Abstract
The molecular mechanisms underlying general anesthesia are unknown. For volatile general anesthetics (VAs), indirect evidence for both lipid and protein targets has been found. However, no in vivo data have implicated clearly any particular lipid or protein in the control of sensitivity to clinical concentrations of VAs. Genetics provides one approach toward identifying these mechanisms, but genes strongly regulating sensitivity to clinical concentrations of VAs have not been identified. By screening existing mutants of the nematode Caenorhabditis elegans, we found that a mutation in the neuronal syntaxin gene dominantly conferred resistance to the VAs isoflurane and halothane. By contrast, other mutations in syntaxin and in the syntaxin-binding proteins synaptobrevin and SNAP-25 produced VA hypersensitivity. The syntaxin allelic variation was striking, particularly for isoflurane, where a 33-fold range of sensitivities was seen. Both the resistant and hypersensitive mutations decrease synaptic transmission; thus, the indirect effect of reducing neurotransmission does not explain the VA resistance. As assessed by pharmacological criteria, halothane and isoflurane themselves reduced cholinergic transmission, and the presynaptic anesthetic effect was blocked by the resistant syntaxin mutation. A single gene mutation conferring high-level resistance to VAs is inconsistent with nonspecific membrane-perturbation theories of anesthesia. The genetic and pharmacological data suggest that the resistant syntaxin mutant directly blocks VA binding to or efficacy against presynaptic targets that mediate anesthetic behavioral effects. Syntaxin and syntaxin-binding proteins are candidate anesthetic targets.
Volatile general anesthetics (VAs) have profound effects on the metazoan nervous system. In humans, these drugs, such as the original general anesthetics chloroform and diethyl ether, produce amnesia, analgesia, and immobility, a behavioral state termed anesthesia. Despite a century-long effort, the molecular mechanisms of VA action are still unknown. However, by using biochemical and electrophysiological techniques, considerable progress has been made toward identifying potential anesthetic targets (1–7). These in vitro techniques have two fundamental limitations. First, they do not assess anesthetic action on an intact, behaving animal. Second, they may be blind to anesthetic targets that do not gate current, are nonabundant, or are difficult to purify. Genetics complements in vitro approaches, starting at a behavioral level and working back toward the gene products responsible for alterations in anesthetic sensitivity. Thus, genetics can uncover behaviorally relevant anesthetic mechanisms, regardless of the nature of the anesthetic targets mediating the effect. However, thus far, only a few genes have been identified that regulate sensitivity to VAs at concentrations similar to those used to produce human anesthesia (8–11), and none have been found that express large allelic differences in VA sensitivity.
Caenorhabditis elegans meets several criteria as a good genetically tractable model for describing VA mechanisms. C. elegans is sensitive to clinical concentrations of VAs (12). Anesthetic concentrations similar to those required for surgery in humans disrupt normal locomotion in C. elegans. Thus, the molecular mechanisms of anesthetic action could be similar in both species. Second, the nervous system is simple and well understood, and it uses evolutionarily conserved presynaptic proteins, neurotransmitters, receptors, and signal transduction systems (13) that might serve as mediators of general anesthetic action in vertebrates (14). Finally, a large number of viable mutations have been isolated in the genes coding for this synaptic machinery (13, 15–21). Here, we describe a set of mutant genes that drastically alter the sensitivity of C. elegans to VAs. These findings suggest a plausible mechanism of VA action that could be responsible for some components of general anesthesia in humans.
METHODS
Behavioral and Aldicarb Assays.
All assays were performed on young adult hermaphrodites at room temperature (21–24°C). The concentration of VA in assay chambers was measured by gas chromatography (12). VA-induced loss of coordinated movement was measured by the radial dispersal assay as described (12), except the assay plates were shaken briefly to encourage movement of the animals at the start of the 45-min assay period. Concentration/response data were fit by nonlinear regression to give EC50 ± SE values of the estimate by using the equation y = min + (max − min)/(1 + {([VA]/[VA]50)−k}). The EC50 was used as the measure of VA sensitivity of a strain. EC50 values were compared statistically by simultaneous curve fitting (22). P < 0.01 was considered significant. For assay of aldicarb sensitivity, 20–30 individuals were placed for 4 h on aldicarb plates (21) that were seeded with a 1- to 2-day-old bacterial lawn and then moved to the middle of a 0.5-cm diameter area on the lawn. The plates were then sealed into a glass chamber that was filled immediately with various concentrations of anesthetic as described (12). After 1 h, the fraction of animals moving out of the circle was scored as the movement index. The effect of anesthetic on aldicarb sensitivity was performed at an aldicarb concentration where on average 80–95% of the animals were paralyzed in the absence of VA (EC80–95). The aldicarb EC80–95 varied for the different mutant strains (see Table 2). Levamisole assays were performed exactly like aldicarb assays, except with 0.24 mM levamisole in the agar for all strains.
Table 2.
Effect of mutations in presynaptic proteins on VA sensitivity
| Strain | Mutant gene | Halothane EC50, % (vol/vol)† | Isoflurane EC50, % (vol/vol)† | Aldicarb EC80–95, mM‡ | Dispersal index (no VA)§ |
|---|---|---|---|---|---|
| N2 | None (wild type) | 0.45 ± 0.02 | 0.74 ± 0.02 | 0.5 | 0.88 ± 0.01 |
| unc-64(md130) | Syntaxin (ref. 21) | 1.29 ± 0.12* | 4.36 ± 0.36* | 1.5 | 0.63 ± 0.03 |
| unc-64(js21) | Syntaxin (ref. 21) | 0.17 ± 0.03* | 0.13 ± 0.04* | 1.5 | 0.64 ± 0.05 |
| unc-64(md1259) | Syntaxin (ref. 21) | 0.17 ± 0.10* | 0.18 ± 0.02* | 1.5 | 0.40 ± 0.07 |
| snb-1(md247) | Synaptobrevin (ref. 19) | 0.11 ± 0.02* | 0.18 ± 0.08* | 2 | 0.55 ± 0.1 |
| ric-4(js20) | SNAP-25 (ref. 13) | 0.22 ± 0.06* | 0.52 ± 0.05 | 1 | 0.62 ± 0.06 |
| rab-3(js49) | rab3 (ref. 18) | 0.37 ± 0.02 | 0.67 ± 0.05 | 1 | 0.78 ± 0.03 |
| aex-3(y255) | rab3 EF (refs. 17 and 32) | 0.41 ± 0.09 | 0.48 ± 0.05 | 1 | 0.89 ± 0.03 |
EF = nucleotide exchange factor. ∗, P < 0.01; different from N2.
Halothane and isoflurane EC50 values are estimated from at least eight concentration/response values (mean ± SEM).
Aldicarb EC80–95 is the aldicarb millimolar concentration at which 80–95% of animals are paralyzed in the absence of anesthetic.
Values are mean ± SEM.
Strains.
The wild-type strain used for comparison was N2 (varient Bristol) (23). unc-64(md130)/+ hermaphrodites were generated in two ways. md130 males were crossed into unc-51(e369) paralyzed hermaphrodites, and non-Unc hermaphrodite progeny were tested. Alternatively, md130 was crossed into bli-5(e518) and non-Bli hermaphrodites were tested. bli-5/+ animals were tested and found to be wild type for isoflurane sensitivity. Both types of md130/+ animals were similarly resistant to isoflurane. unc-64(js115) is genetically and molecularly a null allele of unc-64 syntaxin (21). md130/js115 animals were produced from a cross of md130/bli-5 males with js115/bli-5 hermaphrodites. Severely Unc non-Bli progeny were passaged, and the md130/js115 genotype was confirmed by segregation of js115-like dead larvae, md130-like adults, severely Unc adults, and no Bli progeny. Non-Bli js115/bli-5 animals were used for js115/+ testing.
Mapping and Transformation Rescue.
Non-Bli F2 progeny from a cross of md130 males with bli-5(e518) were picked singly to plates and allowed to self. F3 progeny from plates that segregated no Bli progeny were tested at 1.5–2% (vol/vol) isoflurane. Average dispersal indices greater than 0.3 for the plate were scored as isoflurane-resistant; the dispersal index for wild type for this isoflurane concentration is 0.03 ± 0.01 (range 0–0.09; n = 9). Germ-line transformation (24) of md130 hermaphrodites was accomplished by coinjecting pTX20 (10 μg/ml), which contains a 16.2-kb genomic insert that includes the unc-64 gene and has been shown to rescue unc-64(null) (21); pMR1 (10 μg/ml), which contains an unc-54 promoter∷GFPS65T (green fluorescent protein) construct that dominantly expresses GFPS65T in body-wall and sex muscles (21); and pBluescript SK(+) (100 μg/ml; Stratagene) as carrier DNA. Green transformant F1 progeny were passaged individually to establish independent transformed lines. To measure the isoflurane sensitivity of transformed vs. untransformed animals, dispersal assays were performed as above but with a single isoflurane concentration that ranged between 3% and 4% (vol/vol) depending on the line. Also, the gas phase concentration was measured after 45 min, and after 60 min, the animals that had dispersed were separated from those that had not. These two groups were then scored for the number of green and nongreen animals by using a fluorescent dissecting microscope to calculate the dispersal index of GFP(+) and GFP(−) progeny. Although the isoflurane concentration varied slightly from line to line, for a given transformed line and for the average of all lines, the GFP(+) and GFP(−) siblings were exposed to an identical isoflurane concentration.
cDNA Cloning and Sequencing.
RNA was prepared by LiCl precipitation. Reverse transcription–PCR was performed with primers flanking the splice site mutated in md130, and cDNA was synthesized with random hexanucleotide primers. cDNAs products were separated by size on a 1.2% agarose gel. These products were cloned into pBluescript SK(+) and sequenced with Big Dye terminators (Stratagene).
RESULTS
VA Resistance of a Syntaxin Mutant.
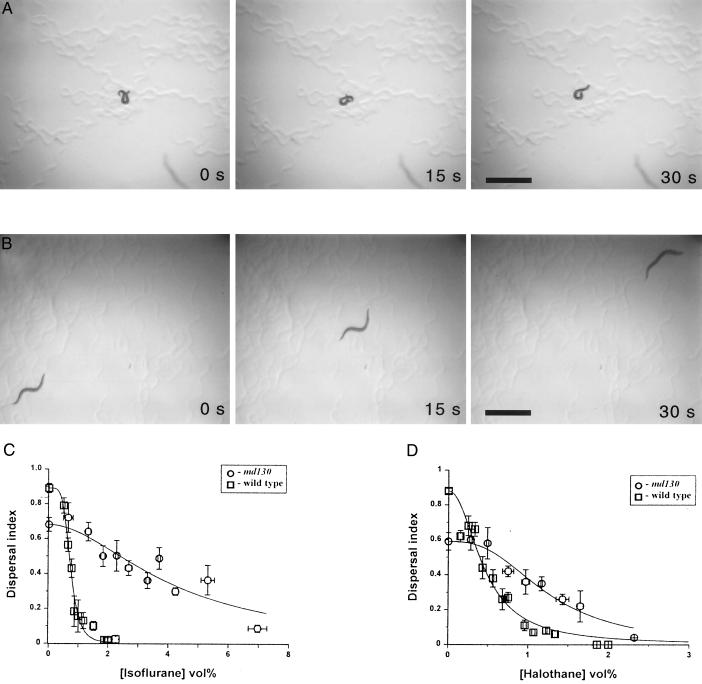
During the course of screening existing C. elegans mutants for VA resistance, we found that md130, which contains a mutation in the neuronal syntaxin gene unc-64 (21), was markedly resistant to the VAs isoflurane and halothane (Fig. 1). VAs produce uncoordinated locomotion of wild-type C. elegans animals so that they adopt abnormal postures and move little (Fig. 1A; ref. 12). However, the md130 mutant moves well at high anesthetic concentrations with a normal sinusoidal wave and nearly normal velocity (Fig. 1B and data not shown). The EC50 for isoflurane-induced loss of coordinated movement of md130 as measured by the fraction of animals able to disperse to the edge of an agar pad (the dispersal index) was 4.36 ± 0.36% (vol/vol), which is 5.9-fold more than the wild-type EC50 [0.74 ± 0.02% (vol/vol)]. For halothane, the resistance was smaller but still quite substantial at 2.9-fold the wild-type EC50 [0.45 ± 0.02% (vol/vol)]. Over 2.5% (vol/vol; ≈1.4 mM at 22°C; ref. 25) isoflurane was required for a significant (P < 0.01; t test, assuming unequal variance) decrease in the dispersal index of md130 compared with 0.7% (vol/vol) for wild type. Likewise, a significant effect of halothane on the dispersal index of md130 was not seen until 1% (vol/vol; ≈0.64 mM) vs. 0.2% (vol/vol) for the wild-type strain. Thus, clinical concentrations of isoflurane and halothane are ineffective in disrupting locomotion in the syntaxin mutant. This phenotype seems to be uncommon. In screening 6,600 haploid genomes that had been exposed to mutagens for resistance to halothane, we found no halothane-resistant mutants. Further, measurement of the EC50 values of 70 existing strains containing mutations in 47 different neuronal genes identified no strains with the level of resistance seen in md130, although mutations in 5 genes do produce significant but substantially smaller degrees of resistance (B.v.S., L.S., and C.M.C., unpublished work). To our knowledge, md130 mutants are the only metazoans known to be highly resistant to VAs.
Figure 1.
VA resistance of the syntaxin mutant md130. (A) Movement of a representative wild-type young adult animal over 30 s in the presence of 2.8% (vol/vol) isoflurane. The animal is on standard nutrient growth medium agar in the absence of a bacterial lawn (23). (B) Movement of a representative md130 young adult animal over 30 s in the presence of 2.8% (vol/vol) isoflurane. (For A and B, bars = 1 mm.) (C) The dispersal index (12) of the fraction of young adult animals moving in 45 min from the center to the edge of a 9.5-cm agar pad rimmed with bacteria is plotted against the percentage of isoflurane concentration (vol%) for both strains. Each data point is mean ± SEM of at least four independent assays performed at the same VA-concentration range. Horizontal error bars represent the SEM of the pooled VA concentrations. Curves were fit to the data by nonlinear regression. The EC50 of isoflurane is 0.74 ± 0.02 for wild type and 4.36 ± 0.36 for md130. (D) Concentration/response curve for halothane. The EC50 is 0.45 ± 0.02 for wild type and 1.29 ± 0.12 for md130.
The Syntaxin Mutation Produces the Anesthetic-Resistance Phenotype.
To confirm that the anesthetic-resistance phenotype of md130 was caused by mutation of the syntaxin gene, we first mapped the isoflurane resistance of md130 against bli-5, a gene closely linked to syntaxin. Linkage to syntaxin was established by detecting no recombinations (n = 60) between bli-5 and the isoflurane-resistance locus. Second, transformation rescue of md130 animals was performed with a plasmid containing a genomic copy of the syntaxin gene along with a dominant transformation marker, a plasmid expressing GFP (21). The dispersal index at a single high isoflurane concentration [ranging from 3% to 4% (vol/vol) for the different lines] was measured for each of 10 independently transformed lines. GFP(+) (i.e., transformed animals) were significantly more sensitive to isoflurane than GFP(−) siblings that had lost the transforming array (Table 1). Transformation with the GFP marker plasmid alone had no effect. Further, the rescuing syntaxin plasmid in a wild-type background (outcrossed from md130) had no effect on isoflurane sensitivity, indicating that the extra copies of wild-type syntaxin did not produce hypersensitivity to isoflurane (data not shown). Thus, the wild-type unc-64 syntaxin gene rescued the isoflurane-resistance phenotype of md130. In addition to anesthetic resistance, md130 has a mild uncoordinated-locomotion phenotype and was isolated originally because of its resistance to the acetylcholinesterase inhibitor aldicarb (26). Aldicarb resistance is produced by a reduction in the release of or response to acetylcholine, and mutations in presynaptic machinery proteins have been found to confer aldicarb resistance (13). Neuronal syntaxin is thought to function normally in neurotransmitter release by mediating fusion of synaptic vesicles with the presynaptic terminal membrane (27); therefore, uncoordinated locomotion and aldicarb resistance are logical phenotypes for reduction-of-function mutants of C. elegans syntaxin. Both the uncoordination and aldicarb-resistance phenotypes of md130 were rescued by transformation with the wild-type syntaxin gene (Table 1). The mapping and rescue data confirm that a mutation in the syntaxin gene is responsible for all of the phenotypes of md130, including its isoflurane resistance.
Table 1.
Transformation rescue of mdl30
| Injected plasmids | GFP expression | Dispersal index†
|
Fraction aldicarb-resistant animals‡ | |
|---|---|---|---|---|
| With 3–4% (vol/vol) isoflurane | With no isoflurane | |||
| Syntaxin + GFP marker | − | 0.45 ± 0.04 | 0.63 ± 0.04 | 0.78 ± 0.06 |
| Syntaxin + GFP marker | + | 0.13 ± 0.02* | 0.98 ± 0.01* | 0.17 ± 0.02* |
| GFP marker | − | 0.59 ± 0.03 | 0.66 ± 0.11 | ND |
| GFP marker | + | 0.48 ± 0.06 | 0.78 ± 0.06 | ND |
The syntaxin plasmid pTX20 contains a full-length genomic syntaxin insert (21). The GFP transformation marker plasmid pMR1 contains a myosin promoter-driven GFP insert (21).
Dispersal index (mean ± SEM) for a given GFP expression of independently transformed lines (n = 10 for syntaxin + GFP; n = 4 for GFP only). For isoflurane sensitivity determinations, each line was tested at a single concentration, ranging from 3–4% (vol/vol).
Fraction of animals moving at 0.5 mM aldicarb.
, P < 0.01 GFP(+) vs. GFP(−); ND, not determined.
Genetic Characteristics of md130.
The isoflurane-resistance phenotype of md130 is semidominant and behaves as a gain-of-function mutation. As determined by the dispersal assay, md130/+ is significantly resistant [EC50 = 2.38 ± 0.2% (vol/vol)] to isoflurane compared with wild-type [EC50 = 0.74 ± 0.02% (vol/vol)] and with null/+ heterozygotes [EC50 = 1.3 ± 0.01% (vol/vol)]. md130/null transheterozygotes were not tested because of their very poor movement in the absence of anesthetic. In contrast, the locomotion phenotype of md130 in the absence of anesthetic is recessive and behaves as a reduction-of-function mutation. The dispersal indices at 45 min for the different genotypes were 0.63 ± 0.03 for md130/md130, 0.99 ± 0.02 for md130/+, 0.12 ± 0.06 for md130/null, 0.96 ± 0.03 for null/+, and 0.88 ± 0.01 for +/+. md130 is also a reduction-of-function mutation for neurotransmitter release as determined by the aldicarb assay (21). Thus, the locomotion and aldicarb-resistance phenotypes can be explained by a reduction in wild-type function of syntaxin. However, the isoflurane-resistance phenotype clearly cannot.
md130 Gene Products.
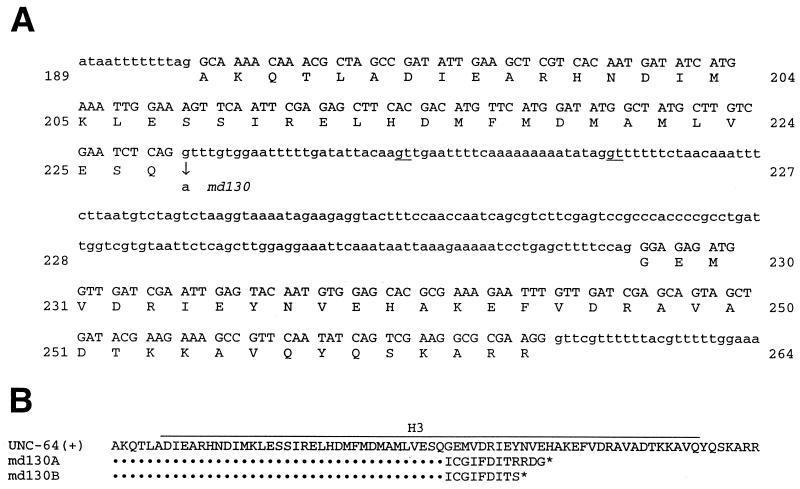
Sequencing of the unc-64 gene in md130 identified a single base change, a G to A at the splice-donor site of the sixth intron (Fig. 2A; ref. 21). The surrounding sixth and seventh exons code for the H3 domain of syntaxin, a helical region shown to interact with several presynaptic proteins (28). Reverse transcription–PCR of md130 RNA with primers flanking the mutation showed products of four sizes. Sequencing of the cloned cDNAs predicts that two of the products use alternative splice sites, adding 25 or 50 bp of intron sequence to the mRNA. (Fig. 2B). Another product is unspliced, and one is properly spliced. Translation of the misspliced and unspliced mRNAs would lead to truncated protein products lacking part of the H3 and all of the transmembrane domains and terminating with additional amino acids normally not found in syntaxin. A dominant effect of one or more of these products on VA action is consistent with the gain-of-function behavior of md130. However, other unidentified md130 products could confer the VA resistance.
Figure 2.
md130 mutant products. (A) The sequence of the wild-type unc-64 gene in the region of the md130 mutation. The G to A md130 mutation at the splice-donor site of the sixth intron is shown. Alternative, downstream splice-donor sites used to produce the cloned md130 cDNA species are underlined. (B) Protein products predicted from the sequence of md130 cDNAs. Some wild-type UNC-64(+) product is predicted. One mutant product, md130A, would be made from mRNA spliced at the upstream alternative donor site; md130B would be produced from mRNA spliced at the downstream site or by unspliced mRNA.
Presynaptic Mechanism of the VA Resistance of md130.
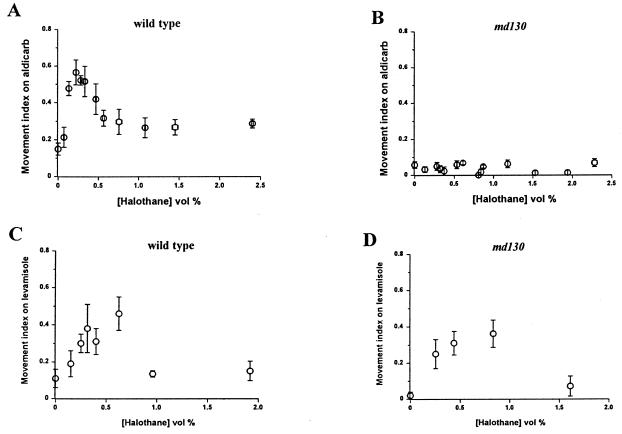
The known function of syntaxin in neurotransmitter release (21, 27) suggests that md130 disrupts a presynaptic action of VA. To test this hypothesis, we measured the effect of VAs on the sensitivity to aldicarb of the wild-type and md130 strains. If VAs were to increase transmitter release, then hypersensitivity to aldicarb-induced paralysis would be observed, because the acetylcholinesterase inhibitor aldicarb potentiates the action of released neurotransmitters (13). If VAs were to decrease release, resistance to aldicarb would be observed. We found the latter result in the wild-type strain (Fig. 3A). Halothane produced an approximately 3-fold increase in the number of animals moving in the presence of aldicarb. The aldicarb antagonism by halothane peaked between 0.1 and 0.6% (vol/vol) and decreased at higher halothane concentrations. Isoflurane also antagonized aldicarb-induced paralysis significantly [the fractions of moving animals were 0.14 ± 0.04 without isoflurane and 0.56 ± 0.03 with 0.92 ± 0.02% (vol/vol) isoflurane; P < 0.01]. The lowest isoflurane concentration [0.54% (vol/vol)] that significantly antagonized aldicarb was higher than that for halothane. For both anesthetics, the anesthetic-induced aldicarb resistance occurs at VA concentrations overlapping those producing the locomotion defects. In contrast to the wild-type strain, md130 was unaffected by halothane in the aldicarb assay (Fig. 3B). The higher concentration of aldicarb needed to immobilize md130 was not the reason for its lack of response to halothane. Wild-type animals tested at this higher aldicarb concentration were still rendered resistant to aldicarb by halothane [40% movement index at 0.2 and 0.36% (vol/vol) halothane vs. 0% movement index without halothane]. Acute resistance to aldicarb can also be produced by reduction in the postsynaptic response to acetylcholine (29).
Figure 3.
Halothane produces aldicarb resistance in wild-type, but not md130, animals. (A) Movement index (fraction of moving animals) for the wild-type strain on 0.5 mM aldicarb at various concentrations of halothane. (B) Movement index for md130 on 1.5 mM aldicarb. (C) Movement index for the wild-type strain on 0.24 mM levamisole at various concentrations of halothane. (D) Movement index for md130 on 0.24 mM levamisole at various concentrations of halothane.
To address this explanation for VA-induced aldicarb resistance, the effect of halothane on sensitivity to levamisole, an acetylcholine receptor agonist in C. elegans, was determined in the wild-type and md130 strains. Between 0.1 and 0.65% (vol/vol) halothane significantly increased the movement index of the wild-type strain on levamisole (Fig. 3C). However, in the same concentration range, halothane also produced a similar and significant degree of antagonism of levamisole in md130 (Fig. 3D). The md130 mutation blocked the effect of halothane on aldicarb sensitivity but not on levamisole sensitivity. Further, md130 and the wild-type strain were similarly sensitive to levamisole (unlike aldicarb) in the absence of anesthetic, suggesting that md130 reduces cholinergic neurotransmission entirely by a presynaptic mechanism. Thus, VAs seem to act both presynaptically and postsynaptically, but given the VA resistance conferred by the md130 mutation, the presynaptic activity is primarily responsible for VA behavioral effects on locomotion in C. elegans. An additional conclusion that can be made from the levamisole data is that the md130 mutation does not block entry of VAs through the cuticle; if it did, md130 would also confer resistance to halothane’s antagonism of levamisole.
Effect of Mutations in Other Presynaptic Machinery Genes.
To examine whether reduction in neurotransmission alone could explain the VA resistance of md130, we tested additional mutants in syntaxin and in genes encoding other presynaptic machinery proteins. Rather than being resistant, two other reduction-of-function syntaxin mutants (21) were markedly hypersensitive to halothane and isoflurane (Table 2). Likewise, reduction-of-function mutants of SNAP-25 (13, 30) and synaptobrevin (19), which form a ternary complex with syntaxin critical for membrane fusion (31), were also hypersensitive to halothane. Mutants in genes encoding homologs of rab3 (18) and of rab3 nucleotide exchange factor (17, 32) were similar to wild type in their halothane and isoflurane sensitivity. By behavioral, physiological, and pharmacological criteria, all of these mutant alleles reduce synaptic transmission (M.L.N. and O.S., unpublished results, and refs. 17–19 and 21). Each is resistant to aldicarb, and some are more behaviorally defective than md130 as indicated by their dispersal indices in the absence of VA (Table 2). Each mutant strain in Table 2 also was tested for sensitivity to halothane in the aldicarb assay (i.e., whether halothane antagonized aldicarb in the strain). Except in the md130 strain as shown above (Fig. 3B), halothane antagonizes aldicarb potency in all of the strains listed in Table 2 (data not shown). Thus, reduction of neurotransmission, even by other mutations in syntaxin, clearly is not sufficient for producing resistance to anesthetics in either the locomotion or aldicarb assays. Rather, decreasing transmission seems generally to increase VA sensitivity, perhaps by acting additively with presynaptic VA action.
DISCUSSION
Vertebrate in vitro experiments have found both presynaptic and postsynaptic effects of anesthetics (14, 33, 34). It is unknown which, if any, of the various cellular and molecular actions of VAs are operant at a behavioral level in vertebrates. However, some effects, such as enhancement of γ-aminobutyric acid type A receptor-mediated neuronal inhibition, do occur at clinically relevant concentrations and seem likely to contribute to at least some aspects of the behavioral effects of anesthetics (1, 3, 11, 14). Although we found evidence for postsynaptic VA effects in C. elegans, the predominant anesthetic mechanism for disrupting locomotion seems to be presynaptic. This conclusion, of course, does not necessarily mean that postsynaptic effects are unimportant in vertebrate anesthesia. However, given the high degree of conservation of presynaptic machinery from nematode to human (13), it is likely that the anesthetic mechanism uncovered here for C. elegans is operant in vertebrates. Indeed, C. elegans syntaxin is 63% identical at the amino acid level with human syntaxin 1A (20, 21). Further, the H3 domain, the portion of the protein disrupted by the md130 mutation, is 80% identical to human syntaxin 1A and contains a perfectly conserved stretch of 22 aa (21), which is truncated in some md130 mRNA species.
How does the md130 mutation confer resistance to VAs? One obvious possibility is that syntaxin is an anesthetic target. Reverse transcription–PCR of md130 RNA found wild-type and truncated products that lack a transmembrane domain. If syntaxin indeed contains a VA-binding pocket, the wild-type protein would be expected to bind and be affected by VAs normally. Thus, the truncated products might either interfere with binding of VAs to wild-type syntaxin, or they might function to mediate synaptic-vesicle release but lack the VA-binding site. However, it is unlikely that the truncated species would be capable of mediating fusion without their transmembrane domains.
Alternatively, other proteins interacting with syntaxin, either alone or in a complex, could be the relevant VA targets. Syntaxin has been shown to form a ternary complex with synaptobrevin and SNAP-25 (31). Formation of the complex requires the syntaxin H3 domain (28) and is thought to produce a four-α-helical bundle mediating vesicle fusion (35, 36). Halothane has been shown to bind with reasonably high affinity to a hydrophobic pocket within a synthetic four-α-helical bundle (37). Thus, the hydrophobic core of this ternary complex is a plausible VA target. Formally, other presynaptic syntaxin-binding proteins also could be VA targets (38–40), although no biochemical data suggest that these proteins might bind VAs.
A final possibility is that the md130 mutation does not affect VA binding; rather, it could indirectly decrease VA efficacy by counteracting the effect of VA binding or regulate the process disrupted by VAs. However, as shown by the VA hypersensitivity of the other syntaxin alleles, simply reducing syntaxin function cannot be the mechanism of the resistance of md130. Rather, a more complex or unknown function of syntaxin and/or the md130 mutant proteins must be invoked. At this point, we cannot rule out this possibility. Whether a VA receptor, effector, or regulator is correct, our data for C. elegans suggest that syntaxin is involved functionally in anesthetic action in vivo. Further, the high-level resistance produced by the md130 mutation indicates that the VAs act specifically through a single major mechanism to disrupt coordinated locomotion in C. elegans.
Acknowledgments
We thank J. Rand for sharing mutant alleles. We also thank A. S. Evers and J. H. Steinbach for comments on the manuscript. This work was supported by grants from the National Institute of General Medical Sciences (to C.M.C., B.v.S., and L.D.S.) and from the National Institute of Neurological Disorders and Stroke (to M.L.N. and O.S.). Some of the C. elegans strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources.
ABBREVIATIONS
- GFP
green fluorescent protein
- md130
the unc-64(md130) mutation
- VA
volatile general anesthetic
References
- 1.Mody I, Tanelian D L, MacIver M B. Brain Res. 1991;538:319–323. doi: 10.1016/0006-8993(91)90447-4. [DOI] [PubMed] [Google Scholar]
- 2.Jones M V, Brooks P A, Harrison N L. J Physiol. 1992;449:279–293. doi: 10.1113/jphysiol.1992.sp019086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mihic S J, Ye Q, Wick M J, Koltchine V V, Krasowski M D, Finn S E, Mascia M P, Valenzuela C F, Hanson K K, Greenblatt E P, et al. Nature (London) 1997;389:385–389. doi: 10.1038/38738. [DOI] [PubMed] [Google Scholar]
- 4.Miao N, Frazer M J, Lynch C., III Anesthesiology. 1995;83:593–603. doi: 10.1097/00000542-199509000-00019. [DOI] [PubMed] [Google Scholar]
- 5.Study R E. Anesthesiology. 1994;81:104–116. doi: 10.1097/00000542-199407000-00016. [DOI] [PubMed] [Google Scholar]
- 6.Flood P, Ramirez-Latorre J, Role L. Anesthesiology. 1997;86:859–865. doi: 10.1097/00000542-199704000-00016. [DOI] [PubMed] [Google Scholar]
- 7.Violet J M, Downie D L, Nakisa R C, Lieb W R, Franks N P. Anesthesiology. 1997;86:866–874. doi: 10.1097/00000542-199704000-00017. [DOI] [PubMed] [Google Scholar]
- 8.Tinklenberg J A, Segal I S, Guo T Z, Maze M. Ann N Y Acad Sci. 1991;625:532–539. doi: 10.1111/j.1749-6632.1991.tb33884.x. [DOI] [PubMed] [Google Scholar]
- 9.Gamo S, Tanaka Y, Yamamoto H, Sakoyama Y. Hiroshima J Anesth. 1992;28:279–285. [Google Scholar]
- 10.Leibovitch B A, Campbell D B, Krishnan K S, Nash H A. J Neurogenet. 1995;10:1–13. doi: 10.3109/01677069509083455. [DOI] [PubMed] [Google Scholar]
- 11.Quinlan J J, Homanics G E, Firestone L L. Anesthesiology. 1998;88:775–780. doi: 10.1097/00000542-199803000-00030. [DOI] [PubMed] [Google Scholar]
- 12.Crowder C M, Shebester L D, Schedl T. Anesthesiology. 1996;85:901–912. doi: 10.1097/00000542-199610000-00027. [DOI] [PubMed] [Google Scholar]
- 13.Rand J B, Nonet M L. In: C. elegans II. Riddle D L, Blumenthal T, Meyer B J, Priess J R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 611–644. [Google Scholar]
- 14.Franks N P, Lieb W R. Nature (London) 1994;367:607–614. doi: 10.1038/367607a0. [DOI] [PubMed] [Google Scholar]
- 15.Gengyo-Ando K, Kamiya Y, Yamakawa A, Kodaira K, Nishiwaki K, Miwa J, Hori I, Hosono R. Neuron. 1993;11:703–711. doi: 10.1016/0896-6273(93)90080-b. [DOI] [PubMed] [Google Scholar]
- 16.Nonet M L, Grundahl K, Meyer B J, Rand J B. Cell. 1993;73:1291–1305. doi: 10.1016/0092-8674(93)90357-v. [DOI] [PubMed] [Google Scholar]
- 17.Iwasaki K, Staunton J, Saifee O, Nonet M, Thomas J H. Neuron. 1997;18:613–622. doi: 10.1016/s0896-6273(00)80302-5. [DOI] [PubMed] [Google Scholar]
- 18.Nonet M L, Staunton J E, Kilgard M P, Fergestad T, Hartwieg E, Horvitz H R, Jorgensen E M, Meyer B J. J Neurosci. 1997;17:8061–8073. doi: 10.1523/JNEUROSCI.17-21-08061.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nonet M L, Saifee O, Zhao H, Rand J B, Wei L. J Neurosci. 1998;18:70–80. doi: 10.1523/JNEUROSCI.18-01-00070.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogawa H, Harada S, Sassa T, Yamamoto H, Hosono R. J Biol Chem. 1998;273:2192–2198. doi: 10.1074/jbc.273.4.2192. [DOI] [PubMed] [Google Scholar]
- 21.Saifee O, Wei L P, Nonet M L. Mol Biol Cell. 1998;9:1235–1252. doi: 10.1091/mbc.9.6.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waud D R. J Pharm Exp Therap. 1972;183:577–607. [PubMed] [Google Scholar]
- 23.Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mello C C, Kramer J M, Stinchcomb D, Ambros V. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franks N P, Lieb W R. Br J Anaesth. 1993;71:65–76. doi: 10.1093/bja/71.1.65. [DOI] [PubMed] [Google Scholar]
- 26.Miller K G, Alfonso A, Nguyen M, Crowell J A, Johnson C D, Rand J B. Proc Natl Acad Sci USA. 1996;93:12593–12598. doi: 10.1073/pnas.93.22.12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sudhof T C. Nature (London) 1995;375:645–653. doi: 10.1038/375645a0. [DOI] [PubMed] [Google Scholar]
- 28.Kee Y, Lin R C, Hsu S C, Scheller R H. Neuron. 1995;14:991–998. doi: 10.1016/0896-6273(95)90337-2. [DOI] [PubMed] [Google Scholar]
- 29.Lewis J A, Wu C H, Berg H, Levine J H. Genetics. 1980;95:905–928. doi: 10.1093/genetics/95.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen M, Alfonso A, Johnson C D, Rand J B. Genetics. 1995;140:527–535. doi: 10.1093/genetics/140.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sollner T, Bennett M K, Whiteheart S W, Scheller R H, Rothman J E. Cell. 1993;75:409–418. doi: 10.1016/0092-8674(93)90376-2. [DOI] [PubMed] [Google Scholar]
- 32.Wada M, Nakanishi H, Satoh A, Hirano H, Obaishi H, Matsuura Y, Takai Y. J Biol Chem. 1997;272:3875–3878. doi: 10.1074/jbc.272.7.3875. [DOI] [PubMed] [Google Scholar]
- 33.Takenoshita M, Takahashi T. Brain Res. 1987;402:303–310. doi: 10.1016/0006-8993(87)90037-0. [DOI] [PubMed] [Google Scholar]
- 34.MacIver M B, Mikulec A A, Amagasu S M, Monroe F A. Anesthesiology. 1996;85:823–834. doi: 10.1097/00000542-199610000-00018. [DOI] [PubMed] [Google Scholar]
- 35.Fasshauer D, Otto H, Eliason W K, Jahn R, Brunger A. J Biol Chem. 1997;272:28036–28041. doi: 10.1074/jbc.272.44.28036. [DOI] [PubMed] [Google Scholar]
- 36.Hanson P I, Roth R, Morisaki H, Jahn R, Heuser J E. Cell. 1997;90:523–535. doi: 10.1016/s0092-8674(00)80512-7. [DOI] [PubMed] [Google Scholar]
- 37.Johansson J, Gibney B, Rabanal F, Reddy K, Dutton P. Biochemistry. 1998;37:1421–1429. doi: 10.1021/bi9721290. [DOI] [PubMed] [Google Scholar]
- 38.Hata Y, Slaughter C A, Sudhof T C. Nature (London) 1993;366:347–351. doi: 10.1038/366347a0. [DOI] [PubMed] [Google Scholar]
- 39.Sheng Z H, Rettig J, Takahashi M, Catterall W A. Neuron. 1994;13:1303–1313. doi: 10.1016/0896-6273(94)90417-0. [DOI] [PubMed] [Google Scholar]
- 40.Betz A, Okamoto M, Benseler F, Brose N. J Biol Chem. 1997;272:2520–2526. doi: 10.1074/jbc.272.4.2520. [DOI] [PubMed] [Google Scholar]