Abstract
Jeune asphyxiating thoracic dystrophy (ATD) is an autosomal-recessive chondrodysplasia characterized by short ribs and a narrow thorax, short long bones, inconstant polydactyly, and trident acetabular roof. ATD is closely related to the short rib polydactyly syndrome (SRP) type III, which is a more severe condition characterized by early prenatal expression and lethality and variable malformations. We first excluded IFT80 in a series of 26 fetuses and children belonging to 14 families diagnosed with either ATD or SRP type III. Studying a consanguineous family from Morocco, we mapped an ATD gene to chromosome 11q14.3-q23.1 in a 20.4 Mb region and identified homozygous mutations in the cytoplasmic dynein 2 heavy chain 1 (DYNC2H1) gene in the affected children. Compound heterozygosity for DYNC2H1 mutations was also identified in four additional families. Among the five families, 3/5 were diagnosed with ATD and 2/5 included pregnancies terminated for SRP type III. DYNC2H1 is a component of a cytoplasmic dynein complex and is directly involved in the generation and maintenance of cilia. From this study, we conclude that ATD and SRP type III are variants of a single disorder belonging to the ciliopathy group.
Main Text
Jeune asphyxiating thoracic dystrophy (ATD [MIM 208500]) is an autosomal-recessive chondrodysplasia characterized by short ribs and respiratory insufficiency and is often fatal in the first year of life. Retinal degeneration, cystic renal disease, and liver involvement occasionally occur in the course of the disease. Mutations in the intraflagellar transport 80 (IFT80 [MIM 611177]) gene have been recently identified in 3/39 families originating from Pakistan and Turkey, ascribing ATD to the ciliopathy group.1 ATD is known to be genetically heterogeneous with another locus mapped on chromosome 15q13.2 ATD is closely related to the short rib polydactyly group, especially to the type III (SRP type III, also called Verma-Naumoff, [MIM 263510]). Both conditions share the same radiological features (including the polydactyly), but SRP type III is more severe, characterized by an early prenatal expression and lethality, variable malformations (cleft lip and/or palate; polycystic kidneys; gastrointestinal, urogenital, brain, and/or cardiac malformations), and severely shortened tubular bones having round metaphyseal ends with lateral spikes.3,4 Here, we report the mapping of a locus on chromosome 11q and the identification of cytoplasmic dynein 2 heavy chain 1 (DYNC2H1 [MIM 603297]) mutations in five families with ATD or SRP type III, supporting the view that ATD is an heterogeneous disorder overlapping with SRP type III and belongs to the ciliopathy group.5
Criteria for inclusion in the study were (1) short ribs and a constricted thoracic cage, (2) trident acetabular roof, (3) small hands and feet, and (4) shortening of the long bones. Included were a total of 15 children (ranging in age from 0 to 19 years of age) and 11 fetuses belonging to 14 families, diagnosed with either ATD (15 living children and four fetuses) or SRP type III (seven fetuses). Among the fetuses, the diagnosis of SRP (rather than ATD) was assigned on the basis of the extreme severity of the thorax narrowness and the long bone shortness with the presence of metaphyseal spikes. Among all families with ATD or SRP type III, six originating from Tunisia, Turkey, Portugal, and Morocco were consanguineous. Blood samples from patients and unaffected relatives were obtained with the appropriate written consent, in accordance with the French ethical standards regarding human subjects.
We first excluded IFT80 either by linkage analysis in consanguineous families with the use of microsatellite markers or by direct sequencing in isolated cases (data not shown).
We then focused on a large consanguineous Moroccan family with two affected children (Figure 1). We performed a genome-wide search, using the GeneChip Human Mapping 250K NspI array (Affymetrix) on DNA samples of the two affected children and one parent. Array experiments were performed according to protocols provided by the manufacturer. The SNP genotypes were analyzed with the software Affymetrix. We found a large region of homozygosity shared by the two children on chromosome 11 (Figure S1, available online). Using the MERLIN program, we found a maximum LOD score (Z = 3, at θ = 0) for the region delimited by SNP markers rs921561 and rs1893996. Further analyses with polymorphic microsatellite markers allowed us to delineate the critical region, the centromeric boundary defined by marker D11S4175 and the telomeric boundary by marker D11S1893 (11q14.3-11q23.1, 20.4 Mb).
Figure 1.
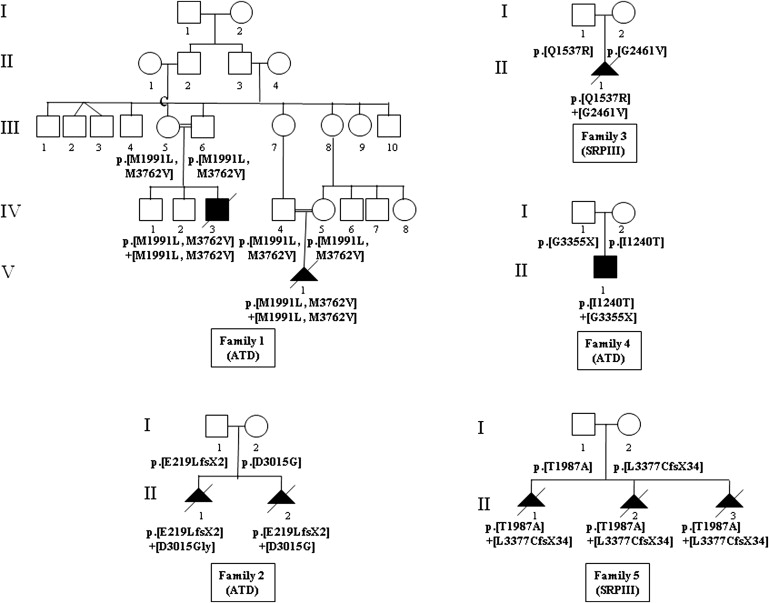
Pedigrees of the Five Affected Families and Segregation of the DYNC2H1 Mutations
This region encompasses 85 genes, and among them, we first considered the genes referenced in the cilia database (see Web Resources), namely KDEL (Lys-Asp-Glu-Leu) containing 2 (KDELC2); Ras-related protein Rab-39A (Rab-39); guanylate cyclase 1, soluble, alpha 2 (GUCY1A2 [MIM 601244]); and cytoplasmic dynein 2 heavy chain 1 (DYNC2H1). After having excluded the first three genes by direct sequencing, we considered DYNC2H1 as a good candidate gene because it encodes a subunit of a cytoplasmic dynein complex.5 This gene is composed of 90 exons and encodes a protein of 4314 amino acids. We performed direct sequencing, using 90 couples of primers (Table S1), and identified two homozygous missense mutations in the two affected children (p.Met1991Leu; p.Met3762Val, Table 1). The mutations cosegregated with the disease and were not identified in 210 control chromosomes from the same ethnic origin (Figure 1). With the use of the PolyPhen web program,6 the consequences of these two mutations were predicted as benign for the p.Met1991Leu mutation and as probably damaging for the p.Met3762Val mutation. However, the p.Met1991Leu mutation is located in the P-loop NTPase domain, whereas the p.Met3762Val mutation is located in a region of unknown function. It is therefore difficult to speculate on the responsibility of one rather than the other mutation. Moreover, whether the disease is the consequence of only one of these two mutations or of their combined effect remains questionable.
Table 1.
DYNC2H1 Mutations Identified in the Five Families with ATD or SRP Type III
| Family (Diagnosis) | Nucleotide Change | Amino Acid Change | Location | Domain |
|---|---|---|---|---|
| Family 1 (ATD) | c.[5971A→T,11284A→G] homozygote | p.[Met1991Leu] | Exon 38 | P-loop NTPase |
| c.[5971A→T, 11284A→G] homozygote | p.[Met1991Leu] | Exon 78 | unknown | |
| Family 2 (ATD) | c.[654_655insTTTATAACTTGGACAGTCTATCCTTACTA]+[9044A→G] | p.[Glu219Leu fsX2]+[Asp3015Gly] | Exon 5+Exon 57 | dynein heavy chain, N-terminal region 1/ coiled-coil domain |
| Family 3 (SRP III) | c.[4610A→G]+[7382G→T] | p.[Gln1537Arg]+ [Gly2461Val] | Exon 30+Exon45 | Walker A motif/ unknown |
| Family 4 (ATD) | c.[3719T→C]+[10063G→T] | p.[Ile1240Thr]+ [Gly3355X] | Exon 25+Exon 66 | Dynein heavy chain, N-terminal region 2/ unknown |
| Family 5 (SRP III) | c.[5959A→G]+ [10130delT] | p.[Thr1987Ala]+[Leu3377CysfsX34] | Exon 38+Exon 67 | P-loop NTPase/ Dynein heavy chain, cytoskeleton region |
The DYNC2H1 locus was excluded in the other five consanguineous families, but direct sequencing in eight nonconsanguineous families identified DYNC2H1 mutations in four of the families (Figure 1). Mutations were present at the compound heterozygote state (Table 1), cosegregated with the disease, and were absent in 210 control chromosomes. Among the ten mutant genotypes, three were premature stop codon mutations and seven were missense mutations. The mutations were located throughout the gene. Of the seven missense mutations, six involved conserved amino acids across species (Met1991, Gln1537, Thr1987, Gly2461, Asp3015, Met3762; Figure 2). Apart from the Met1991 change, all of the mutations were predicted as probably damaging by the PolyPhen program.6
Figure 2.
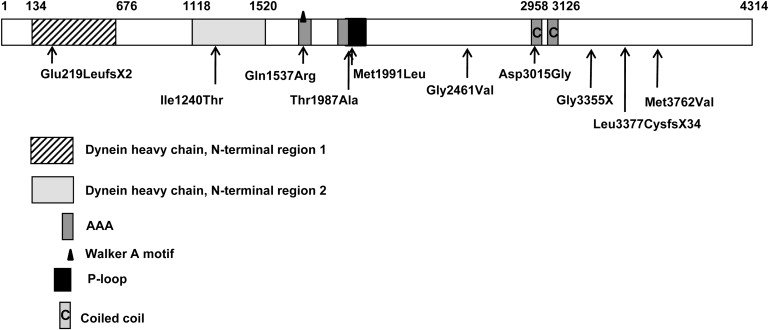
Location of the DYNC2H1 Mutations
Although no tridimensional structural templates were available, we modelized a short region of DYNC2H1 (residues Ile2967 to Val3137), using the microtubule-binding domain (MTBD) from mouse cytoplasmic dynein 1 heavy chain 1 (dync1h1) as a template.7 The 34% degree of alignment sequence identity between DYNC2H1 and the MTBD of dync1h1 was sufficient for homology modeling. The modelized region of DYNC2H1 is located between AAA5 (a non-nucleotide-binding AAA+ domain) and the dynein heavy chain domain. This region corresponds to the MTBD, part of the flexible coiled-coil region of the template, suggesting a possibly important functional role of this region. No experimental evidence was available for analysis of the functional impact of the p.Asp3015Gly mutation. We therefore used the program Swiss-Pdb Viewer 3.7 to compute possible hydrogen bonds on the bases of atom distances, atom angle, and atom types (O. Lund et al., 2002, CASP 5 conferenceA102 abstract; ref. 8) and found that Asp3015 can interact with Asn3070 (Figure 3). The p.Asp3015Gly mutation is expected to disrupt this hydrogen bond and to compromise the stability of the two α helices. We therefore speculate that this conformational change may alter the DYNC2H1 microtubule-binding capacity. To predict the putative impact of the other missense mutations on DYNC2H1 structure and function, we used the secondary structure prediction methods available on the Network Protein Sequence Analysis (NPS@) web server.9 The p.Ile1240Thr mutation localized in the dynein heavy chain, N-terminal region 2 and the p.Thr1987Ala and p.Met1991Leu mutations localized in the P loop NTPase are predicted to be in α helices. The p.Gln1537Arg and p.Gly2461Val mutations are localized, respectively, in the Walker A motif and in a nonconserved domain, and both are predicted to be in a random coil. Taken altogether, these data suggest that the six missense mutations may induce local conformational changes altering the function of DYNC2H1.
Figure 3.
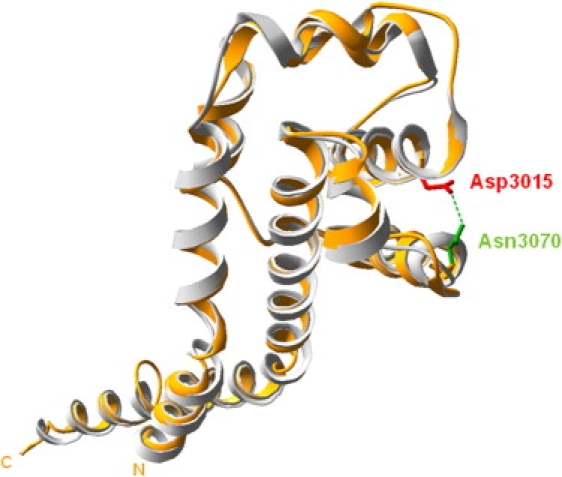
Tridimensional Structure Model of a DYNC2H1 Putative Microtubule-Binding Domain Via Swiss-Pdb Viewer 3.7 Representation
The modeled protein is represented in orange and is superimposed with the template (PDB code: 3err) shown in gray. Between Asp3015 (in red) and Asn3070 (in green), a putative hydrogen may be computed. The mutation p.Asp3015Gly disrupts this stabilizing hydrogen bond, inducing a local conformational change altering the function of this putative microtubule-binding domain.
We report here the identification of DYNC2H1 mutations in five distinct families with either ATD or SRP type III (Table 2 and Figures 4 and 5). Interestingly, while the submission of our manuscript was being processed, a similar paper was published by Merrill et al., reporting DYNC2H1 mutations in three families with SRP type III.10
Table 2.
Clinical Features of the Five Families with DYNC2H1 Mutations
| Family | Origin | CS | Age of Affected Patient(s) | Diagnosis | Polydactyly | Kidney Anomaly | Liver/Pancreas Microscopic Changes | Other Features |
|---|---|---|---|---|---|---|---|---|
| 1 | Morocco | Yes | Case 1: Death at day 2 | ATD | No | No | No | No |
| 1 | Morocco | Yes | Case 2 : TP at 28 wg | ATD | No | No | No | No |
| 2 | France | No | Case 1: TP at 28 wg | ATD | No | No | No | No |
| 2 | France | No | Case 2: TP at 24 wg | ATD | No | No | No | No |
| 3 | France | No | Case 1: TP at 25 wg | SRP type III | Postaxial and Bilateral | Tubular microcysts | Hepatic biliary, hyperplasia, periduodenal pancreas | Anal anteposition, micropenis |
| 4 | France | No | Case 1: 19 years | ATD | No | No | No | No |
| 5 | Madagascar | No | Case 1: TP at 24 wg | SRP type III | No | No | No | Ascites |
| 5 | Madagascar | No | Case 2 : TP at 22 wg | SRP type III | No | No | No | No |
| 5 | Madagascar | No | Case 3 : TP at 16 wg | SRP type III | No | No | No | No |
CS denotes consanguinity, TP denotes terminated pregnancy, wg denotes weeks of gestation.
Figure 4.
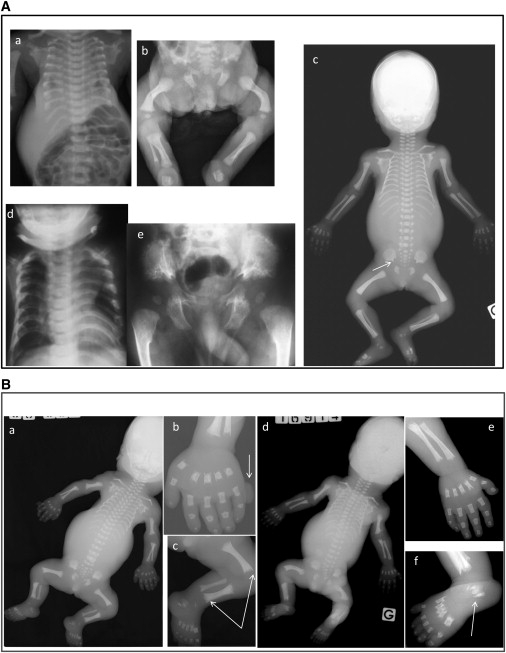
Radiological Findings in the Families with DYNC2H1 Mutations
(A) ATD cases. a and b: case 1 (day 2) from family 1; c: case 1 (28 wg) from family 2; d and e: affected child (4 mo) from family 4. Note the short bones, narrow thorax, and trident aspect of the acetabular roof (c, arrow). Note also the advanced proximal femoral ossification in the family 4 case (e).
(B) SRP cases. a–c: terminated pregnancy at 25 wg from family 3; d–f: case 1 from family 5. Note the severe shortness of the long bones, the narrow thorax, and the trident aspect of the acetabular roof. Note also the round metaphyseal ends with lateral spikes (c, arrows), the triplicate calcaneum (d, arrow), and the postaxial polydactyly (a, arrow).
Note the bowing of the femora observed in the cases from families 1–3 and 5 (Ab, Ac, Bc, Bd).
Figure 5.
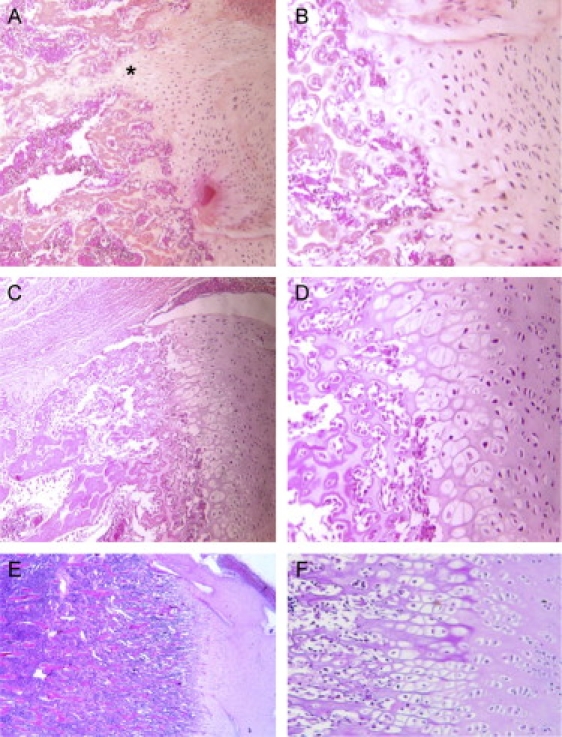
Femoral Histology in the Two SRP Type III Patients and in a Control Fetus
Physeal growth zone shows short and irregular columns in the proliferative zone in the family 5 case (A×10, B×20) and in the family 3 case (C×10, D×20), compared to a control of the same gestational age (E×10, F×20). Note also the more severe disorganization in the family 5 case, with associated elongated tongues of hypertrophic cartilage (asterisks) and patchy cellular proliferation.
Our findings further support the view that ATD and SRP type III are allelic disorders and belong to the same spectrum. ATD and SRP type III have been previously reported in the same family.4 Moreover, the same histological anomalies at the growth plate level have been reported in the two disorders, also suggesting that these conditions are variants of a single disorder, with SRP type III being at the more severe end of the spectrum (Figure 5).11 In the SRP cases (four fetuses belonging to families 3 and 5), the diagnosis was made antenatally (before 20 weeks of gestation [wg]). Postaxial polydactyly of the hands was present in one case (family 3 case), and other malformations included renal tubular microcysts, hepatic biliary hyperplasia (case 3), and unexplained ascites (case 1, family 5). The three ATD families included a total of five affected cases. In family 1, one child died of respiratory distress and pregnancy of her aunt was terminated at 28 wg for severe narrowing of the thorax. In family 2, two pregnancies were terminated for severe narrowing of the thorax, one at 28 wg and on at 26 wg. Finally, the family 4 case, the only survivor, is now 19 years old, and no eye, liver, or kidney manifestations have been hitherto detected.
This wide clinical variability prompted us to search for genotype-phenotype correlation. We identified a majority of missense mutations (7/10) in both ATD and SRP, as well as three nonsense mutations, also occurring in both ATD and SRP, with no obvious correlation between genotype and phenotype. The absence of homozygous nonsense mutations supports a partial loss of DYNC2H1 function. The same findings have been observed in the three ATD cases as a result of IFT80 mutations—two missense mutations and one in-frame deletion mutation have been found so far.1
The finding of DYNC2H1 mutations in ATD and SRP type III gives strong support to the view that these conditions belong to the ciliopathy group, as previously suggested by the discovery of IFT80 mutations in ATD. DYNC2H1 is a component of the cytoplasmic dynein complex, DYNC2, in association with the light intermediate chain (DYNC2LI1) and is directly involved in the contacts and translocation of the dynein complex along microtubules via its large motor domain.12 Moreover, in mammalian tissues, the colocalization of DYNC2H1, DYNC2LI1, and IFT pathway homologs supports a specific role in the generation and maintenance of mammalian cilia. Primary cilia and IFT proteins have been shown to be components of morphogenetic pathways (including Hedgehog and Wnt pathways) essential for skeletal development, and IFT proteins have been shown to be involved in chondrocyte maturation through bone morphogenetic protein (BMP) signaling.13,14 The findings of DYNC2H1 mutations in ATD and SRP type III give support to the role of cilia and IFT proteins in endochondral bone formation. The presence of polydactyly also suggests a requirement for DYNC2H1 in Sonic Hedgehog signaling.15
In conclusion, this study demonstrates that ATD and SRP type III belong to the same heterogeneous spectrum of conditions caused by either IFT80 or DYNC2H1 mutations and suggests a role for altered primary cilium function in these disorders. Yet, the presence of nine families with ATD and/or SRP type III unlinked to IFT80 and DYNC2H1 supports the genetic heterogeneity of this condition. Ongoing studies will hopefully lead to identification of other disease gene(s) presumably also involved in primary cilia function.
Acknowledgments
Part of this work has been supported by a national grant from Programme Hospitalier de Recherche Clinique (PHRC AOM06031).
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
Cilia database: ExPASy, http://www.expasy.ch/sprot/; Zebrafish and comparative genomics blast, http://danio.mgh.harvard.edu/blast/blast_grp.html
Ensembl Human Genome server, http://ensembl.org/
MultAlin, http://bioinfo.genotoul.fr/multalin/
Network Protein Sequence Analysis (NPS@), http://npsa-pbil.ibcp.fr/
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
PolyPhen, http://genetics.bwh.harvard.edu/pph/
Primer3, http://frodo.wi.mit.edu/
University of California, Santa Cruz (UCSC) Genome Browser, http://genome.ucsc.edu/
References
- 1.Beales P.L., Bland E., Tobin J.L., Bacchelli C., Tuysuz B., Hill J., Rix S., Pearson C.G., Kai M., Hartley J. IFT80, which encodes a conserved intraflagellar transport protein, is mutated in Jeune asphyxiating thoracic dystrophy. Nat. Genet. 2007;39:727–729. doi: 10.1038/ng2038. [DOI] [PubMed] [Google Scholar]
- 2.Morgan N.V., Bacchelli C., Gissen P., Morton J., Ferrero G.B., Silengo M., Labrune P., Casteels I., Hall C., Cox P. A locus for asphyxiating thoracic dystrophy, ATD, maps to chromosome 15q13. J. Med. Genet. 2003;40:431–435. doi: 10.1136/jmg.40.6.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Superti-Furga A., Unger S. Nosology and classification of genetic skeletal disorders: 2006 revision. Am. J. Med. Genet. 2007;143:1–18. doi: 10.1002/ajmg.a.31483. [DOI] [PubMed] [Google Scholar]
- 4.Ho N.C., Francomano C.A., van Allen M. Jeune asphyxiating thoracic dystrophy and short-rib polydactyly type III (Verma-Naumoff) are variants of the same disorder. Am. J. Med. Genet. 2000;90:310–314. doi: 10.1002/(sici)1096-8628(20000214)90:4<310::aid-ajmg9>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 5.Pfister K.K., Shah P.R., Hummerich H., Russ A., Cotton J., Annuar A.A., King S.M., Fisher E.M. Genetic analysis of the cytoplasmic dynein subunit families. PLoS Genet. 2006;2:e1. doi: 10.1371/journal.pgen.0020001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramensky V., Bork P., Sunyaev S. Human non-synonymous SNPs: Server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carter A.P., Garbarino J.E., Wilson-Kubalek E.M., Shipley W.E., Cho C., Milligan R.A., Vale R.D., Gibbons I.R. Structure and functional role of dynein's microtubule-binding domain. Science. 2008;322:1647–1648. doi: 10.1126/science.1164424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arnold K., Bordoli L., Kopp J., Schwede T. The SWISS-MODEL Workspace: A web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 9.Combet C., Blanchet C., Geourjon C., Deléage G. NPS@: Network protein sequence analysis. Trends Biochem. Sci. 2000;25:147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 10.Merrill A.E., Merriman B., Farrington-Rock C., Camacho N., Sebald E.T., Funari V.A., Schibler M.J., Firestein M.H., Cohn Z.A., Priore M.A., Thompson A.K. Ciliary abnormalities due to defects in the retrograde transport protein DYNC2H1 in short-rib polydactyly syndrome. Am. J. Hum. Genet. 2009;84:542–549. doi: 10.1016/j.ajhg.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang S.S., Langer L.O., Jr., Cacciarelli A., Dahms B.B., Unger E.R., Roskamp J., Dinno N.D., Chen H. Three conditions in neonatal asphyxiating thoracic dysplasia (Jeune) and short rib-polydactyly syndrome spectrum: A clinicopathologic study. Am. J. Med. Genet. 1987;3:191–207. doi: 10.1002/ajmg.1320280523. [DOI] [PubMed] [Google Scholar]
- 12.Porter M.E., Bower R., Knott J.A., Byrd P., Dentler W. Cytoplasmic dynein heavy chain 1b is required for flagellar assembly in Chlamydomonas. Mol. Biol. Cell. 1999;10:693–712. doi: 10.1091/mbc.10.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haycraft C.J., Zhang Q., Song B., Jackson W.S., Detloff P.J., Serra R., Yoder B.K. Intraflagellar transport is essential for endochondral bone formation. Development. 2007;134:307–316. doi: 10.1242/dev.02732. [DOI] [PubMed] [Google Scholar]
- 14.Gouttenoire J., Valcourt U., Bougault C., Aubert-Foucher E., Arnaud E., Giraud L., Mallein-Gerin F. Knockdown of the intraflagellar transport protein IFT46 stimulates selective gene expression in mouse chondrocytes and affects early development in zebrafish. J. Biol. Chem. 2007;282:30960–30973. doi: 10.1074/jbc.M705730200. [DOI] [PubMed] [Google Scholar]
- 15.Ocbina P.J., Anderson K.V. Intraflagellar transport, cilia, and mammalian Hedgehog signaling: Analysis in mouse embryonic fibroblasts. Dev. Dyn. 2008;237:2030–2038. doi: 10.1002/dvdy.21551. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


