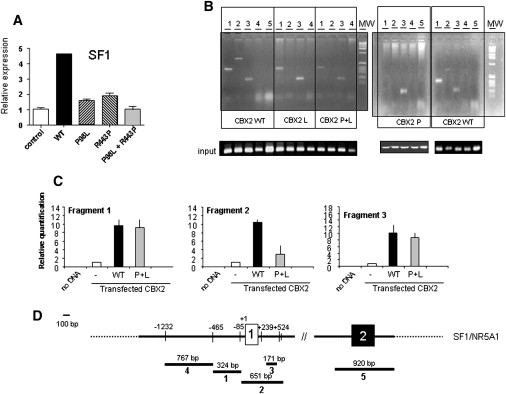
Binding of CBX2 to SF1/NR5A1 Promoter and Influence on Expression
(A) Role of CBX2 variants on the expression of the putative target gene SF1/NR5A1 and proof of a direct influence of CBX2 by its direct binding to promoter target sequences. Expression of SF1 in H295R cells nontransfected (control) or transfected with WT or mutant CBX2.
(B–D) Qualitative (B) and quantitative (C) chromatine immunoprecipitation (ChIP) assay. Crosslinked extracts from H295R cells with or without transfection with WT or mutated CBX2 were sonicated and immunoprecipitated with anti-myc (tag) antibody. After reverse cross-linking, PCR was performed with primers specific for the regions depicted in (D). The numbers on top of the gel in (B) correspond to fragment numbers . Quantitative RT-PCR was performed with SYBR Green incorporation. The data are expressed as mean ± SD.
In silico search for possible Polycomb responsive elements (PRE) was carried out with GeneACT potential binding sites online facility with PRE/TRE motifs17,18 as possible binding site sequences.
Three different constructs containing fragments of the SF1 promoter upstream of a firefly luciferase reporter (in pGL4, Promega), a kind gift of Serdar E. Bulun (Northwest University, Chicago, IL), were used as a putative target for CBX2. The constructs encompass sequence −465/+524 relative to the start site of transcription (NG_008176 4563-5550), −85/+524, and + 293/+524, respectively.19 The two longest constructs contain the first noncoding SF1/NR5A1 exon. The fragment −85/+524 was scrambled and inserted in the pGL4 vector in the inverse direction by substituting KpnI with EcoRV on the direct primer (−85 GATATCGTGGGGGCAGAGACCAAT) and vice versa on the reverse primer (+524 GGTACCAGAGAGAGCCACAGAGACAAC). This experiment substituted the classical site-directed mutagenesis, because of the lack of clear polycomb recognition sites in our sequence of interest.
3 × 103 H295R or HepG2 cells were seeded in 24-well plates and let grow to 60% confluence. These cell lines were chosen because of the expression of SF1/NR5A1 in both and of CBX2 in one (H295R) and not in the other (HepG2). After optimization, a 2:1 reporter:CBX2 plus 1/20 Renilla luciferase (pRL-SV40) construct mixture (total 5 μg DNA/well) was added to a 6-fold FuGene (Roche) and used for transfection. Mock transfection was used as basal (0) level. The empty vector pGL3 was used as internal negative control. 48 hr after transfection, cells were prepared for the Dual-Luciferase Reporter Assay (Promega). Relative luciferase units (Firefly/Renilla) were obtained with a Lumat 9017 luminometer (Berthold Technologies).
To assess the effect of CBX2 directly on transcription, we assayed transactivation of a luciferase reporter driven by deletion constructs of the SF1 promoter.19 The longest construct (−465/+524) contains ChIP fragments 1, 2, and 3; the −85/+524 construct is fragment 2 and encompasses fragment 3; and construct +239/+524 contains fragment 3 (Figure 2D). Wild-type CBX2 had an inhibitory effect on the −465/+524 construct, a stimulatory effect on the −85/+524, and no effect on the shorter construct in transfected H295R cells (Figure 3A). That hints to the presence of a negative regulatory element between −465 and −85 and of an activating element between −85 and + 239, the latter being dominant over the former, as indicated by the fact that overexpression of WT CBX2 increases SF1 mRNA in quantitative RT-PCR experiments (Figures 2A and 3B). CBX2 mutants were significantly less efficient in stimulating luciferase expression when the −85/+524 construct was used. This sequence is the ChIP fragment that the mutant CBX2 failed to bind (fragment 2). The insertion of the same scrambled sequence in the inverse direction abolished all these differences, indicating that the effect is specific (Figure 3A). The differences in luciferase activity between WT and mutant CBX2 for the other two constructs were not significant.
Figure 3.
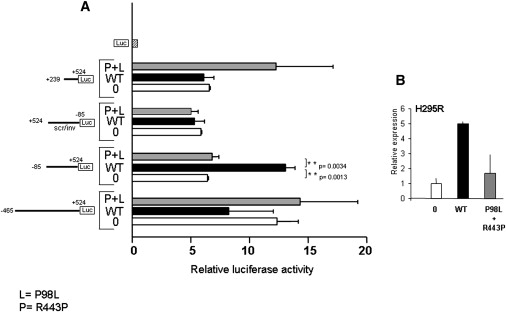
Effect of CBX2 Variants on Gene Target Promoters
(A) Transactivation of luciferase reporter driven by deletion construct of SF1 promoter in absence (0) or presence of overexpression of WT or both CBX2 mutants (P+L). Luciferase activity is expressed as relative to empty vector (luc). Construct +524/85 scr/inv contains the same sequence as in −85/+524 that was scrambled and inserted in the opposite direction. For reference to the ChIP experiments, construct −465/+524 contains ChIP fragments 1, 2, and 3; −85/+524 is fragment 2 and contains fragment 3; +239/+524 contains fragment 3.
(B) Real-time PCR of endogenous SF1 in H295R without (0) or with transfection of wild-type (WT) or mutant CBX2. The image represents the results of mean ± SD of three independent experiments.
The comparison between (A) and (B) suggests that the stimulatory effect of WT CBX2 on the −85/+524 sequence is dominant over the inhibitory effect of the bigger construct (−465/+524).
The present case indicates that CBX2 might now be added to the list of genes involved in the molecular pathogenesis of DSD. As in the mouse model, it appears that CBX2 is upstream of SRY in the sex determination cascade in humans, and its deficiency represents an autosomal-recessive cause of 46,XY DSD. That CBX2 stimulates expression of target genes (SF1/NR5A1) is in agreement with previous data,16 but in contrast to the dogma that polycomb proteins are silencing factors.20 However, some examples have emerged in which polycomb proteins are stimulators of transcription in Drosophila21–23 and in mouse,24 showing that there may be exceptions to the rule. As elegantly discussed by Katoh-Fukui and coworkers,16 CBX2 might have a role (as transactivator) distinct from its known function as chromatin modifier. Accordingly, our experiments suggest a possible model in which normal CBX2 acts preferentially on regions of the promoter of the representative target gene SF1/NR5A1 that are related to activation of transcription. The mutants bind to a different sequence with consequent lack of stimulation. Future studies should establish whether these differences are due to differences in interaction with the partners in the PRC1 complex.
In contrast to the mouse, human skeletal development, adrenal, and splenic growth do not seem to be grossly influenced by these CBX2 mutations. This suggests the presence of alternative pathways or functions for CBX2 in humans. Targeted specific knockin animal experiments might provide means for the direct comparison between human and mouse phenotypes.
Our case was discovered because the prenatal karyotype and the phenotype at birth were discrepant. In contrast to most cases of 46,XY DSD, this girl had normal female internal and external genitalia and, more importantly, instead of dysgenetic gonads, normal bilateral ovaries at histology. Our observations might be important for ovarian development as well as for male sexual differentiation. CBX2 seems to function by actively repressing ovarian development in XY gonads, because mutations in this gene cause male-to-female sex reversal with ovaries in SRY-positive mice12 and humans (present work). As suggested by preliminary data, CBX2 might influence sex differentiation by regulating SRY expression directly or via other factors such as WT1 (unpublished data).
Although generalization should await the description of additional cases, the identification of such cases might be difficult, given the completely normal female phenotype. Given the young age of our patient, even the high FSH levels cannot reflect accurately the potential for pubertal function and fertility. If the similarity between mouse and human phenotype remains throughout life, unexplained sterility in women might be a unique sign of CBX2 abnormalities in the human population.
Acknowledgments
This work was supported by Research Grant of the University of Zurich (54181801) and Grant 32-116636 of the Swiss National Science Foundation. We thank Christiane Donner, Rene Scheiden, Annemarie Tanous, and Paul Philippe for their contribution in the clinical characterization of the patient and Serdar E. Bulun for the generous gift of the SF1 promoter constructs. The authors deny any conflict of interest.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
ExPASy Proteomics Server, http://www.expasy.org/
GeneACT, http://promoter.colorado.edu/geneact/potentialBS.html
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
Promega siRNA designer, http://www.promega.com/siRNADesigner/program/
References
- 1.Hughes I.A., Houk C., Ahmed S.F., Lee P.A. Consensus statement on management of intersex disorders. Arch. Dis. Child. 2006;91:554–563. doi: 10.1136/adc.2006.098319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Polanco J.C., Koopman P. Sry and the hesitant beginnings of male development. Dev. Biol. 2007;302:13–24. doi: 10.1016/j.ydbio.2006.08.049. [DOI] [PubMed] [Google Scholar]
- 3.Kent J., Wheatley S.C., Andrews J.E., Sinclair A.H., Koopman P. A male-specific role for SOX9 in vertebrate sex determination. Development. 1996;122:2813–2822. doi: 10.1242/dev.122.9.2813. [DOI] [PubMed] [Google Scholar]
- 4.Raymond C.S., Murphy M.W., O'Sullivan M.G., Bardwell V.J., Zarkower D. Dmrt1, a gene related to worm and fly sexual regulators, is required for mammalian testis differentiation. Genes Dev. 2000;14:2587–2595. doi: 10.1101/gad.834100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colvin J.S., Green R.P., Schmahl J., Capel B., Ornitz D.M. Male-to-female sex reversal in mice lacking fibroblast growth factor 9. Cell. 2001;104:875–889. doi: 10.1016/s0092-8674(01)00284-7. [DOI] [PubMed] [Google Scholar]
- 6.Bitgood M.J., Shen L., McMahon A.P. Sertoli cell signaling by Desert hedgehog regulates the male germline. Curr. Biol. 1996;6:298–304. doi: 10.1016/s0960-9822(02)00480-3. [DOI] [PubMed] [Google Scholar]
- 7.Chaboissier M.C., Kobayashi A., Vidal V.I., Lutzkendorf S., van de Kant H.J., Wegner M., de Rooij D.G., Behringer R.R., Schedl A. Functional analysis of Sox8 and Sox9 during sex determination in the mouse. Development. 2004;131:1891–1901. doi: 10.1242/dev.01087. [DOI] [PubMed] [Google Scholar]
- 8.Meeks J.J., Weiss J., Jameson J.L. Dax1 is required for testis determination. Nat. Genet. 2003;34:32–33. doi: 10.1038/ng1141. [DOI] [PubMed] [Google Scholar]
- 9.Biason-Lauber A., Konrad D., Navratil F., Schoenle E.J. A WNT4 mutation associated with Mullerian-duct regression and virilization in a 46,XX woman. N. Engl. J. Med. 2004;351:792–798. doi: 10.1056/NEJMoa040533. [DOI] [PubMed] [Google Scholar]
- 10.Wedeen C., Harding K., Levine M. Spatial regulation of Antennapedia and bithorax gene expression by the Polycomb locus in Drosophila. Cell. 1986;44:739–748. doi: 10.1016/0092-8674(86)90840-8. [DOI] [PubMed] [Google Scholar]
- 11.Kuziora M.A., McGinnis W. Autoregulation of a Drosophila homeotic selector gene. Cell. 1988;55:477–485. doi: 10.1016/0092-8674(88)90034-7. [DOI] [PubMed] [Google Scholar]
- 12.Katoh-Fukui Y., Tsuchiya R., Shiroishi T., Nakahara Y., Hashimoto N., Noguchi K., Higashinakagawa T. Male-to-female sex reversal in M33 mutant mice. Nature. 1998;393:688–692. doi: 10.1038/31482. [DOI] [PubMed] [Google Scholar]
- 13.Cui K.H., Warnes G.M., Jeffrey R., Matthews C.D. Sex determination of preimplantation embryos by human testis-determining-gene amplification. Lancet. 1994;343:79–82. doi: 10.1016/s0140-6736(94)90815-x. [DOI] [PubMed] [Google Scholar]
- 14.Collins J.S., Schwartz C.E. Detecting polymorphism and mutations in candidate genes. Am. J. Hum. Genet. 2002;71:1251–1252. doi: 10.1086/344344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biason-Lauber A., Schoenle E.J. Apparently normal ovarian differentiation in a prepubertal girl with transcriptionally inactive steroidogenic factor 1 (NR5A1/SF-1) and adrenocortical insufficiency. Am. J. Hum. Genet. 2000;67:1563–1568. doi: 10.1086/316893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katoh-Fukui Y., Owaki A., Toyama Y., Kusaka M., Shinohara Y., Maekawa M., Toshimori K., Morohashi K. Mouse Polycomb M33 is required for splenic vascular and adrenal gland formation through regulating Ad4BP/SF1 expression. Blood. 2005;106:1612–1620. doi: 10.1182/blood-2004-08-3367. [DOI] [PubMed] [Google Scholar]
- 17.Mihaly J., Mishra R.K., Karch F. A conserved sequence motif in Polycomb-response elements. Mol. Cell. 1998;1:1065–1066. doi: 10.1016/s1097-2765(00)80107-0. [DOI] [PubMed] [Google Scholar]
- 18.Ringrose L., Rehmsmeier M., Dura J.-M., Paro R. Genome-wide prediction of polycomb/trythorax elements in Drosophila melanogaster. Dev. Cell. 2003;5:759–771. doi: 10.1016/s1534-5807(03)00337-x. [DOI] [PubMed] [Google Scholar]
- 19.Xue Q., Lin Z., Yin P., Milad M.P., Cheng Y.H., Confino E., Reierstad S., Bulun S.E. Transcriptional activation of steroidogenic factor-1 by hypomethylation of the 5′ CpG island in endometriosis. J. Clin. Endocrinol. Metab. 2007;92:3261–3267. doi: 10.1210/jc.2007-0494. [DOI] [PubMed] [Google Scholar]
- 20.Orlando V. Polycomb, epigenomes, and control of cell identity. Cell. 2003;112:599–606. doi: 10.1016/s0092-8674(03)00157-0. [DOI] [PubMed] [Google Scholar]
- 21.Fauvarque M.O., Zuber V., Dura J.M. Regulation of polyhomeotic transcription may involve local changes in chromatin activity in Drosophila. Mech. Dev. 1995;52:343–355. doi: 10.1016/0925-4773(95)00412-t. [DOI] [PubMed] [Google Scholar]
- 22.LaJeunesse D., Shearn A. E(z): A polycomb group gene or a trithorax group gene? Development. 1996;122:2189–2197. doi: 10.1242/dev.122.7.2189. [DOI] [PubMed] [Google Scholar]
- 23.Milne T.A., Sinclair D.A., Brock H.W. The Additional sex combs gene of Drosophila is required for activation and repression of homeotic loci, and interacts specifically with Polycomb and super sex combs. Mol. Gen. Genet. 1999;261:753–761. doi: 10.1007/s004380050018. [DOI] [PubMed] [Google Scholar]
- 24.Shirai M., Osugi T., Koga H., Kaji Y., Takimoto E., Komuro I., Hara J., Miwa T., Yamauchi-Takihara K., Takihara Y. The Polycomb-group gene Rae28 sustains Nkx2.5/Csx expression and is essential for cardiac morphogenesis. J. Clin. Invest. 2002;110:177–184. doi: 10.1172/JCI14839. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


