Abstract
Asthma, a chronic airway disease with known heritability, affects more than 300 million people around the world. A genome-wide association (GWA) study of asthma with 359 cases from the Childhood Asthma Management Program (CAMP) and 846 genetically matched controls from the Illumina ICONdb public resource was performed. The strongest region of association seen was on chromosome 5q12 in PDE4D. The phosphodiesterase 4D, cAMP-specific (phosphodiesterase E3 dunce homolog, Drosophila) gene (PDE4D) is a regulator of airway smooth-muscle contractility, and PDE4 inhibitors have been developed as medications for asthma. Allelic p values for top SNPs in this region were 4.3 × 10−07 for rs1588265 and 9.7 × 10−07 for rs1544791. Replications were investigated in ten independent populations with different ethnicities, study designs, and definitions of asthma. In seven white and Hispanic replication populations, two PDE4D SNPs had significant results with p values less than 0.05, and five had results in the same direction as the original population but had p values greater than 0.05. Combined p values for 18,891 white and Hispanic individuals (4,342 cases) in our replication populations were 4.1 × 10−04 for rs1588265 and 9.2 × 10−04 for rs1544791. In three black replication populations, which had different linkage disequilibrium patterns than the other populations, original findings were not replicated. Further study of PDE4D variants might lead to improved understanding of the role of PDE4D in asthma pathophysiology and the efficacy of PDE4 inhibitor medications.
Introduction
Asthma (MIM 600807), a chronic respiratory disease resulting from the complex interaction of multiple genetic and environmental factors, affects more than 20 million Americans and 300 million people worldwide.1,2 In candidate-gene association and linkage studies, more than 40 genes have been associated with asthma and replicated in at least one independent population.3 Recently, the first genome-wide association (GWA) study of asthma found that ORMDL3 (MIM 610075) variants contribute to the risk of childhood-onset asthma,4 and its results have been successfully replicated in at least eight populations.5–10 Another GWA study in the Hutterites, a founder population of European descent, found that CHI3L1 (MIM 601525) variants are associated with asthma and related phenotypes.11 Its findings were replicated with partial success in two independent cohorts.11 The results of these studies have added to the growing credibility of the GWA approach for uncovering novel disease variants in complex diseases.12 In this work, we describe the results of a GWA study in which we found association between asthma and single-nucleotide polymorphisms (SNPs) in the phosphodiesterase 4D, cAMP-specific (phosphodiesterase E3 dunce homolog, Drosophila) gene (PDE4D [MIM 600129]). PDE4D is a compelling asthma candidate gene because its protein products are involved in the regulation of airway smooth-muscle contractility.13,14
Material and Methods
Subjects
Cases were 422 non-Hispanic white subjects from CAMP, a clinical trial that followed 1,041 asthmatic children for 4 years and nearly 80% of the original participants for 12 years.15 Stringent inclusion criteria ensured that participants had mild to moderate asthma, which was defined as asthmatic symptoms at least twice per week and either use of asthma medication daily or use of an inhaled bronchodilator twice per week for six or more months of the year prior to recruitment. CAMP subjects had increased airway responsiveness, as established by a bronchoprovocation test in which there was 20% or greater FEV1 reduction after administration of up to 12.5 mg/dl of methacholine. CAMP participants and their parents provided DNA for genetic studies. Although CAMP was designed for family-based genetic studies, this design was underpowered for the measurement of associations to asthma. We sought to increase the ability to find such associations by comparing CAMP probands to publicly available controls. Genotype data for 1,533 white control subjects were obtained from Illumina's iControlDB resource.
Genotyping and Quality Control
Genome-wide SNP genotyping for CAMP subjects, their families, and iControlDB controls was performed on Illumina's HumanHap550 Genotyping BeadChip (Illumina, Inc., San Diego, CA). CAMP samples and markers passed stringent quality-control standards; 6,257 markers were removed as a result of low clustering scores. Markers whose flanking sequences did not map to a unique position on the HG17 reference genome sequence were removed (n = 1,329). Further quality control was performed with PLINK version 1.03.16 The average completion rate for each marker was more than 99%. Monomorphic markers (n = 3,790) and those with five or more Mendel errors (n = 2,445) were removed. We assessed genotype reproducibility by plating four subjects once on each of 14 genotyping plates. All of these replicates had at least 99.8% concordance. The average genotyping completion rate for each subject was 99.75%.
In addition to undergoing quality-control procedures at Illumina (Illumina, Inc., San Diego, CA), controls from iControlDB were processed with equal quality-control standards as CAMP cases, except for searches of Mendelian errors because family data were not available. CAMP cases and iControlDB controls had 547,497 overlapping SNPs that passed these initial filters.
Additional filtering of subjects (cases plus controls) and SNPs was performed with PLINK. Subjects were dropped from further analysis for the following reasons: they were siblings of other subjects (23 cases), more than 5% of genotype data was missing (19 cases), there was evidence of identity by descent (IBD) with Pi-hat > 0.01 (57 controls), or there was discordance between reported and observed sex (three controls). SNPs were dropped from further analysis for the following reasons: they were missing in more than 5% of subjects (n = 3,837), minor allele frequency (MAF) was less than 1% (n = 17,088), Hardy-Weinberg equilibrium p values among controls were less than 0.001 (n = 2,046), the missing rate in cases and controls was significantly different as determined by a p value less than 10−5 (n = 6,642). Evidence of population stratification was present in the remaining 1,913 subjects (380 cases and 1,533 controls) and 518,230 SNPs, as indicated by a genomic inflation factor of 1.33 obtained in PLINK from unadjusted Chi-square values for allelic association of a subset of 28,189 independent SNPs (r2 < 0.07).
In order to control for population stratification, we used genetic matching (GEM).17 This approach removes genetic outliers and matches subjects on the basis of genetic similarities derived from eigenvector decomposition. A subset of 22,828 independent SNPs, which all had a pairwise r2 less than 0.07 among controls in PLINK, were selected for use in GEM. The initial matrix created with data on the independent SNPs had three significant eigenvectors, and the initial outlier-removal step in GEM, which removed subjects who were 6 SD or further along any of the significant eigenvector directions, resulted in the exclusion of three cases and 29 controls. Next, we selected a GEM clustering coefficient of 0.01, which excluded an additional 18 cases and 658 controls for whom distances from the case to the nearest control and vice-versa in the eigenspace composed of the three significant eigenvectors was larger than the 0.01 cutoff. The genomic inflation factor for the remaining 1,205 subjects (359 cases and 846 controls) was 1.03, demonstrating minimal population stratification. To further verify that population stratification among these subjects was minimal, we used EIGENSTRAT to obtain unadjusted Cochran-Armitage trend test and EIGENSTRAT-corrected association statistics.18 Comparison of the corrected and uncorrected association statistics demonstrated that uncorrected test statistics changed minimally with further correction for population stratification. Therefore, EIGENSTRAT was not used for adjusting statistics in subsequent analyses. The 1,205 subjects (359 cases, 846 controls) that were used in subsequent statistical analyses are referred to as the CAMP/Illumina study.
Statistical Analysis
Figure 1 is an overview of our study design. First, we performed a case-control analysis with CAMP/Illumina (Figure 1A). We selected SNPs for further consideration by choosing a nominal p value cutoff (1× 10−6) based on the allelic p value distribution. After identifying top case-control associations, we performed family-based association tests to ensure that associations remained in parent-offspring trios (Figure 1B). Case-control associations were measured in PLINK16; allelic tests were used for screening, and Cochran-Armitage trend tests were used for comparison with family-based studies. Family-based association statistics for 387 CAMP trios under an additive model were calculated with Golden Helix PBAT version 6.4.0.19 Replication of association results was attempted in ten independent populations (Figure 1C). Haploview20 was used for inferring linkage disequilibrium (LD) among SNPs from r2 measures in family data. We measured joint evidence for association across populations by using Fisher's combined probability method21 to combine p values (Figure 1D). In the combining of p values, all hypothesis tests in replication populations had one-sided alternatives (based on the direction of the association in the testing population) so that SNPs with association tests in opposite directions would not produce inappropriately small p values. Effect estimates were calculated with allelic odds ratios (ORs) for case-control data and transmitted-to-untransmitted (T:U) ratios for family data. T:U ratios were calculated in Haploview. Summary ORs, based on allelic ORs for case-control and population-based studies, were estimated with the DerSimonian-Laird22 random-effects meta-analysis approach as implemented in the rmeta package in R.23 Power calculations were performed with the genetic power calculator of Purcell S, et al.24 with a high-risk allele frequency of 0.30, prevalence of 0.10, D-prime of 1, marker allele frequency of 0.30, an assumption of the use of unselected controls, and default error rates (alpha = 0.05, power = 0.80). In CAMP families, the association between SNPs and the log-transformed methacholine concentration that caused a 20% decrease in FEV1 was measured under an additive model via generalized estimating equations in Golden Helix PBAT version 6.4.0,19 in which adjustments were made for age and gender.
Figure 1.
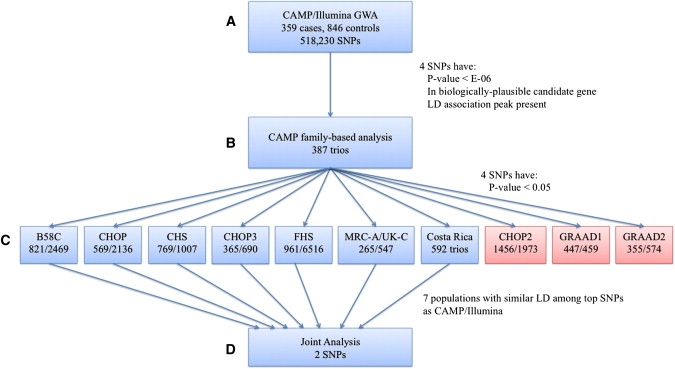
Study Overview
(A) 359 CAMP cases and 846 Illumina controls were used in tests for the genome-wide association of individual SNPs to asthma status. Four SNPs with p values less than 10−6 in a biologically plausible candidate gene and in an LD association peak were analyzed further.
(B) Using genome-wide association results of family-based statistical analysis in CAMP probands and their parents, we analyzed the four SNPs that met the criteria described in (A). These four SNPs had p values less than 0.05.
(C) Generalizability of the findings was tested through replication attempts in ten independent populations. Numbers under each name are cases/controls for case-control designs; affected/unaffected for population-based designs and GRAAD2; and number of trios for Costa Rica. Data for two SNPs were available in the ten populations.
(D) Joint association analysis for the two SNPs in seven populations with similar LD patterns among the top CAMP/Illumina SNPs was performed. Because LD patterns among the most significant SNPs were very different in black populations (red boxes), these were analyzed separately.
Replication Studies
(1) B58C. The British 1958 Birth Cohort is a nationally representative sample of people who were born during one week in 1958 and who have been followed periodically through childhood and adult life. Blood samples obtained at a medical examination at age 44–45 years25 were used as the basis of a DNA reference collection for genetic case-control studies. Information on wheezing illness and asthma was gathered through parental interviews at ages 7, 11, and 16 years and through cohort-member interviews at ages 23, 33, and 42 years, as described in detail elsewhere.26 For this study, cases were 821 individuals who had asthma or wheezing by age 16 and controls were 2,469 individuals who did not have asthma or wheezing by the last cohort interview. Cases and controls were determined to be white by self-report as adults and by school medical examinations in childhood. Genotype data was obtained with the Illumina HumanHap550 Genotyping BeadChip. Association between individual SNPs and asthma status was measured with the Cochran-Armitage trend test implemented in R.23
(2) CHOP. North American white children of Northern European descent were recruited at the Children's Hospital of Philadelphia (CHOP) between 2006 and 2008.5 Cases included 569 patients with physician-diagnosed persistent asthma necessitating regular administration of inhaled glucocorticoid medications for symptom control. Disease severity matched steps 2–6 as reported in the Asthma Expert Panel-3 guidelines.27 Controls included 2,136 subjects who were determined to have no history of asthma or reactive airway disease by questionnaire and who had never been prescribed asthma medications according to their medical records. Mean age of cases was 8.7 ± 5.7 SD years, and 58.7% were male; the mean age of the controls was 8.4 ± 6.1 SD years, and 49.4% were male. Genome-wide genotyping was performed at the Center for Applied Genomics on the Illumina HumanHap550 Genotyping BeadChip. Patients and controls were screened at ancestry-informative markers, and the corresponding genomic inflation factor, 1.07, demonstrated minimal background population stratification. Association between individual SNPs and asthma status was measured with the Cochran-Armitage trend test implemented in PLINK.
(3) CHOP2. African American children were recruited on the basis of criteria similar to those used for CHOP subjects.5 Cases included 1,456 patients with physician-diagnosed persistent asthma. Controls included 1,973 subjects who were determined to have no history of asthma or reactive airway disease by questionnaire and who had never been prescribed asthma medications according to their medical records. The mean age of cases was 7.5 ± 5.7 SD years, and 57% were male; the mean age of controls was 6.7 ± 5.2 SD years, and 49% were male. Genome-wide genotyping was performed at the Center for Applied Genomics on the Illumina HumanHap550 Genotyping BeadChip. Patients and controls were screened at ancestry-informative markers, and the corresponding genomic inflation factor, 1.1, demonstrated minimal background population stratification. Association between individual SNPs and asthma status was measured with the Cochran-Armitage trend test implemented in PLINK.
(4) CHOP3. This cohort consisted of 365 white cases who were of Northern European descent and who were recruited at CHOP. Cases had asthma according to physician diagnosis and used beta-agonist medications. Controls consisted of 690 white subjects who were recruited at CHOP, who were of Northern European descent, and who were determined to have no history of asthma or reactive airway disease by questionnaire and had never been prescribed asthma medications according to their medical records. CHOP cases and controls used in this cohort did not overlap with those in the CHOP population described in (2). Genome-wide genotyping was performed at the Center for Applied Genomics on the Illumina HumanHap550 Genotyping BeadChip. Subjects were screened at ancestry-informative markers, and the corresponding genomic inflation factor, 1.05, demonstrated minimal background population stratification. Association between individual SNPs and asthma status was measured with the Cochran-Armitage trend test implemented in PLINK.
(5) CHS. The Children's Health Study (CHS) is an ongoing cohort study in Southern California and investigates both genetic and environmental factors related to childhood asthma and lung function growth.28 The CHS GWAS was based on a nested case-control sample of 769 asthmatics and 1,007 controls selected from within the cohort. All subjects in this GWAS sample were either Hispanic white (n = 817) or non-Hispanic white (n = 959). On the basis of questionnaire responses, children were characterized as having doctor-diagnosed asthma at study entry or during active follow-up (cases) or as never having a diagnosis of asthma (controls). Buccal cells were collected from study subjects, and genotyping was performed by the USC Genomics Center with the Illumina HumanHap550 Genotyping BeadChip. Association between individual SNPs and asthma status was measured in R23 via logistic regression with additive genetic models; adjustments were made for age, sex, Hispanic status, community, and q factors obtained from STRUCTURE.29
(6) FHS. The Framingham Heart Study (FHS) conducted clinical examinations that include spirometry and collection of smoking history data on three generations of white adults of European descent, and research participants provided DNA samples that have recently been genotyped for genome-wide association studies.30,31 Asthma was classified on the basis of self-report of physician diagnosis, and according to this definition, there were 961 cases and 6,516 controls. In FHS subjects, genotyping was performed with the Affymetrix GeneChip Human Mapping 500K Array Set and an additional Affymetrix 50K Array (HuGeneFocused50K). Because data from these assays did not include that of the most associated SNPs in the CAMP/Illumina analysis, those genotypes were inferred via imputation with the Markov Chain Haplotyping software (MaCH).32 The ratio of the empirically observed dosage variance to the expected (binomial) dosage variance for these imputed SNPs was greater than 0.9, indicating good quality of imputation. Association to asthma was measured with logistic regression models, in which robust variance was estimated via generalized estimating equations with each pedigree as a cluster; adjustments were made for age, former smoking, current smoking, pack-years, sex, BMI, and membership in one of the three recruited generations. The genomic inflation factor for the imputed genome-wide results was 1.048, indicating minimal population stratification.
(7) MRC-A/UK-C. Study subjects include 378 children from the MRC-A cohort (265 cases and 113 controls) and 434 nonasthmatic white UK controls (UK-C) studied according to the same protocols.4 The MRC-A cohort was recruited through a proband with severe childhood onset asthma and consisted of 295 sibling pairs, 11 half-sibling pairs, and three singletons. Asthmatic subjects were defined as those who gave a positive response to the question, “Has your doctor ever told you that you have asthma”? Additionally, probands were determined to have severe asthma if they had step 3 or worse asthma according to British Thoracic Society guidelines (i.e., if they were on high-dose inhaled steroids or if they were on low-dose inhaled steroids and a long-acting beta-agonist). In MRC-A/UK-C subjects, genotyping was performed with the Illumina Sentrix HumanHap300 BeadChip.4 In MRC-A/UK-C subjects, association between individual SNPs and asthma status was measured via logistic regression models, and robust sandwich estimation of the variance as implemented in the Stata logit function was used for modeling the clustering of siblings' genotypes.4
(8) Costa Rica. This cohort consists of 592 probands from the Genetics of Asthma in Costa Rica Study, which is comprised of Costa Rican schoolchildren with asthma and their parents.33,34 Children had a high probability of having at least six great-grandparents born in the Central Valley of Costa Rica and were defined as having asthma if they had a doctor's diagnosis of asthma and at least two respiratory symptoms or asthma attacks in the year prior to enrollment in the study. Methacholine-challenge testing was performed in probands as described previously.35 Genotype data intended to replicate CAMP/Illumina findings was obtained with Taqman real-time PCR with an ABI Prism 7900 machine (Applied Biosystems, Foster City, CA). Standard PCR conditions, as recommended by the manufacturer, were used. Family-based association statistics for asthma affection status under an additive model were calculated with Golden Helix PBAT version 6.4.0.19 The association between SNPs and the log-transformed methacholine dose that caused a 20% decrease in FEV1 was measured under an additive model via generalized estimating equations in PBAT; adjustments were made for age and gender. T:U ratios of alleles were calculated in Haploview.20
(9) GRAAD1. The Genomic Research on Asthma in the African Diaspora (GRAAD) African American cohort consisted of 447 asthma cases and 459 nonasthmatic controls. Asthmatic subjects were defined as those who had both a reported history of asthma and a history of physician-diagnosed asthma (past/current). All controls except 50 were determined to be negative for a history of asthma; asthma status on the 50 controls participating in a study on the genetics of human pigmentation was unknown.36 Genotyping was performed with the Illumina HumanHap650Y BeadChip. Association between individual SNPs and asthma status was measured with the Cochran-Armitage trend test in PLINK.
(10) GRAAD2. The GRAAD Barbados cohort consisted of 929 subjects (355 asthmatics) from 163 families. These African Caribbean asthma probands and their nuclear and extended family members were recruited in Barbados through referrals at local polyclincs or the Accident and Emergency Department of the Queen Elizabeth Hospital.37 Asthmatic subjects were defined as those who had both a reported history of asthma and a documented history of physician-diagnosed asthma (past/current), plus a history of wheezing without an upper-respiratory infection (URI) or two out of four hallmark symptoms (wheezing with a URI, cough without a URI, shortness of breath, and tightness in the chest). Genotyping was performed with the Illumina HumanHap650Y BeadChip. Family-based association statistics under an additive model were calculated with FBAT.38 T:U ratios of alleles were calculated in Haploview.20
Each study was approved by the institutional review board of the corresponding Institution, which ensured that all procedures were in accordance with the ethical standards of the responsible committee on human experimentation. Informed consent was obtained for all study participants.
Results
After QC filters, 518,230 SNPs genotyped in 359 CAMP cases and 846 Illumina controls were used for testing the association of SNPs to asthma. There was little evidence of population stratification in these subjects, who were matched via GEM, as indicated by a genomic inflation factor of 1.03. Furthermore, EIGENSTRAT-corrected association statistics were highly correlated (r2 = 0.98) with unadjusted statistics (Figure 2). A quantile-quantile plot comparing allelic-association p values to those expected for a null distribution revealed some deviation of measures at the tail, indicating that true associations between SNPs and asthma may be present (Figure 3). The top five SNPs, whose p values deviated most from the distribution of all p values and had nominal p values less than 1 × 10−6, were located on Chromosomes 5 and 8 (Table 1; Figure 4).
Figure 2.
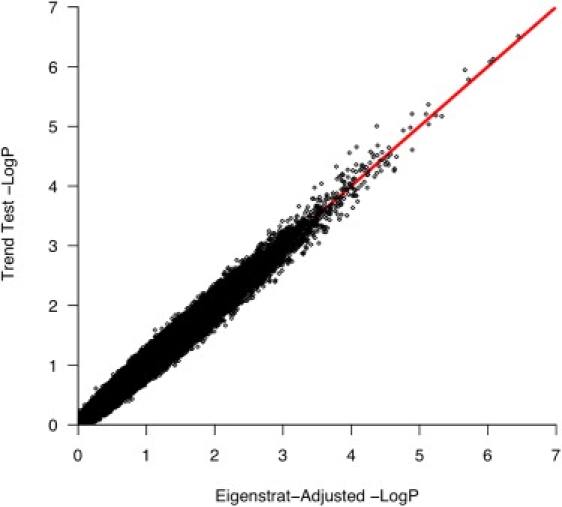
EIGENSTRAT-Corrected Association Statistics
CAMP/Illumina GWA statistics as adjusted by EIGENSTRAT are highly correlated (r2 = 0.98) with unadjusted statistics, demonstrating minimal residual population stratification.
Figure 3.
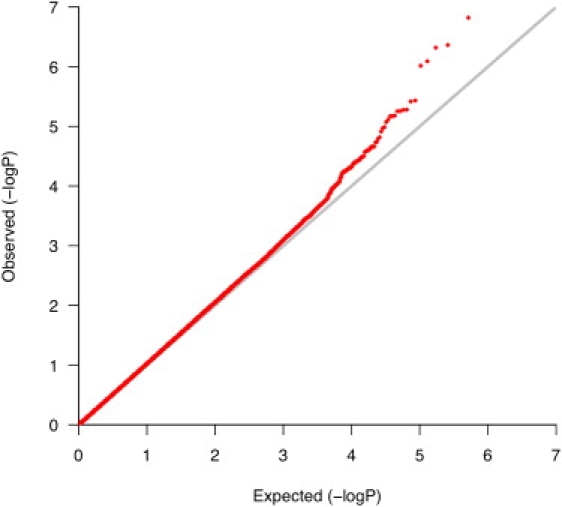
Quantile-Quantile Plot
Comparison of CAMP/Illumina allelic p values to those expected for a null distribution. Deviation of measures at the tail may indicate true associations between some SNPs and asthma.
Table 1.
Top 5 SNPs that Were Individually Associated with Asthma According to Allelic-Model p Values in CAMP/Illumina GWA Study
| SNP | CHR | BP | HWE p Value | Minor Allele | MAF Cases | MAF Controls | Allelic p Value | OR (95% CI) |
|---|---|---|---|---|---|---|---|---|
| rs2548659 | 5 | 59419643 | 0.45 | G | 0.23 | 0.34 | 1.5 × 10−7 | 0.58 (0.48–0.71) |
| rs1588265 | 5 | 59405551 | 0.68 | G | 0.23 | 0.34 | 4.3 × 10−7 | 0.60 (0.49–0.73) |
| rs983280 | 5 | 59480894 | 0.63 | G | 0.23 | 0.34 | 4.8 × 10−7 | 0.60 (0.49–0.73) |
| rs11778371 | 8 | 27375822 | 1 | A | 0.09 | 0.04 | 8.1 × 10−7 | 2.32 (1.65–3.27) |
| rs1544791 | 5 | 59474839 | 0.63 | A | 0.24 | 0.34 | 9.7 × 10−7 | 0.61 (0.54–0.78) |
Minor allele corresponds to Illumina's top-strand convention. CHR: chromosome. BP: base pairs along corresponding chromosome. HWE: Hardy-Weinberg equilibrium. MAF: minor-allele frequency. OR: odds ratio.
Figure 4.
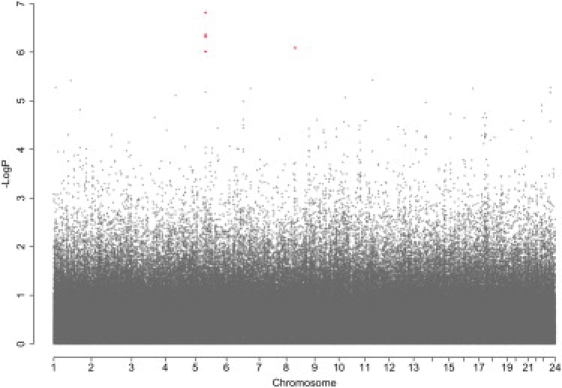
GWA Results for CAMP/Illumina
The x axis denotes the position along each chromosome. The y axis denotes −Log10(P), corresponding to allelic p values. Five SNPs have nominal p values less than 1 × 10−6 (shown in red); four are located on chromosome 5, and one is on chromosome 8.
Initially, we substantiated case-control results by comparing them to family-based statistical analysis by using CAMP probands and their parents. The top case-control SNPs had significant (i.e., < 0.05) PBAT additive-model p values in the CAMP families, although they did not rank highest on the list (Table 2). Instead, they ranked between positions 2,947 and 10,852 out of more than 500,000 SNPs. Comparison of CAMP-family T:U ratios to allelic ORs obtained in CAMP/Illumina reveals that the effect estimate is consistent but smaller in the family-based analysis. Although the trio results are not entirely independent of the CAMP/Illumina results because CAMP probands were used for both analyses, they add credibility to the CAMP/Illumina findings. We proceeded by attempting to replicate the chromosome 5, rather than chromosome 8, SNP findings because (1) these four SNPs, which are in nearly complete LD with each other, are in an LD peak, whereas other SNPs are more weakly associated with asthma (Figure 5), and (2) the chromosome 5 LD peak is in PDE4D, which is a plausible biological candidate gene for asthma because it is an important regulator of airway smooth-muscle contractility.13,14
Table 2.
Family-Based Analysis of Top PDE4D SNPs from CAMP/Illumina GWA Study in CAMP Trios
| SNP | CHR | PBAT Additive Model p Value | Family-Based Analysis Ranking | Minor-Allele T:U Ratio |
|---|---|---|---|---|
| rs2548659 | 5 | 0.006 | 3,122 | 0.73 |
| rs1588265 | 5 | 0.009 | 5,000 | 0.75 |
| rs983280 | 5 | 0.019 | 10,253 | 0.77 |
| rs11778371 | 8 | 0.006 | 2,947 | 1.76 |
| rs1544791 | 5 | 0.020 | 10,852 | 0.77 |
Family-based analysis ranking of SNPs among the GWA set based on PBAT additive-model p values shows that the SNPs were not highest on the list, as they were in CAMP/Illumina. The minor-allele T:U ratio demonstrates that the direction of the effect was equal to, but the magnitude was lower than, that measured in CAMP/Illumina.
Figure 5.
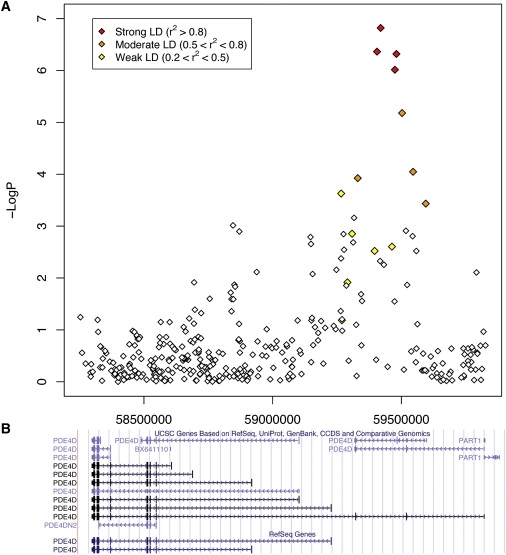
Region of Association of PDE4D SNPs to Asthma in CAMP/Illumina GWAS
(A) The x axis denotes the position along chromosome 5. The y axis denotes –Log10(P), corresponding to allelic p values. LD between the SNP with the lowest p value (rs1544791) to each SNP in the plot is denoted in colors (red, strong LD; orange, moderate LD; and yellow, weak LD).
(B) Corresponding location of genomic sequences available in the UCSC genome browser.
The generalizability of CAMP/Illumina chromosome 5 findings was tested by replication attempts in ten independent populations with diverse ethnicities. LD patterns of the top SNPs were obtained for family-based populations and for European American (CEU), Chinese and Japanese (CHB+JPT), and Yoruban (YRI) HapMap39 populations (Figure 6). Because Costa Rica had LD patterns in the top PDE4D SNPs that were very similar to those of CAMP and CEU, the Hispanic populations were analyzed in conjunction with the white populations. However, because the LD patterns in YRI and GRAAD2 were extremely different from those of other populations, the black populations were analyzed separately. We focused our results on two PDE4D SNPs that had genotype data in all populations.
Figure 6.
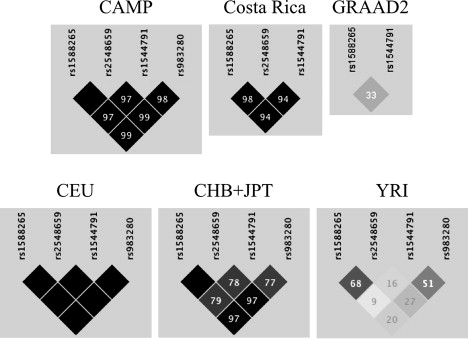
Linkage Disequilibrium among Top Chromosome 5 CAMP/Illumina SNPs
Panels are for six family-based populations: CAMP, Costa Rica, GRAAD2, HapMap European American (CEU), Chinese and Japanese (CHB+JPT), and Yoruban (YRI). Values in boxes are r2 measures on a decimal scale (i.e., 97 represents r2 = 0.97). Boxes without numbers have r2 = 1. The pattern of LD is very similar in CAMP, Costa Rica, and CEU. There are moderate differences in LD between CHB+JPT and each of the CAMP, Costa Rica, and CEU populations. The patterns in GRAAD2 and YRI are extremely different from those of the other populations. Genotype data for three of the four SNPs were available in Costa Rica. Genotype data for two of the four SNPs were available in GRAAD2.
Among white and Hispanic populations, all replication attempts had effects in the same direction as CAMP/Illumina (Table 3). Two of these (FHS, CHOP) had individual p values less than 0.05, and five (CHS, CHOP3, Costa Rica, B58C and MRCA/UKC) had p values that were not less than 0.05. The combined p values for 18,891 white and Hispanic individuals (4,342 cases) in these replication populations were 4.1 × 10−4 for rs1588265 and 9.2 × 10−4 for rs1544791. Overall, if CAMP/Illumina are included in the joint calculation, the p values were 2.5 × 10−8 for rs1588265 and 1.2 × 10−7 for rs1544791. Summary ORs for the effects of these two SNPs in case-control and population-based studies were 0.85 (95% confidence interval [CI]: 0.77, 0.93) among all populations and 0.88 (95% CI: 0.83, 0.94) among replication populations for rs1588265, and they were 0.85 (95% CI: 0.78, 0.94) among all populations and 0.89 (95% CI: 0.84, 0.94) among replication populations for rs1544791 (Figure 7). The Costa Rica effect estimate, which was a T:U ratio, was not amenable to being combined with the ORs in the meta-analysis. In three black replication populations, original findings were not replicated (Table 4). Association of rs1588265 and rs1544791 to airway responsiveness was measured in CAMP and Costa Rica families, the only subjects with methacholine-challenge test data available, and neither group had significant p values (Table 5).
Table 3.
Association Results of PDE4D SNPs in Eight Independent Populations
| Study |
Sample Size |
Analysis |
rs1588265 |
rs1544791 |
Replication |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MAF Affected |
MAF Unaffected |
Effect Estimate |
p Value |
MAF Affected |
MAF Unaffected |
Effect Estimate |
p Value |
||||
| Case-Control Designs | |||||||||||
| CAMP/Illumina | 359; 846 | Trend test | 0.23 | 0.34 | 0.60 (0.48–0.71) | 5.10 × 10−7 | 0.24 | 0.34 | 0.61 (0.54–0.78) | 1.20 × 10−6 | Initial finding |
| B58C | 821; 2469 | Trend test | 0.3 | 0.31 | 0.93 (0.82–1.05) | 0.12 | 0.3 | 0.31 | 0.94 (0.84–1.07) | 0.18 | Nonreplication, same direction |
| CHOP | 569; 2136 | Trend test | 0.28 | 0.32 | 0.85 (0.73–0.98) | 0.016 | 0.29 | 0.32 | 0.86 (0.75–0.99) | 0.024 | Replication |
| CHOP3 | 365; 690 | Trend test | 0.31 | 0.32 | 0.95 (0.78–1.15) | 0.29 | 0.3 | 0.32 | 0.91 (0.75–1.10) | 0.27 | Nonreplication, same direction |
| CHS | 769; 1007 | LR | 0.28 | 0.3 | 0.86 (0.70–1.08) | 0.07 | 0.28 | 0.31 | 0.82 (0.65–1.02) | 0.07 | Nonreplication, same direction |
| Population-Based Designs | |||||||||||
| FHS | 961; 6516 | LR | 0.26 | 0.29 | 0.86 (0.76–0.97) | 0.006 | 0.26 | 0.29 | 0.85 (0.75–0.96) | 0.006 | Replication |
| MRC-A/UK-C | 265; 547 | LR | 0.29 | 0.32 | 0.87 (0.67–1.13) | 0.14 | 0.3 | 0.32 | 0.91 (0.70–1.18) | 0.24 | Nonreplication, same direction |
| Family-Based Designs | |||||||||||
| Costa Rica | 353/355 | PBAT | 0.32 | 0.33 | 0.9 | 0.12 | 0.32 | 0.33 | 0.91 | 0.12 | Nonreplication, same direction |
Sample sizes refer to the number of cases; controls in the case-control designs, affected; unaffected individuals in the population-based designs, and the number of informative families for SNPs rs1588265 and rs1544791, respectively, in the family-based designs. In the Analysis column, the method used for obtaining p values is described: “Trend test” refers to the Cochran-Armitage trend test; “LR” to logistic regression; and “PBAT” to the pedigree-based association test. Logistic regressions and PBAT used additive models. MAF unaffected in Costa Rica is the parental allele frequency. Effect estimates are as follows: allelic OR and corresponding 95% CI for studies that used trend tests; OR calculated from beta coefficient and corresponding 95% CI for studies that used LR models; and minor-allele T:U ratio for family-based designs. The overall p values for the independent populations were 4.1 × 10−4 for rs1588265 and 9.2 × 10−4 for rs1544791.
Figure 7.
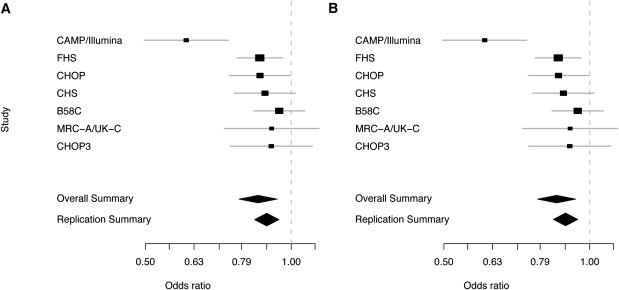
Summary ORs for Effects of PDE4D SNPs in Seven Populations
(A) Summary ORs for rs1588265 are 0.85 (95% CI: 0.77, 0.93) among all populations and 0.88 (95% CI: 0.83, 0.94) among replication populations.
(B) Summary ORs for rs1544791 are 0.85 (95% CI: 0.78, 0.94) among all populations and 0.89 (95% CI: 0.84, 0.94) among replication populations. Boxes are at the mean OR for each study, and their size is proportional to study size. Gray bars represent 95% CIs. The summary OR (diamond) was estimated with the DerSimonian-Laird random-effects model.
Table 4.
Association Results of PDE4D SNPs in Three Black Populations
| Population | Design | Sample Size | Analysis | rs1588265 |
rs1544791 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MAF Affected | MAF Unaffected | Effect Estimate | p Value | MAF Affected | MAF Unaffected | Effect Estimate | p Value | ||||
| CHOP2 | Case-control | 1456, 1973 | Trend test | 0.22 | 0.22 | 1.00 (0.89, 1.12) | 0.50 | 0.36 | 0.34 | 1.09 (0.99, 1.20) | 0.95 |
| GRAAD1 | Case-control | 447, 459 | Trend test | 0.20 | 0.21 | 0.94 (0.75, 1.19) | 0.22 | 0.35 | 0.35 | 1.00 (0.83–1.22) | 0.38 |
| GRAAD2 | Family-based | 77/88 | FBAT | 0.20 | 0.23 | 1.16 | 0.88 | 0.38 | 0.38 | 1.14 | 0.84 |
In the analysis column, “trend test” refers to the Cochran-Armitage trend test, and “FBAT” refers to the family-based association test. FBAT used additive models. MAF unaffected in GRAAD2 is the allele frequency among all unaffected family members. Sample size in GRAAD2 is that of informative families for each SNP. Effect estimates are as follows: allelic OR and corresponding 95% CI for studies that used trend tests and T:U ratio of the minor allele for GRAAD2 trios.
Table 5.
Family-Based Analysis of Airway Responsiveness Association to PDE4D SNPs in CAMP and Costa Rica Trios
| SNP | Minor Allele | CAMP p Value | Costa Rica p Value |
|---|---|---|---|
| rs1588265 | G | 0.42 | −0.74 |
| rs1544791 | A | 0.38 | −0.74 |
Association between SNPs and the log-transformed methacholine concentration (in CAMP) and dose (in Costa Rica) that caused a 20% decrease in FEV1 were measured under an additive model, in which adjustments were made for age and gender. No significant associations were found. The negative sign indicates association direction.
The association between SNPs and asthma at variants reported to modify asthma risk in previous GWA studies4,11 was measured in CAMP/Illumina (Table 6).
Table 6.
Attempted Replication of GWAS Results Initially Reported by Moffatt et al.4 for Chromosome 17 and Ober et al.11 for Chromosome 1 in CAMP/Illumina
| SNP | CHR | BP | Initial Report |
CAMP/Illumina p Value | |
|---|---|---|---|---|---|
| Primary GWA Study p Value | Replication Study p Value | ||||
| rs9303277 | 17 | 35229995 | 1.58 × 10−9 | 1.58 × 10−3 | 1.03 × 10−2 |
| rs11557467 | 17 | 35282160 | 7.94 × 10−10 | 7.94 × 10−4 | 6.03 × 10−3 |
| rs8067378 | 17 | 35304874 | 1.00 × 10−9 | 6.77 × 10−3 | |
| rs2290400 | 17 | 35319766 | 1.58 × 10−10 | 6.31 × 10−4 | 4.87 × 10−3 |
| rs7216389 | 17 | 35323475 | 1.00 × 10−10 | 7.94 × 10−4 | 1.60 × 10−3 |
| rs4795405 | 17 | 35341943 | 2.00 × 10−9 | 2.51 × 10−4 | 7.17 × 10−4 |
| rs8079416 | 17 | 35346239 | 6.31 × 10−9 | 1.26 × 10−2 | 1.28 × 10−2 |
| rs3894194 | 17 | 35375519 | 2.00 × 10−8 | 6.31 × 10−2 | 7.69 × 10−3 |
| rs3859192 | 17 | 35382174 | 5.01 × 10−8 | 3.98 × 10−1 | 2.79 × 10−3 |
| rs946263 | 1 | 201432004 | 0.008 | 0.31 | |
Discussion
The CAMP population was designed for and has often been used in family-based candidate-gene association studies.40–42 Recently, GWA data has been acquired for a portion of the cohort, namely, the white probands and their parents. Although the intention behind gathering these data was to conduct family-based genetic-association studies, preliminary analysis found that the data were gravely underpowered for the measurement of associations to asthma (data not shown). We sought to increase the ability to find associations to asthma by creating a case-control study with CAMP probands. The availability of population controls provided by Illumina through the iControlDB resource and the design of programs such as GEM to genetically match such populations to cases has made our analysis possible. Although it is optimal to design case-control genetic studies by carefully selecting cases and controls at the outset, gathering the same data for all subjects, and ensuring that controls do not share the same phenotype as the cases, the approach we have used is an alternative that may yield useful results for population designs that lack controls. In ongoing work, we are more thoroughly comparing the CAMP/Illumina results to the CAMP family-based ones.
In CAMP/Illumina, the top five p values, which are all smaller than our nominal cutoff of 1 × 10−6, do not reach genome-wide significance as defined by a traditional Bonferroni correction threshold near 1 × 10−7 (= 0.05/500,000). Despite not reaching this threshold, we proceeded to test for replication of PDE4D SNPs because (1) CAMP/Illumina was composed of relatively few subjects for a GWA study and therefore, although better powered than the family-based design, was not of high enough power to measure effect sizes near 1.0, which are expected for complex diseases12, and (2) PDE4D is a highly plausible candidate gene, which adds to the credibility of the association result. If the empirical values for CAMP/Illumina genotype-relative risks (i.e., 0.7 for heterozygotes and 0.5 for minor-allele homozygotes) are reflective of a true process, then despite its small size, CAMP/Illumina has a power of 0.96 to detect the effect measured. If, however, the true effects are more modest and closer to those measured in many of the replication populations (i.e., 0.82–0.95), then CAMP/Illumina would have nearly no power to measure the association, as reflected by power values as low as 0.10–0.25. The combined p values of our replication populations were less than 0.05, but only two out of seven of these populations have individual p values less than 0.05. As was the case for CAMP/Illumina, most of the populations used in this study are individually too small to be powered to find associations with SNPs whose effect sizes are in the range expected for a complex disease. Although there are many subjects overall, combining the populations to make overall conclusions about asthma is subject to error because of the different recruitment criteria of each study, the different study designs used (e.g., family-based versus case-control designs), and the wide range of geographic and environmental differences among populations. Even though PDE4D is a highly plausible asthma candidate gene, in the absence of biological evidence, we cannot exclude the possibility that the associations we measured were due to chance alone.
PDE4D, a gene whose sequence spans a 1.5 Mb region on chromosome 5q12, encodes several isoforms that play a role in tailoring cyclic adenosine monophosphate (cAMP) signaling.43,44 Due to the ubiquitous role of cAMP signaling in all cell types, it is likely that polymorphisms of PDE4D influence a wide variety of diseases. Previous studies have reported that polymorphisms of this gene are associated with stroke (MIM 601367), COPD (MIM 606963), and bone mineral density (MIM 601884). The initial report that a PDE4D haplotype consisting of one SNP (rs12188950) and a microsatellite (AC008818-1) were associated with stroke45 has been followed by at least 15 studies making similar inquiries.44,46 Because these studies, which differ considerably in stroke subtypes and genotypic polymorphisms investigated, have provided varying results for associations between PDE4D and stroke in populations of various origins, there is currently no conclusive evidence that PDE4D variants are associated with stroke.44,46 A small study reported the associations of one PDE4D SNP (rs829259) and a haplotype consisting of one PDE4D (rs10075508) and one interleukin 13 SNPs to COPD in a Japanese population, but neither association replicated in an Egyptian population.47 One PDE4D SNP (rs1498608) was found to be associated with bone mineral density in women of a discovery and independent populations, whereas a second independent cohort of women had an association in the opposite direction.48 To the best of our knowledge, PDE4D polymorphisms have not been previously associated with asthma.
The importance of PDE4D in airway contractility has been demonstrated in studies showing that it is the predominant phosphodiesterase expressed in mouse lung and that PDE4D knockout mice have a complete absence of airway response to methacholine.13,14 Clinical trials have tested PDE4 inhibitors for asthma and COPD treatment.49,50 Most PDE4 inhibitors reduce airway responsiveness and the recruitment of eosinophils to airways, two characteristic features of asthma.49,50 We found that four PDE4D SNPs were associated with asthma in a GWA study of 359 CAMP probands and 846 publicly available Illumina controls. Two of these SNPs had allelic p values of 4.3 × 10−7 (rs1588265) and 9.7 × 10−7 (rs1544791). After attempts to replicate the initial CAMP/Illumina findings in seven white and Hispanic independent populations, combined p values in the replication populations were 4.1 × 10−4 (rs1588265) and 9.2 × 10−4 (rs1544791). Including CAMP/Illumina in the joint calculation resulted in overall p values of 2.5 × 10−8 for rs1588265 and 1.2 × 10−7 for rs1544791.
Based on comparisons with the PDE4D sequence reported in the paper originally associating SNPs to stroke,45 the PDE4D SNPs associated to asthma are located near the 5′ end of PDE4D, as was the SNP originally associated with stroke, but more than 340 Kb away from it in the 3′ direction, whereas the asthma PDE4D SNPs are more than 1 Mb away from the SNPs reported to be associated to COPD and bone mineral density, which are at the 3′ end of PDE4D. The top four CAMP/Illumina PDE4D SNPs are located between two exons, referred to as D7-2 and D7-3 in Gretarsdottir et al.45, whereas the SNPs in moderate LD with them span these exons (Figure 5). The D7-2 and D7-3 exons are part of a long-form splice variant of PDE4D. Based on another PDE4D sequence, RefSeq version NM_001104631.1, the region of highest association is upstream of the 5′ UTR. If the association involving these SNPs is truly indicative of a process that affects asthma, then they could be in linkage disequilbrium with a sequence variant that affects the function or transcription of a particular PDE4D isoform. For instance, a PDE4D variant could result in a version of the protein that preferentially constricts airway smooth-muscle cells, leading to asthma. Alternatively, a sequence variant could lead to preferential transcription of a PDE4D isoform that is more effective at constricting airway smooth-muscle cells, thereby leading to asthma. Understanding the biological role that these associations may play in asthma will be challenging because of the large size of PDE4D (1.5 Mb) and the variety of PDE4D isoforms. Furthermore, as experiments in mice suggest, the cells where PDE4D variants are most likely to play an important role are airway smooth-muscle cells. Obtaining and performing experiments in such cells in humans is particularly challenging. Nonetheless, relating our specific PDE4D polymorphisms to biological function is critical for corroborating our association results and potentially increasing our knowledge of asthma pathophysiology and of the heterogeneity in efficacy of PDE4 inhibitor medications.
The observed effects corresponding to the PDE4D SNPs associated with asthma are small (Table 3). The summary ORs in all of the white and Hispanic populations, 0.85 (95% CI: 0.77, 0.93) for rs1588265 and 0.85 (95% CI: 0.78, 0.94) for rs1544791, reveal an overall protective effect of the minor alleles (Figure 7). The effect sizes measured in CAMP/Illumina, 0.60 (95% CI: 0.48-0.71) for rs1588265 and 0.61 (95% CI: 0.54-0.87) for rs1544791, were much smaller than those of the independent populations, which have overall ORs of 0.88 (95% CI: 0.83, 0.94) for rs1588265 and 0.89 (95% CI: 0.84, 0.94) for rs1544791. One important distinction between CAMP/Illumina and each of the other populations that could partially account for the difference in effect estimates is that the selection of CAMP cases included a methacholine-challenge test, whereas most other cases were selected on the basis of self-reported or doctor-diagnosed asthma only. Because PDE4D is involved in airway responsiveness and increased airway responsiveness was a selection criterion for CAMP participants, it is likely that the CAMP/Illumina population was biased toward the association of variants related to this phenotype. One could test this hypothesis by measuring the association of the PDE4D variants to airway responsiveness directly. Among the populations used, measures of airway responsiveness are available in CAMP and Costa Rica probands only. Family-based association analyses in CAMP and Costa Rica did not produce statistically significant results for airway responsiveness in either of these populations (Table 5). In CAMP, the minor alleles seem to be associated with less airway responsiveness (i.e., higher concentration of methacholine required for 20% drop in FEV1), which is expected for subjects with less severe asthma and is consistent with our finding that these SNPs have a protective effect in asthma affection status. However, the Costa Rica results have an opposite trend. These outcomes do not support our hypothesis that PDE4D variants are directly associated with airway responsiveness, but they are not sufficient to disprove it. Ideally, we would be able to test for the association of airway responsiveness over a range that is typical of both asthmatics and nonasthmatics. Neither with CAMP nor Costa Rica are we able to do this. Furthermore, because CAMP probands were selected to have hyperresponsiveness, we are unable to measure the association to airway responsiveness over a range that is reflective of all asthmatics in CAMP families. This illustrates a disadvantage of using unselected controls and comparing them to more thoroughly phenotyped cases: we are unable to test all of the phenotypes available in cases in the case/control design. In future studies, we hope to obtain a population in which we can more carefully test whether PDE4D variants are associated with airway responsiveness.
Three black populations were analyzed separately from the white and Hispanic populations because the LD pattern among the top PDE4D SNPs was different in these three populations (Table 4). The CAMP, Costa Rica, and CEU populations had very similar LD patterns in this region, which makes the information at any of the top four PDE4D SNPs nearly redundant (Figure 6). In contrast, the GRAAD2 and YRI LD patterns demonstrate that information at each of the four PDE4D SNPs is nearly independent of the others. The results from the two PDE4D SNPs that are available for the three black populations are therefore not informative about the two nongenotyped SNPs. This suggests that the two genotyped SNPs do not directly modify the asthma phenotype in any of the populations but rather that they are in LD with variants that directly modify asthma susceptibility. More extensive genotyping may help in the determination of whether other SNPs in PDE4D are associated with asthma, especially among black populations.
In addition to studying top CAMP/Illumina SNPs, we attempted to replicate findings from previous asthma GWA studies. Specifically, we measured association between asthma and ORMDL34 and CHI3L111 variants by looking at SNPs that passed QC filters in CAMP/Ilumina. Nine of the original ORMDL3 SNPs reported by Moffatt et al.4 were significant in CAMP/Illumina (p value < 0.05), whereas one CHI3L1 SNP reported by Ober et al.11 did not replicate (Table 6). These results further strengthen the hypothesis that ORMDL3 variants, or those in LD to them, are related to asthma susceptibility. Further genotype data are necessary to allow a more definitive statement about whether the Ober et al.11 findings hold in CAMP/Illumina.
In summary, we have found in a GWA study that SNPs in PDE4D seem to be associated with asthma, as indicated by allelic p values of 4.3 × 10−7 for rs1588265 and 9.7 × 10−7 for rs1544791. Combined p values across seven white and Hispanic independent populations were 4.1 × 10−4 for rs1588265 and 9.2 × 10−4 for rs1544791, suggesting that further study of PDE4D may lead to improved understanding of its role in asthma pathophysiology and the efficacy of PDE4 inhibitor medications.
Web Resources
The URLs for data presented herein are as follows:
British 1958 Birth Cohort, http://www.b58cgene.sgul.ac.uk/collection.php
Family Based Association Tests (FBAT), http://biosun1.harvard.edu/∼fbat/fbat.htm
Genetic Power Calculator, http://pngu.mgh.harvard.edu/∼purcell/gpc/
Illumina iControlDB resource, http://www.illumina.com/pages.ilmn?ID = 231
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim
Acknowledgments
We thank all CAMP subjects for their ongoing participation in this study. We acknowledge the CAMP investigators and research team, supported by the National Heart, Lung, and Blood Institute (NHLBI), for collection of CAMP Genetic Ancillary Study data. All work on data collected from the CAMP Genetic Ancillary Study was conducted at the Channing Laboratory of the Brigham and Women's Hospital under appropriate CAMP policies and human subject protections. The CAMP Genetics Ancillary Study is supported by U01 HL075419, U01 HL65899, P01 HL083069, R01 HL086601, and T32 HL07427 from the NHLBI, National Institutes of Health (NIH). Additional support was provided by the following NIH grants: R37 HL066289, 5R01HL087680 (NHLBI); 5P01ES011627, and 5P30ES007048 (National Institute of Environmental Health Sciences); and 2T15LM007092-16 (National Library of Medicine). We acknowledge use of genotype data from the B58C DNA collection, funded by the Medical Research Council grant G0000934 and the Wellcome Trust grant 068545/Z/02. We are grateful to John Todd, Diabetes & Inflammation Laboratory, University of Cambridge, and Panos Deloukas, Wellcome Trust Sanger Institute, for depositing genotype data on the B58C. K.C.B. was supported in part by the Mary Beryl Patch Turnbull Scholar Program. R.A.M. was supported by the Intramural Research Program of the National Human Genome Research Institute, NIH. We thank Georgia Dunston and Mezbah Faruque from Howard University, Washington, DC, for their vital contribution in the recruitment of the GRAAD1 cohort.
References
- 1.American Lung Association . Epidemiology and Statistics Unit, Research and Program Services, American Lung Association; New York, NY: 2006. Trends in Asthma Morbidity and Mortality. [Google Scholar]
- 2.Global Initiative for Asthma Management and Prevention . US Department of Health and Human Services; Bethesda, MD: 1995. NHLBI/WHO Workshop Report. National Institutes of Health. [Google Scholar]
- 3.Ober C., Hoffjan S. Asthma genetics 2006: The long and winding road to gene discovery. Genes Immun. 2006;7:95–100. doi: 10.1038/sj.gene.6364284. [DOI] [PubMed] [Google Scholar]
- 4.Moffatt M.F., Kabesch M., Liang L., Dixon A.L., Strachan D., Heath S., Depner M., von Berg A., Bufe A., Rietschel E. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 5.Sleiman P.M., Annaiah K., Imielinski M., Bradfield J.P., Kim C.E., Frackelton E.C., Glessner J.T., Eckert A.W., Otieno F.G., Santa E. ORMDL3 variants associated with asthma susceptibility in North Americans of European ancestry. J. Allergy Clin. Immunol. 2008;122:1225–1227. doi: 10.1016/j.jaci.2008.06.041. [DOI] [PubMed] [Google Scholar]
- 6.Galanter J., Choudhry S., Eng C., Nazario S., Rodriguez-Santana J.R., Casal J., Torres-Palacios A., Salas J., Chapela R., Watson H.G. ORMDL3 gene is associated with asthma in three ethnically diverse populations. Am. J. Respir. Crit. Care Med. 2008;177:1194–1200. doi: 10.1164/rccm.200711-1644OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tavendale R., Macgregor D.F., Mukhopadhyay S., Palmer C.N. A polymorphism controlling ORMDL3 expression is associated with asthma that is poorly controlled by current medications. J. Allergy Clin. Immunol. 2008;121:860–863. doi: 10.1016/j.jaci.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 8.Leung T.F., Sy H.Y., Ng M.C., Chan I.H., Wong G.W., Tang N.L., Waye M.M., Lam C.W. Asthma and atopy are associated with chromosome 17q21 markers in Chinese children. Allergy. 2009;64:621–628. doi: 10.1111/j.1398-9995.2008.01873.x. [DOI] [PubMed] [Google Scholar]
- 9.Wu H., Romieu I., Sienra-Monge J.J., Li H., Del Rio-Navarro B.E., London S.J. Genetic variation in ORM1-like 3 (ORMDL3) and gasdermin-like (GSDML) and childhood asthma. Allergy. 2008;64:621–628. doi: 10.1111/j.1398-9995.2008.01912.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirota T., Harada M., Sakashita M., Doi S., Miyatake A., Fujita K., Enomoto T., Ebisawa M., Yoshihara S., Noguchi E. Genetic polymorphism regulating ORM1-like 3 (Saccharomyces cerevisiae) expression is associated with childhood atopic asthma in a Japanese population. J. Allergy Clin. Immunol. 2008;121:769–770. doi: 10.1016/j.jaci.2007.09.038. [DOI] [PubMed] [Google Scholar]
- 11.Ober C., Tan Z., Sun Y., Possick J.D., Pan L., Nicolae R., Radford S., Parry R.R., Heinzmann A., Deichmann K.A. Effect of variation in CHI3L1 on serum YKL-40 level, risk of asthma, and lung function. N. Engl. J. Med. 2008;358:1682–1691. doi: 10.1056/NEJMoa0708801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manolio T.A., Brooks L.D., Collins F.S. A HapMap harvest of insights into the genetics of common disease. J. Clin. Invest. 2008;118:1590–1605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansen G., Jin S., Umetsu D.T., Conti M. Absence of muscarinic cholinergic airway responses in mice deficient in the cyclic nucleotide phosphodiesterase PDE4D. Proc. Natl. Acad. Sci. USA. 2000;97:6751–6756. doi: 10.1073/pnas.97.12.6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mehats C., Jin S.L., Wahlstrom J., Law E., Umetsu D.T., Conti M. PDE4D plays a critical role in the control of airway smooth muscle contraction. FASEB J. 2003;17:1831–1841. doi: 10.1096/fj.03-0274com. [DOI] [PubMed] [Google Scholar]
- 15.Childhood Asthma Management Program Research Group The Childhood Asthma Management Program (CAMP): Design, rationale, and methods. Control. Clin. Trials. 1999;20:91–120. [PubMed] [Google Scholar]
- 16.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luca D., Ringquist S., Klei L., Lee A.B., Gieger C., Wichmann H.E., Schreiber S., Krawczak M., Lu Y., Styche A. On the use of general control samples for genome-wide association studies: Genetic matching highlights causal variants. Am. J. Hum. Genet. 2008;82:453–463. doi: 10.1016/j.ajhg.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 19.Lange C., DeMeo D., Silverman E.K., Weiss S.T., Laird N.M. PBAT: Tools for family-based association studies. Am. J. Hum. Genet. 2004;74:367–369. doi: 10.1086/381563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 21.Fisher R.A. Combining independent tests of significance. Am. Stat. 1948;2:30. [Google Scholar]
- 22.DerSimonian R., Laird N. Meta-analysis in clinical trials. Control. Clin. Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 23.R Development Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2007. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 24.Purcell S., Cherny S.S., Sham P.C. Genetic Power Calculator: Design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 25.Strachan D.P., Rudnicka A.R., Power C., Shepherd P., Fuller E., Davis A., Gibb I., Kumari M., Rumley A., Macfarlane G.J. Lifecourse influences on health among British adults: Effects of region of residence in childhood and adulthood. Int. J. Epidemiol. 2007;36:522–531. doi: 10.1093/ije/dyl309. [DOI] [PubMed] [Google Scholar]
- 26.Butland B.K., Strachan D.P. Asthma onset and relapse in adult life: The British 1958 birth cohort study. Ann. Allergy Asthma Immunol. 2007;98:337–343. doi: 10.1016/S1081-1206(10)60879-4. [DOI] [PubMed] [Google Scholar]
- 27.National Asthma Education Program . US Department of Health and Human Services; Bethesda, MD: 2007. Expert Panel Report 3: Guidelines for the Diagnosis and Management of Asthma. National Institutes of Health. [Google Scholar]
- 28.Peters J.M., Avol E., Navidi W., London S.J., Gauderman W.J., Lurmann F., Linn W.S., Margolis H., Rappaport E., Gong H. A study of twelve Southern California communities with differing levels and types of air pollution. I. Prevalence of respiratory morbidity. Am. J. Respir. Crit. Care Med. 1999;159:760–767. doi: 10.1164/ajrccm.159.3.9804143. [DOI] [PubMed] [Google Scholar]
- 29.Pritchard J.K., Stephens M., Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cupples L.A., Arruda H.T., Benjamin E.J., D'Agostino R.B., Demissie S., DeStefano A.L., Dupuis J., Falls K.M., Fox C.S., Gottlieb D.J. Framingham Heart Study 100K SNP genome-wide association study resource: Overview of 17 phenotype working group reports. BMC Med. Genet. 2007;8(Suppl 1):S1. doi: 10.1186/1471-2350-8-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Splansky G.L., Corey D., Yang Q., Atwood L.D., Cupples L.A., Benjamin E.J., D'Agostino R.B., Fox C.S., Larson M.G., Murabito J.M. The Third Generation Cohort of the National Heart, Lung, and Blood Institute's Framingham Heart Study: Design, recruitment, and initial examination. Am. J. Epidemiol. 2007;165:1328–1335. doi: 10.1093/aje/kwm021. [DOI] [PubMed] [Google Scholar]
- 32.Willer C.J., Sanna S., Jackson A.U., Scuteri A., Bonnycastle L.L., Clarke R., Heath S.C., Timpson N.J., Najjar S.S., Stringham H.M. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hunninghake G.M., Soto-Quiros M.E., Avila L., Ly N.P., Liang C., Sylvia J.S., Klanderman B.J., Silverman E.K., Celedon J.C. Sensitization to Ascaris lumbricoides and severity of childhood asthma in Costa Rica. J. Allergy Clin. Immunol. 2007;119:654–661. doi: 10.1016/j.jaci.2006.12.609. [DOI] [PubMed] [Google Scholar]
- 34.Hunninghake G.M., Soto-Quiros M.E., Avila L., Su J., Murphy A., Demeo D.L., Ly N.P., Liang C., Sylvia J.S., Klanderman B.J. Polymorphisms in IL13, total IgE, eosinophilia, and asthma exacerbations in childhood. J. Allergy Clin. Immunol. 2007;120:84–90. doi: 10.1016/j.jaci.2007.04.032. [DOI] [PubMed] [Google Scholar]
- 35.Ly N.P., Soto-Quiros M.E., Avila L., Hunninghake G.M., Raby B.A., Laskey D., Sylvia J.S., Celedon J.C. Paternal asthma, mold exposure, and increased airway responsiveness among children with asthma in Costa Rica. Chest. 2008;133:107–114. doi: 10.1378/chest.07-2130. [DOI] [PubMed] [Google Scholar]
- 36.Bonilla C., Boxill L.A., Donald S.A., Williams T., Sylvester N., Parra E.J., Dios S., Norton H.L., Shriver M.D., Kittles R.A. The 8818G allele of the agouti signaling protein (ASIP) gene is ancestral and is associated with darker skin color in African Americans. Hum. Genet. 2005;116:402–406. doi: 10.1007/s00439-004-1251-2. [DOI] [PubMed] [Google Scholar]
- 37.Barnes K.C., Neely J.D., Duffy D.L., Freidhoff L.R., Breazeale D.R., Schou C., Naidu R.P., Levett P.N., Renault B., Kucherlapati R. Linkage of asthma and total serum IgE concentration to markers on chromosome 12q: Evidence from Afro-Caribbean and Caucasian populations. Genomics. 1996;37:41–50. doi: 10.1006/geno.1996.0518. [DOI] [PubMed] [Google Scholar]
- 38.Laird N.M., Horvath S., Xu X. Implementing a unified approach to family-based tests of association. Genet. Epidemiol. 2000;19(Suppl 1):S36–S42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 39.The International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 40.Silverman E.K., Kwiatkowski D.J., Sylvia J.S., Lazarus R., Drazen J.M., Lange C., Laird N.M., Weiss S.T. Family-based association analysis of beta2-adrenergic receptor polymorphisms in the childhood asthma management program. J. Allergy Clin. Immunol. 2003;112:870–876. doi: 10.1016/s0091-6749(03)02023-2. [DOI] [PubMed] [Google Scholar]
- 41.DeMeo D.L., Lange C., Silverman E.K., Senter J.M., Drazen J.M., Barth M.J., Laird N., Weiss S.T. Univariate and multivariate family-based association analysis of the IL-13 ARG130GLN polymorphism in the Childhood Asthma Management Program. Genet. Epidemiol. 2002;23:335–348. doi: 10.1002/gepi.10182. [DOI] [PubMed] [Google Scholar]
- 42.Randolph A.G., Lange C., Silverman E.K., Lazarus R., Silverman E.S., Raby B., Brown A., Ozonoff A., Richter B., Weiss S.T. The IL12B gene is associated with asthma. Am. J. Hum. Genet. 2004;75:709–715. doi: 10.1086/424886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Houslay M.D., Baillie G.S., Maurice D.H. cAMP-Specific phosphodiesterase-4 enzymes in the cardiovascular system: a molecular toolbox for generating compartmentalized cAMP signaling. Circ. Res. 2007;100:950–966. doi: 10.1161/01.RES.0000261934.56938.38. [DOI] [PubMed] [Google Scholar]
- 44.Munshi A., Kaul S. Stroke genetics—Focus on PDE4D gene. Int. J. Stroke. 2008;3:188–192. doi: 10.1111/j.1747-4949.2008.00199.x. [DOI] [PubMed] [Google Scholar]
- 45.Gretarsdottir S., Thorleifsson G., Reynisdottir S.T., Manolescu A., Jonsdottir S., Jonsdottir T., Gudmundsdottir T., Bjarnadottir S.M., Einarsson O.B., Gudjonsdottir H.M. The gene encoding phosphodiesterase 4D confers risk of ischemic stroke. Nat. Genet. 2003;35:131–138. doi: 10.1038/ng1245. [DOI] [PubMed] [Google Scholar]
- 46.Rosand J., Bayley N., Rost N., de Bakker P.I. Many hypotheses but no replication for the association between PDE4D and stroke. Nat. Genet. 2006;38:1091–1092. doi: 10.1038/ng1006-1091. [DOI] [PubMed] [Google Scholar]
- 47.Homma S., Sakamoto T., Hegab A.E., Saitoh W., Nomura A., Ishii Y., Morishima Y., Iizuka T., Kiwamoto T., Matsuno Y. Association of phosphodiesterase 4D gene polymorphisms with chronic obstructive pulmonary disease: Relationship to interleukin 13 gene polymorphism. Int. J. Mol. Med. 2006;18:933–939. [PubMed] [Google Scholar]
- 48.Reneland R.H., Mah S., Kammerer S., Hoyal C.R., Marnellos G., Wilson S.G., Sambrook P.N., Spector T.D., Nelson M.R., Braun A. Association between a variation in the phosphodiesterase 4D gene and bone mineral density. BMC Med. Genet. 2005;6:9. doi: 10.1186/1471-2350-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lipworth B.J. Phosphodiesterase-4 inhibitors for asthma and chronic obstructive pulmonary disease. Lancet. 2005;365:167–175. doi: 10.1016/S0140-6736(05)17708-3. [DOI] [PubMed] [Google Scholar]
- 50.Spina D. PDE4 inhibitors: Current status. Br. J. Pharmacol. 2008;155:308–315. doi: 10.1038/bjp.2008.307. [DOI] [PMC free article] [PubMed] [Google Scholar]


