Abstract
Although oxidative damage has long been associated with ageing and neurological disease, mechanistic connections of oxidation to these phenotypes have remained elusive. Here we show that the age-dependent somatic mutation associated with Huntington’s disease occurs in the process of removing oxidized base lesions, and is remarkably dependent on a single base excision repair enzyme, 7,8-dihydro-8-oxoguanine-DNA glycosylase (OGG1). Both in vivo and in vitro results support a ‘toxic oxidation’ model in which OGG1 initiates an escalating oxidation–excision cycle that leads to progressive age-dependent expansion. Age-dependent CAG expansion provides a direct molecular link between oxidative damage and toxicity in post-mitotic neurons through a DNA damage response, and error-prone repair of single-strand breaks.
Huntington’s disease is one of several progressive neurodegenerative disorders caused by CAG expansion in the coding region of the Huntington’s disease gene (HD)1–3. Disease severity and onset depend on the number of CAG repeats, which expand in germ cells during differentiation. Recently, CAG expansion has also been detected in brains of Huntington’s disease patients; large increases in CAG length are observed in striatum, the region most affected in disease4. Age-dependent somatic expansion in brain cells cannot be monitored in humans; however, it is well documented in mouse models for Huntington’s disease5,6 and myotonic dystrophy7. In R6/1 mice, which harbour a transgene containing exon 1 of the human HD gene and include the CAG repeat, the inherited repeat tracts are stably maintained from birth until roughly 11 weeks of age, but begin to expand at midlife and continue to increase in length as these animals age6. The expanding CAG tract serves as a template for synthesis of an increasingly toxic HD protein in brain1. Thus, in addition to the inherited expansion, somatic changes in repeat tracts may contribute to toxicity. Indeed, published experiments demonstrate that expression of the expanded HD gene is toxic in somatic cells, and that cell death is accelerated and directly proportional to repeat length1,2. These data suggest that somatic expansion may modulate onset and progression of toxicity, and that blocking somatic expansion in the brain would be beneficial. However, the mechanism by which CAG expansions might occur in post-mitotic neurons remains unclear.
Expansion correlates with DNA oxidation in vivo
We investigated whether age-dependent expansion in young (7–15-week-old) and older (15–52-week-old) R6/1 transgenic mice had any relationship to oxidation, a major factor in ageing (Fig. 1). Somatic tissues of R6/1 mice vary in the degree of expansion. However, tail, brain and liver were particularly informative because they represented tissues with very different degrees of age-dependent instability6 (Fig. 1a).
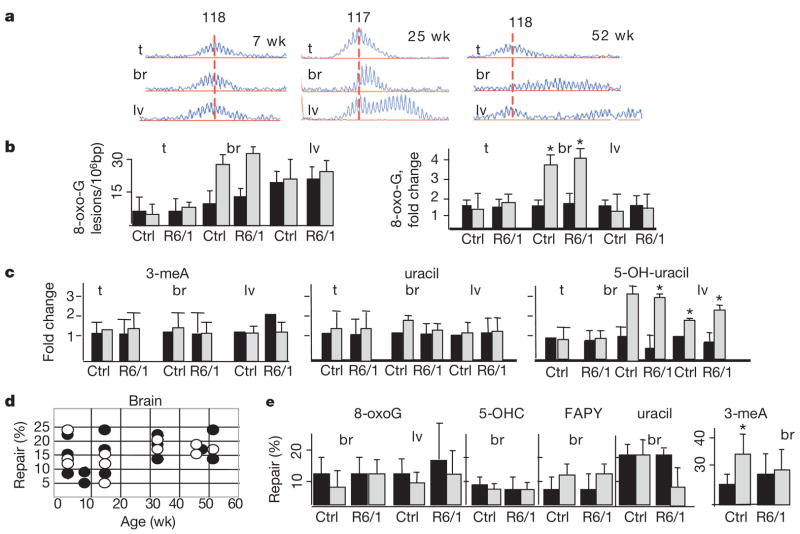
Figure 1. Oxidative lesions accumulate in tissues of ageing mice.
a, Age-dependent CAG repeat distribution in the tissues of R6/1 transgenic mice at indicated ages. The vertical line designates the midpoint length of the CAG repeat distribution in the tail of tested animals. Expansion is an increase in the number of CAG repeats indicated by the shift of distributions to the right (x axis is length in base pairs). b, Left panel, level of oxidative lesions in the tail (t), brain (br) (cortex) and liver (lv) for 8-oxo-G in control (Ctrl) and R6/1 animals at 7 (black) and 52 (grey) weeks. Right panel, accumulation (fold change) of the number of lesions from 7 to 52 weeks. Error bars, s.d. c, Accumulation as in b for 5-OH-uracil, 3-meA and uracil. ★P < 0.01 for b and c. d, Repair activity (Methods) of 8-oxo-G DNA lesion in R6/1 animals (black circles) and wild-type littermate control (open circles) does not change with age (weeks) e, Quantified repair activity (%) of 8-oxo-G, 5-OHC, 3-meA, FAPY and uracil as in d for the indicated tissues at 7 (black) and 52 (grey) weeks. Reported are the mean repair activity (%) and the s.d. The limit of the s.d. is 50 (3-meA).
We found that the level and accumulation of oxidative DNA damage correlated well with the degree of expansion (Fig. 1a, b). For example, 7,8-dihydro-8-oxoguanine (8-oxo-G) in the tail was low and expansion was modest at all ages tested, whereas in liver and in brain, the lesion level was high and expansion continued to progress with age (ref. 6; Fig. 1b). Oxidative lesions including 8-oxo-G, 5-hydroxyuracil (5-OH-uracil), 5-hydroxycytosine (5-OHC), and formamidopyrimidine (FAPY) tended to accumulate in brain and liver of R6/1 animals as they age from 7 to 52 weeks (Fig. 1c, Supplementary Fig. 1a). Neither uracil nor 3-methyladenine (3-meA) accumulated in any tissue at any age tested (Fig. 1c). Thus, the age-dependent accumulation in DNA damage seemed to be somewhat restricted to oxidative lesions. Elevation of oxidative damage was not limited to R6/1 animals. Control animals of equivalent ages accumulated the same degree of oxidative lesions in all tissues tested (Fig. 1b, c). Thus, the level of oxidation was not due to the presence of the transgene, but occurred during the process of ageing.
No reduction in repair of DNA damage in R6/1 mice
The rise in oxidative DNA damage might reflect a decrease in the capacity to repair these lesions or an increase in endogenous oxidation state with age. To distinguish between these two possibilities, we directly measured the repair activity in tissue extracts from ageing control and R6/1 animals. Repair of oxidized bases is typically initiated by cleavage of the C1 glycosidic bond by the action of a DNA glycosylase, followed by ribose-phosphate removal and generation of a single-strand break (SSB)8. To evaluate repair activity, we synthesized a DNA oligonucleotide containing a single base lesion, and measured generation of a 22 nucleotide cleavage product (Supplementary Fig. 1b) after incubation with tissue extracts of young or old animals (Fig. 1d, illustrated is 8-oxo-G). For 8-oxo-G, the most common lesion in DNA, we found no differences in repair activity between control or R6/1 mice at any age tested (Fig. 1d). Moreover, repair was not significantly decreased with age for other lesions tested (5-OHC, 3-meA, FAPY) with the exception of uracil (Fig. 1e). Thus, in vivo, accumulation of oxidative lesions did not reflect a loss of repair, but rather an increase in endogenous oxidative damage during normal ageing.
Expansion occurs during normal repair of SSB
Although trinucleotide expansion in the brain has been observed in vivo, it has not yet been determined whether the expansions are present in the terminally differentiated neurons in R6/1 animals. To address this issue, we isolated pure populations of neurons or astrocytes from the cortex of R6/1 animals using fluorescence-activated cell sorting (FACS)9 (Supplementary Fig. 2a) and tracked CAG expansion (Fig. 2a, b). CAG somatic expansions were, indeed, present in the terminally differentiated neurons from ageing animals. The CAG tracts in pure sorted neurons were larger than those in the tail at 3 weeks, and were similar in size to the CAG repeats in whole-cortex suspension before sorting (Fig. 2a, sorted neurons versus cortex mixture, 27 weeks).
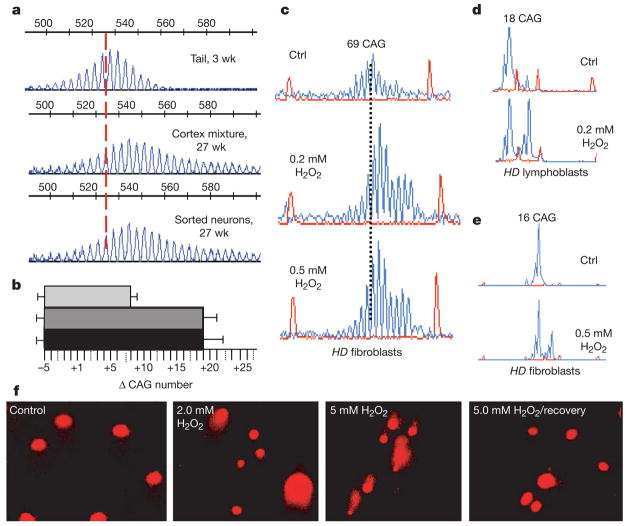
Figure 2. Direct exposure to oxidizing agents causes expansion at the human HD locus in vitro.
aa, CAG repeat distribution of pure sorted cortical neurons at 27 weeks as compared with that of the tail at 3 weeks and to whole-cortex cell suspensions of the same animal before sorting. Vertical dashed line designates the midpoint length of the CAG repeat tract in the tail. Numbers represent size standards. b, The quantified sizes of the repeat tract in tail at 3 weeks (light grey), cortex mixture (dark grey) and pure sorted neurons at 27 weeks (black). Error bars, s.d.; Δ, mean length change of CAG repeats. c–e, Fibroblasts (c and e) and lymphoblasts (d) from Huntington’s disease patients treated in culture with indicated concentrations of hydrogen peroxide. The expanded allele is 69 repeats and the normal allele is 16 CAG repeats. Expansion in peroxide-treated cells versus untreated cells (Ctrl) was determined as in Fig. 1. f, Comet assay for SSBs. Increasing peroxide treatment as indicated induces SSB as detected by comet tails. SSBs were repaired in cells by 2 h post treatment (5 mM H2O2/recovery) as judged by the loss of the comet tails.
Because the accumulation of oxidative damage in the brain correlated with age-dependent expansion in vivo, we tested whether base oxidation could directly lead to CAG expansion. CAG repeats posed no impediment to nicking and repair in vitro. In the presence of protein extract and radiolabelled nucleotides, the CAG tracts within the human HD allele were heavily labelled when purified plasmid DNA carrying the repeats was exposed to oxidizing conditions in vitro (Supplementary Fig. 3).
We next tested the effects of base oxidation on the human HD allele in cultured cells. We treated human Huntington’s disease fibroblasts with non-physiological levels of H2O2, to artificially induce DNA oxidation and evaluated CAG repeat length. Peroxide was chosen as the oxidizing agent because a major fraction of endogenous DNA damage arises during mitochondrial respiration when the superoxide anion radical (O2•−) is converted into hydrogen peroxide10. We have previously shown in these cells that repeats are stably maintained when DNA was replicated over multiple cell divisions11. In the same cells, however, acute treatment with H2O2 uniformly led to CAG expansion of medium-length and disease-length alleles11 (Fig. 2c–e). Indeed, peroxide treatment caused a dose-dependent increase in SSB as measured by the comet assay (Fig. 2f). Cells remained viable during the treatment (Supplementary Fig. 4) and repaired the SSBs within 2 h (Fig. 2f). Thus, oxidative DNA damage induced CAG expansion, which occurred in the process of repairing SSB.
DNA expansion in human disease is length- and sequence-dependent. To test whether oxidative damage in vitro caused expansion in a length- and sequence-dependent manner, we examined the stability of other repetitive sites. We failed to observe expansions at other sites. TTC/GAA repeats have been reported to be the longest tandem repeating array in the genome of humans12. Despite the fact that this sequence contained C and G nucleotides every third base, we were unable to detect expansion at this locus in peroxide-treated cells (Supplementary Fig. 2b). The results were similar for other sites tested (data not shown). Only CAG repeats at the long HD locus expanded in vitro under conditions of oxidative damage, whereas non-CAG sites seemed to be repaired faithfully.
Age-dependent CAG expansion in vivo depends on OGG1
In vivo, oxidized bases are repaired predominantly by the BER pathway13. The significant level of SSB in Huntington’s disease fibroblasts after peroxide treatment was consistent with such a mechanism. Thus, we tested whether DNA glycosylases, the enzymes which initiate BER, contributed to CAG expansion. We first examined the role of OGG1, the primary enzyme that recognizes and removes 8-oxoG from opposite C in DNA14,15. We crossed R6/1 mice with mice lacking OGG1 (ref. 16) and measured the effects on age-dependent somatic expansion in vivo (Fig. 3a, b). Surprisingly, we found that loss of this single glycosylase significantly suppressed or delayed age-dependent expansion in vivo (Fig. 3a, b). Although loss of OGG1 inhibited expansion overall, the effect was not absolute (Fig. 3b). Expansion was prevented in roughly 70% of R6/1/OGG−/− animals. In the other 30%, tissue-specific, age-dependent expansion could be observed in either brain or liver as in age and gender matched R6/1 littermates (Fig. 3b).
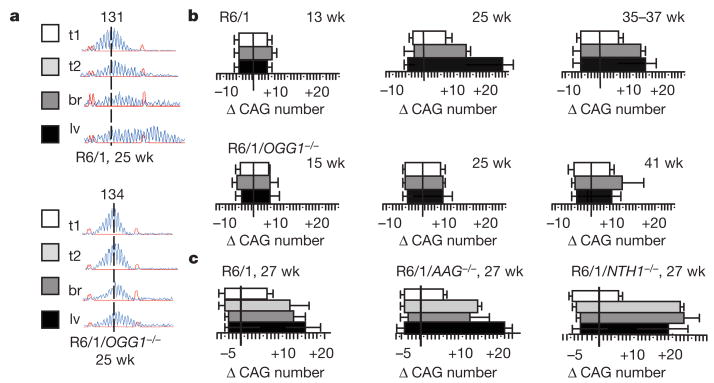
Figure 3. Age-dependent expansion is suppressed in mice lacking OGG1 glycosylase.
a, b, Representative CAG repeat distributions in the tissues of R6/1 transgenic or R6/1/OGG1−/− mice. a, Vertical dashed line designates the midpoint of the CAG repeat tract length in the tail at 3 weeks (t1, white box). The CAG tract lengths in the tail (t2, light grey), brain (br, dark grey) (cortex) and liver (lv, black) at 25 weeks in the same animal are shown. b, The mean length change of CAG repeats (Δ) at the indicated ages in the tissues of R6/1 mice and R6/1/OGG1−/− mice. Each value is expressed as the mean change and the s.d. Tissues are indicated by colour: tail at 3 weeks in white; brain and liver at indicated ages in dark grey and in black respectively. c, Quantified data for mean length change of CAG repeats from R6/1 mice, R6/1/AAG−/− or R6/1/NTH−/−. Tissues are indicated by colour: tail at 3 weeks in white; tail, brain and liver at 27 weeks in light grey, dark grey and black respectively. Analysis is the same as in a, b.
Many glycosylases have preferred but overlapping substrate specificity8,13–19. Therefore, the dependence of CAG expansion for OGG1 was unexpected because, in vivo, any particular glycosylase can be ‘backed-up’. To test further the specificity of OGG1 in the expansion process, we created two additional lines by crossing R6/1 mice with mice lacking alkyladenine glycosylase (AAG)20 or NTH1 (homologue of Escherichia coli endonuclease III)21 (Supplementary Fig. 5a). NTH1 prefers to excise thymine glycol17,19,21 whereas AAG excises a variety of alkylated bases with the highest affinity for 3-methyladenine18,20. In contrast to R6/1/OGG1−/− animals, deletion of either AAG or NTH1 in R6/1/AGG−/− and R6/1/NTH1−/− mice did not reduce somatic expansion relative to their R6/1 littermate controls at any age tested (Fig. 3c; Supplementary Fig. 5a). Thus, loss of the single glycosylase, OGG1, was a dominant factor contributing to age-dependent expansion in vivo, despite the fact that other DNA glycosylases were present and the tissues were competent for SSB repair.
Somatic expansion in R6/1 mice is length- and sequence-dependent
We tested whether somatic expansion in vivo in R6/1 animals recapitulated the features that characterize the mutation in humans. First, it is widely accepted that the expansion mutation occurs primarily at repeats capable of forming secondary structure2,3,22–24, and is not accompanied by general microsatellite instability2,25 Second, in human disease, there is a threshold CAG length, below which there is little probability of observing expansion. In humans, CAG expansion is typically observed at tract lengths above 36 repeats26,27, whereas in mouse, the threshold is around 100 (refs 5, 6, 28). Therefore, we tested in vivo whether age-dependent expansion in R6/1 mice was (i) length-dependent, (ii) sequence-dependent, (iii) occurred at other repetitive sequences, and (iv) whether loss of OGG1 altered these properties.
Expansion did not occur at short CAG repeats in R6/1 and in R6/1/OGG1−/− animals—it was not observed in endogenous mouse HD gene (6 CAG repeats) in either line (not shown). We next tested unrelated alleles containing longer CAG tracts. We found that CAG tracts on chromosome 9 (19 repeats) and chromosome 7 (31 repeats) were stable in both R6/1 animals and in R6/1/OGG1−/− mice (Supplementary Fig. 5b). Thus, loss of OGG1 in vivo blocked expansion only at the long CAG tract (135 repeats) within the human HD transgene, the length of which was above the threshold reported for the mouse homologue. We observed no age-dependent expansion in R6/1 and in R6/1/OGG1−/− animals at sequences that lacked structure-forming capability. Shown is polyA microsatellite (Supplementary Fig. 5b) but the same result was observed for other microsatellites tested (data not shown). Thus, age-dependent somatic expansion in R6/1 mice showed the properties observed in human disease.
OGG1 initiates CAG expansion during BER reconstituted in vitro
The in vivo requirement of OGG1 for expansion of CAG repeats implicated a BER mechanism. Therefore, we tested whether expansion could be regenerated in vitro using purified human OGG1 and the BER machinery. DNA polymerases can generate expansion on CAG-containing templates by primer extension29–32 and on substrates that mimic BER intermediates32,33. However, no experiments have tested the entire BER process beginning with the glycosylase incision of a lesion within CAG duplex DNA. We synthesized two DNA templates, each 100 base pairs in length, in which a single 8-oxoG base was positioned 23 nucleotides from the 5′ end of one strand (Supplementary Fig. 6a, b, top panels). In the random template control (Supplementary Fig. 6a), the base lesion was flanked by random sequence DNA of roughly equal CG/AT content, whereas in the CAG template, the sequence was identical except that the base lesion was flanked on the 3′ side by 19 CAG repeats (Supplementary Fig. 6b).
We evaluated the BER pathway using a step-by-step addition of OGG1, apurinic/apirimidinic endonuclease (APE1) and polymerase β (Polβ) to the reconstituted in vitro system. Removal of the damaged base by OGG1/APE cleavage generates a fragment of 22 nucleotides (Supplementary Fig. 6a, b). Polβ was chosen as the gap-filling polymerase because it is the major BER polymerase in mammalian cells, and because it favours single nucleotide additions34. If the CAG sequences did not promote expansion, the CAG templates should behave as a random template. Polβ would restore the initial 23 nucleotide length and the template to 100 nucleotides after ligation (Supplementary Fig. 6a). On the other hand, if the CAG sequences promoted expansion, then some strand displacement/slippage should occur. In this case, Polβ should yield multi-nucleotide additions (n > 1 nucleotide) and templates longer than 100 nucleotides following ligation of looped intermediates (Supplementary Fig. 6b).
With both templates, OGG1 activity was strong, and no difference was observed in the products between the CAG and random templates (Fig. 4a, lanes 1 and 4). Addition of APE1 resulted in nicking of the phosphodiester backbone and the production of the 22 nucleotide fragment (Fig. 4a, lanes 2 and 5). In the absence of APE, Polβ was unable to carry out strand extension of the 22 nucleotide fragment, as expected (data not shown).
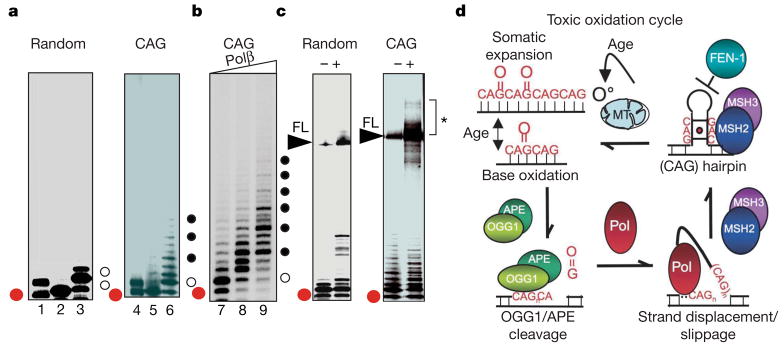
Figure 4. OGG1 excision of 8-oxo-G within CAG repeat DNA can initiate strand displacement and expansion in vitro during BER.
a, Reaction products after step-wise addition of OGG1 (lanes 1 and 4), OGG1 + APE (lanes 2 and 5), and OGG1 + APE + Polβ (lanes 3 and 6) to random (left) or CAG (right) templates. Red dots indicate 22 nucleotide excision/incision product, open and filled circles are 1 nucleotide and 3 nucleotide additions, respectively. b, Triplet pattern on CAG template as a function of increasing Polβ concentration indicated by the triangle: 0 nM (lane 7); 0.5 nM (lane 8); and 1 nM (lane 9). c, Products of the BER reactions for random (left) or CAG (right) templates in the absence (−) or presence (+) of DNA ligase. FL, 100 nucleotide full-length product after ligation. The bracket and asterisk depict the 100 nucleotide FL template and larger expansion products. Red dot as in a. d, ‘Toxic oxidation cycle’ model for age-dependent somatic expansion. Endogenous oxidative radicals (O°) arising from mitochondrial (MT) respiration creates oxidative DNA lesions. Under conditions of normal BER, OGG1/APE cleavage produces a nick, and polymerase (Pol) facilitates hairpin formation during gap-filling synthesis. CAG hairpins are stabilized by MSH2/MSH3 binding (red dot is a mismatch in the stem) and escape FEN-1 loading and cleavage owing to a hidden 5′ end. The hairpin intermediate is processed to restore duplex DNA generating a longer CAG template, which is again subject to oxidative DNA damage. The cycle continues with age.
We found that gap filling by Polβ on the random template resulted in a single nucleotide addition as the major product, and restored the base at the position 23 nucleotides from the end (Fig. 4a, lane 3). In contrast, on CAG templates, OGG1/APE cleavage and gap filling synthesis by Polβ generated longer addition products (Fig. 4a, lane 6; black dots). Three nucleotide additions were favoured (Fig. 4b, black dots). On random templates, the addition of ligase primarily restored the 100 nucleotide full-length starting material (Fig. 4c, lane 2), whereas on the CAG template, ligation resulted in appearance of fragments corresponding to the starting material as well as expanded products (shown in starred bracket Fig. 4c). Thus, OGG1-mediated BER was able to initiate expansion through strand displacement/slippage during the gap-filling step of BER, and both displacement and expansion depended on the CAG sequence.
Discussion
Here we demonstrate that, in vivo, age-dependent somatic CAG expansion is initiated in the process of removing oxidized DNA bases, and is dependent on a single DNA glycosylase, OGG1. The unexpected specificity for OGG1 is not due to a specific rise of its preferred 8-oxo-G lesions because many oxidative DNA lesions accumulate in the brains of ageing R6/1 animals. Further, the requirement for OGG1 in CAG expansion does not seem to arise from lack of functional back-up systems. All DNA glycosylases are present in R6/1 animals and the tissues were competent for lesion repair (Fig. 1, Supplementary Fig. 1). Because only OGG1 suppressed the mutation, we favour a model in which the unusual dependence of expansion on OGG1 arises from its interaction with the CAG tract in a manner that is not shared by other glycosylases. Thus, redundant biochemical activities for removing oxidative DNA damage in vitro may promote distinct biological consequences in vivo, which are relevant to the mechanism for human neurodegenerative disease.
The specificity of OGG1 may indicate that 8-oxoG lesions are favoured within CAG sites and occur frequently. Alternatively, OGG1 might bind better to CAG sequences or their DNA tertiary structures, or binding may induce conformations that confer specificity. CAG sequences can form DNA hairpins with mismatched bases every third nucleotide. We have previously shown that binding of the MSH2/MSH3 complex to the CAG hairpin alters properties needed for recognition and repair of mispaired bases35. Moreover, we and others have demonstrated that loss of mismatch repair proteins, MSH2 (refs 6, 35–37), MSH3 (refs 35, 38) and PMS2 (ref. 39) abrogates age-dependent somatic expansion in vivo in a number of mouse models for Huntington’s disease and myotonic dystrophy. Because loss of OGG1 also suppresses the mutation, these data imply that the mismatch repair complex physically or functionally cooperates with OGG1 to promote expansion of the CAG repeat sequences.
Although many models have been proposed for trinucleotide expansion on the basis of in vitro systems, the results presented here support the hypothesis that, in vivo, somatic expansion in mammalian cells initiates from an OGG1-mediated BER mechanism. Somatic expansion does not require cell division. We find that somatic age-dependent expansion occurs in neurons well after these cells are terminally differentiated and mitotic replication has ceased. Likewise, it has been demonstrated that loss of RAD52, RAD54 or Ku did not suppress CTG expansion in the mouse model for myotonic dystrophy36. Thus, enzymes generally needed for repair of double-strand breaks are not required for expansion in vivo. Our data indicate that a SSB mechanism such as BER is more likely. Highly processive polymerases, such as Polδ or Polε, can copy thousands of base pairs, and are often used to complete long-patch BER40. Although we have not yet tested this hypothesis, strand displacement and gap-filling synthesis over a long stretch of triplet repeat DNA might facilitate formation of larger structural intermediates. Thus, long patch BER might also be relevant for large expansions associated with non-coding regions of genes1,2.
Inheritance of an expanded CAG tract is the underlying cause for Huntington’s disease toxicity. However, we propose a ‘toxic oxidation cycle’ model in which somatic mutations contribute to onset and progression. The oxidation–excision–expansion cycle escalates with age as oxidative lesions in the brain accumulate (Fig. 4d). The increase in oxidative lesions elicits a DNA damage response and increases the need for OGG1 repair through a SSB mechanism. Expansion would result from ‘error-prone repair’ in the process of gap-filling synthesis by a strand displacement/slippage mechanism (Fig. 4d). Loop entrapment on one strand can place constraints on the other strand and initiate restoration of duplex DNA. Because the ‘toxic oxidation cycle’ is iterative with age, the resulting mutation would be predicted to grow progressively (Fig. 4d). Oxidative damage has been recognized as a major cause of ageing and neurological disease in all aerobic organisms11,14. CAG expansion at the disease allele in Huntington’s disease provides a glimpse of the underlying molecular mechanism for at least one neurodegenerative disease. However, the accumulation of oxidized bases in the brain overwhelms the repair machinery throughout the genome, thereby increasing the probability of unrepaired SSBs, which can be fatal41. Thus, the importance of oxidative DNA damage and SSB intermediates probably extends beyond Huntington’s disease and has general relevance to late onset neurodegeneration.
METHODS SUMMARY
Transgenic male mice B6CBA-TgN R6/1 (ref. 5) were crossed with C57BL/6J female partners that lacked one of the glycosylases—AAG (ref. 20), OGG1 (ref. 16) or NTH1 (gift from L. Samson). Cell lines from Huntington’s disease patients used in the study are described in Methods. For the comet assay, SK-N cells untreated or treated with H2O2 were used. For in vitro oxidation, a plasmid DNA that contained a truncated version of human HD complementary DNA with 40 CAG repeats42 was treated with methylene blue, exposed to visible light and used as a substrate in in vitro repair assay. Statistical analysis for CAG repeat change was performed, and change was expressed as mean ± standard deviation (s.d.). Statistical significance was determined by two-way analysis of variance (ANOVA), followed by Fisher’s exact test. Details of FACS analysis, preparation of nuclear extracts and enzyme cleavage assays, in vitro reconstitution of BER, preparation of nuclear DNA and lesion detection by HPLC can be found in Methods.
Supplementary Material
Acknowledgments
This work was supported by the Mayo Foundation, the National Institutes of Health (C.T.M.), and, in part, by the Intramural Research Program of the National Institute of Environmental Health Sciences. The authors wish to dedicate this work to Erling Seeberg. We thank J. Hoeijmakers for NTH−/− and L. Samson for AAG−/− mice, and N. Kinzel for help in cortex dissection.
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions C.T.M. oversaw the entire project. I.V.K. and C.T.M. conceived the experiments, wrote the manuscript, and prepared all Figures. I.V.K. carried out in vitro and in vivo experiments in cell lines and in mice (animal breeding, oxidation, comet assay, FACS, analysis of repeat size, dissected animal tissue) and prepared tissue extracts for all testing of repair activity in vitro, and performed analysis of the results. Y.L. with S.H.W. supervision performed base excision repair reconstitution experiments, M.B. and A.K. carried out DNA repair assays. All co-authors contributed to the manuscript with their comments.
Author Information Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.Cummings CJ, Zoghbi HY. Trinucleotide repeats: mechanisms and pathophysiology. Annu Rev Genom Hum Genet. 2000;1:281–328. doi: 10.1146/annurev.genom.1.1.281. [DOI] [PubMed] [Google Scholar]
- 2.Pearson CE, Edamura KN, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nature Rev Genet. 2005;10:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- 3.Kovtun IV, McMurray CT. In: Genetic Instabilities and Hereditary Neurological Diseases. Wells RD, editor. Academic Press; 2006. pp. 679–690. [Google Scholar]
- 4.Kennedy L, et al. Dramatic tissue-specific mutation length increases are an early molecular event in Huntington disease pathogenesis. Hum Mol Genet. 2003;12:3359–3367. doi: 10.1093/hmg/ddg352. [DOI] [PubMed] [Google Scholar]
- 5.Mangiarini L, et al. Instability of highly expanded CAG repeats in mice transgenic for the Huntington’s disease mutation. Nature Genet. 1997;15:197–200. doi: 10.1038/ng0297-197. [DOI] [PubMed] [Google Scholar]
- 6.Kovtun IV, McMurray CT. Trinucleotide expansion in haploid germ cells by gap repair. Nature Genet. 2001;27:407–411. doi: 10.1038/86906. [DOI] [PubMed] [Google Scholar]
- 7.Seznec H, et al. Transgenic mice carrying large human genomic sequences with expanded CTG repeat mimic closely the DM CTG repeat intergenerational and somatic instability. Hum Mol Genet. 2000;9:1185–1194. doi: 10.1093/hmg/9.8.1185. [DOI] [PubMed] [Google Scholar]
- 8.Dizdaroglu M. Substrate specificities and excision kinetics of DNA glycosylases involved in base-excision repair of oxidative DNA damage. Mutat Res. 2003;531:109–126. doi: 10.1016/j.mrfmmm.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 9.Sergent-Tanguy S, Chagneau C, Neveu I, Naveilhan P. Fluorescent activated cell sorting (FACS): a rapid and reliable method to estimate the number of neurons in a mixed population. J Neurosci Methods. 2003;129:73–79. doi: 10.1016/s0165-0270(03)00210-3. [DOI] [PubMed] [Google Scholar]
- 10.Brand MD, et al. Mitochondrial superoxide: production, biological effects, and activation of uncoupling proteins. Free Radic Biol Med. 2004;37:755–767. doi: 10.1016/j.freeradbiomed.2004.05.034. [DOI] [PubMed] [Google Scholar]
- 11.Kovtun IV, Thornhill AR, McMurray CT. Somatic deletion events occur during early embryonic development and modify the extent of CAG expansion in subsequent generations. Hum Mol Genet. 2004;13:3057–3068. doi: 10.1093/hmg/ddh325. [DOI] [PubMed] [Google Scholar]
- 12.Chauhan C, Dash D, Grover D, Rajamani J, Mukerji M. Origin and instability of GAA repeats: insights from Alu elements. J Biomol Struct Dyn. 2002;20:253–263. doi: 10.1080/07391102.2002.10506841. [DOI] [PubMed] [Google Scholar]
- 13.Bjelland S, Seeberg E. Mutagenicity, toxicity and repair of DNA base damage induced by oxidation. Mutat Res. 2003;531:37–80. doi: 10.1016/j.mrfmmm.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 14.Auffret van der Kemp P, Thomas D, Barbey R, de Oliveira R, Boiteux S. Cloning and expression in Escherichia coli of the OGG1 gene of Saccharomyces cerevisiae, which codes for a DNA glycosylase that excises 7,8-dihydro-8-oxoguanine and 2,6-diamino-4-hydroxy-5-N-methylformamido pyrimidine. Proc Natl Acad Sci USA. 1996;93:5197–5202. doi: 10.1073/pnas.93.11.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zharkov DO, Rosenquist TA, Gerchman SE, Grollman AP. Substrate specificity and reaction mechanism of murine 8-oxoguanine-DNA glycosylase. J Biol Chem. 2000;275:28607–28617. doi: 10.1074/jbc.M002441200. [DOI] [PubMed] [Google Scholar]
- 16.Klungland A, et al. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc Natl Acad Sci USA. 1999;96:13300–13305. doi: 10.1073/pnas.96.23.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dizdaroglu M, Karahalil B, Senturker S, Buckley TJ, Roldan-Arjona T. Excision of products of oxidative DNA base damage by human NTH1 protein. Biochemistry. 1999;38:243–246. doi: 10.1021/bi9819071. [DOI] [PubMed] [Google Scholar]
- 18.O’Brien PJ, Ellenberger T. Dissecting the broad substrate specificity of human 3-methyladenine-DNA glycosylase. J Biol Chem. 2003;279:9750–9757. doi: 10.1074/jbc.M312232200. [DOI] [PubMed] [Google Scholar]
- 19.Marenstein DR, et al. Substrate specificity of human endonuclease III (hNTH1) Effect of human APE1 on hNTH1 activity J Biol Chem. 2003;278:9005–9012. doi: 10.1074/jbc.M212168200. [DOI] [PubMed] [Google Scholar]
- 20.Engelward BP, et al. Base excision repair deficient mice lacking the Aag alkyladenine DNA glycosylase. Proc Natl Acad Sci USA. 1997;94:13087–13092. doi: 10.1073/pnas.94.24.13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hazra TK, et al. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc Natl Acad Sci USA. 2002;99:3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kovtun IV, Goellner G, McMurray CT. Structural features of trinucleotide repeats associated with DNA expansion. Biochem Cell Biol. 2001;279:325–336. [PubMed] [Google Scholar]
- 23.Gacy AM, et al. GAA instability in Friedreich’s Ataxia shares a common, DNA-directed and intraallelic mechanism with other trinucleotide diseases. Mol Cell. 1998;1:583–593. doi: 10.1016/s1097-2765(00)80058-1. [DOI] [PubMed] [Google Scholar]
- 24.Spiro C, et al. Inhibition of FEN-1 processing by DNA secondary structure at trinucleotide repeats. Mol Cell. 1999;4:1079–1085. doi: 10.1016/s1097-2765(00)80236-1. [DOI] [PubMed] [Google Scholar]
- 25.Goellner GM, et al. Different mechanisms underlie DNA instability in Huntington disease and colorectal cancer. Am J Hum Genet. 1997;60:879–890. [PMC free article] [PubMed] [Google Scholar]
- 26.Kremer B, et al. Sex-dependent mechanisms for expansions and contractions of the CAG repeat on affected Huntington disease chromosomes. Am J Hum Genet. 1995;57:343–350. [PMC free article] [PubMed] [Google Scholar]
- 27.Chong SS, et al. Contribution of DNA sequence and CAG size to mutation frequencies of intermediate alleles for Huntington disease: evidence from single sperm analysis. Hum Mol Genet. 1997;6:301–309. doi: 10.1093/hmg/6.2.301. [DOI] [PubMed] [Google Scholar]
- 28.Wheeler VC, et al. Length-dependent gametic CAG repeat instability in the Huntington’s disease knock-in mouse. Hum Mol Genet. 1999;8:115–122. doi: 10.1093/hmg/8.1.115. [DOI] [PubMed] [Google Scholar]
- 29.Wilson SH, et al. In: Genetic Instabilities and Hereditary Neurological Diseases. Wells RD, Warren ST, editors. Academic Press; 1998. pp. 493–698. [Google Scholar]
- 30.Petruska J, Hartenstine MJ, Goodman MF. Analysis of strand slippage in DNA polymerase expansions of CAG/CTG triplet repeats associated with neurodegenerative disease. J Biol Chem. 1998;273:5204–5210. doi: 10.1074/jbc.273.9.5204. [DOI] [PubMed] [Google Scholar]
- 31.Hartenstine MJ, Goodman MF, Petruska J. Base stacking and even/odd behavior of hairpin loops in DNA triplet repeat slippage and expansion with DNA polymerase. J Biol Chem. 2000;275:18382–18390. doi: 10.1074/jbc.275.24.18382. [DOI] [PubMed] [Google Scholar]
- 32.Hartenstine MJ, Goodman MF, Petruska J. Weak strand displacement activity enables human DNA polymerase β to expand CAG/CTG triplet repeats at strand breaks. J Biol Chem. 2002;277:41379–41389. doi: 10.1074/jbc.M207013200. [DOI] [PubMed] [Google Scholar]
- 33.Lyons-Darden T, Topal MD. Abasic sites induce triplet-repeat expansion during DNA replication in vitro. J Biol Chem. 1999;274:25975–25978. doi: 10.1074/jbc.274.37.25975. [DOI] [PubMed] [Google Scholar]
- 34.Beard WA, Wilson SH. Structure and mechanism of DNA polymerase β. Chem Rev. 2006;106:361–382. doi: 10.1021/cr0404904. [DOI] [PubMed] [Google Scholar]
- 35.Owen BA, et al. (CAG)n-hairpin DNA binds to Msh2-Msh3 and changes properties of mismatch recognition. Nature Struct Mol Biol. 2005;12:663–670. doi: 10.1038/nsmb965. [DOI] [PubMed] [Google Scholar]
- 36.Savouret C, et al. CTG repeat instability and size variation timing in DNA repair-deficient mice. EMBO J. 2003;22:2264–2273. doi: 10.1093/emboj/cdg202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manley K, Shirley TL, Flaherty L, Messer A. MSH2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nature Genet. 1999;23:471–473. doi: 10.1038/70598. [DOI] [PubMed] [Google Scholar]
- 38.van den Broek WJ, et al. Somatic expansion behaviour of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum Mol Genet. 2002;11:191–198. doi: 10.1093/hmg/11.2.191. [DOI] [PubMed] [Google Scholar]
- 39.Gomes-Pereira M, Fortune MT, Ingram L, McAbney JP, Monckton DG. Pms2 is a genetic enhancer of trinucleotide CAG. CTG repeat somatic mosaicism: implications for the mechanism of triplet repeat expansion Hum Mol Genet. 2004;13:1815–1825. doi: 10.1093/hmg/ddh186. [DOI] [PubMed] [Google Scholar]
- 40.Stucki M, et al. Mammalian base excision repair by DNA polymerases δ and ε. Oncogene. 1998;17:835–843. doi: 10.1038/sj.onc.1202001. [DOI] [PubMed] [Google Scholar]
- 41.Ahel I, et al. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature. 2006;443:713–716. doi: 10.1038/nature05164. [DOI] [PubMed] [Google Scholar]
- 42.Trushina E, et al. Microtubule destabilization and nuclear entry are sequential steps leading to toxicity in Huntington’s disease. Proc Natl Acad Sci USA. 2003;100:12171–12176. doi: 10.1073/pnas.2034961100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.