Abstract
AIM: To investigate the effects of Gli-1 small interference RNA (siRNA) on Huh7 cells, and the change of Bcl-2 expression in Huh7 cells.
METHODS: Human hepatocellular carcinoma cells Huh7 were used. Cell viability was analyzed by 3-(4, 5-Dimethylthiazol-2-yl)-2, 5-diphenyl tetrazolium bromide (MTT) assay. The expressions of Gli-1 and Bcl-2 family members were detected by RT-PCR and Western blot. Apoptosis was detected by Flow cytometry using propidium iodide, measured by Hoechst 33258 staining using Advanced Fluorescence Microscopy and caspase-3 enzymatic assay. Cell growth was analyzed after treatment with Gli-1 siRNA and 5-fluorouracil (5-Fu).
RESULTS: Inhibition of Gli-1 mRNA in Huh7 cells through Gli-1 siRNA reduced cell viability. Gli-1 siRNA treatment also induced apoptosis by three criteria, increase in the sub-G1 cell cycle fraction, nuclear condensation, a morphologic change typical of apoptosis, and activation of caspase-3. Gli-1 siRNA was also able to down-regulate Bcl-2. However, Gli-1 siRNA resulted in no significant changes in Bcl-xl, Bax, Bad, and Bid. Furthermore, Gli-1 siRNA increased the cytotoxic effect of 5-Fu on Huh7 cell.
CONCLUSION: Down-regulation of Bcl-2 plays an important role in apoptosis induced by Gli-1 siRNA in HCC cells. Combination Gli-1 siRNA with chemotherapeutic drug could represent a more promising strategy against HCC. The effects of the strategies need further investigation
in vivo and may have potential clinical application.
Keywords: Gli-1 transcription factor, Small interference RNA, Apoptosis, Hepatocellular carcinoma, 5-fluorouracil
INTRODUCTION
Hedgehog (Hh) gene was first identified in a Drosophila genetic screen for mutations affecting body patterning[1]. This pathway contains Hh proteins (Sonic Hh, Indian Hh, and Desert Hh), the twelve transmembrane protein Patched (Ptc), the seven transmembrane protein Smoothened (Smo), and the five-zinc finger transcription factor Glis (Gli-1, Gli-2, and Gli-3)[2–4]. When Hh binds to Ptc, Smo is released and the downstream transcription factors such as Gli-1 are activated. Conversely, when Hh unbinds to Ptc, Ptc prevents Smo from activating the pathway[5]. Glis proteins are regarded as nuclear executors of hedgehog signaling[6,7]. Evidence from several groups suggests that Gli-1 and Gli-2 represent the main activators of Hh-target genes, while Gli3 acts mainly as repressor[8,9]. Gli-1 has been demonstrated to represent a direct transcriptional Hh target gene and the marker of Hh pathway activation[10–13].
Constitutive activation of the Hh pathway has been shown to contribute to the growth and maintenance of various cancers[14–21]. We have previously shown that Hh pathway activation was an important event for the development of human hepatocellular carcinoma (HCC)[22]. Our results corroborated and extended the findings of Sicklick et al[23] about disregulation of Hh signaling in human hepatocarcinogenesis. The fact that the Hh pathway is active in human HCC suggests a significance of novel targeted strategy for HCC therapy.
Sustained application of specific Hh-pathway inhibitors has been proven effective in preventing growth of many tumors with activating Hh signaling in vitro and in vivo[21–26], suggesting a promising approach of Hh signaling interference. Current strategies focus on the use of small molecule inhibitors of Smo. Blocking Smo with antagonist, e.g. cyclopamine or KAAD-cyclopamine[27] is a reasonable yet limited approach, since it is only effective for tumors with Hh pathway activation occurring upstream or at the level of Smo. However, tumors with inactivating mutations in Smo or downstream components will most likely not respond to Smo antagonists[23,28]. It has been reported that Gli-1 is the last and essential step of the pathway and plays a central role in HCC proliferation[24]. Thus, blocking Gli-1 transcription factor would be an ideal strategy to combat HCC. In this paper, we investigated the effects of Gli-1 small interfering RNA (siRNA) on the human liver cancer cells. Our results indicated that Gli-1 siRNA induced apoptosis through down-regulation of Bcl-2 in HCC cells.
MATERIALS AND METHODS
Reagents
The primary goat polyclonal anti-Gli-1 and nonspecific control siRNA were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Rabbit monoclonal anti-Bcl-2, SABC kit, and horseradish peroxidase-conjugated mouse anti-goat/rabbit IgG secondary antibody were obtained from Boster (Wuhan, China). 5-fluorouracil (5-Fu), TRIzol reagent, and propidium iodide (PI) were obtained from Sigma Chemical Co (St. Louis, MO, USA). 5-Fu was dissolved in phosphate buffered solution (PBS).
Cell culture
The human HCC cell line Huh7 and human hepatocyte cell line L02 were kindly donated by professor Guan Xin-yuan (Hong Kong University). Huh7 and L02 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Gibco BRL, Grand Island, NY, USA) supplemented with 10% (v/v) fetal bovine serum (Bio-Whittaker, Walkersville, MD, USA) at 37°C in a humidified atmosphere with 5% CO2.
Immunocytochemistry assay
Immunocytochemical staining was performed as described by the manufacturer on cell slides by the streptavidin-biotin-peroxidase complex method using a SABC kit. For negative control, the primary anti-Gli-1 antibody was replaced with PBS.
Treatment with siRNA
The Gli-1 and Bcl-2 siRNA reagent kits were purchased from RiboBio Co. LTD (Guangzhou, China). siRNAs were transfected using the LipofectAMINE 2000 reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions after cells were grown to 50% confluence. Briefly, 1 μL LipofectAMINE was diluted in 50 μL OptiMEMI Reduced Serum Medium (Invitrogen, Carlsbad, CA, USA), mixed gently, and incubated for 5 min at room temperature. Subsequently, a mixture of siRNA was added and incubated for 20 min. The mixture was diluted with medium and the final concentration of siRNA was 100 nmol/L. After treatment for the indicated time period, cells were collected for further investigation.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay for cell viability
Cells were seeded onto 96-well plates at 5000 cells/well overnight. After incubating with the test compounds for the indicated time period, 20 μL MTT (5 g/L MTT in PBS) was added to each well and incubated at 37°C for 4 h. The cells were then centrifuged (700 × g for 15 min), and the supernatant was removed. DMSO (200 μL/well) was added to dissolve the cell pellets. Absorbance was measured at a wavelength of 570 nm with enzyme-linked immunity implement (BIO-RAD 2550, USA).
Flow cytometry assay
Huh7 cells were incubated with each experimental treatment. Cells were collected, washed twice with pre-cold PBS, and fixed with 70% ethanol overnight at 4°C. Cell pellets were resuspended in PBS after being washed twice with pre-cold PBS. Then 50 μL of RNase (10 μg/mL) and 25 μL of propidium iodide (1 mg/mL) were added to the cells for 30 min at room temperature, and then flow cytometry (EPICS-XL, Beckman Coulter, Fullerton, USA) analysis was performed. Data were analyzed using CellQuest Software (Becton Dickinson, Mountain View, CA, USA).
Chromatin staining with Hoechst 33258
Apoptosis was observed by chromatin staining with Hoechst 33258 as previously described[29]. In brief, after 48 h transfection with Gli-1 siRNA, Huh7 cells were washed with ice-cold PBS, fixed with 4% paraformaldehyde in PBS for 10 min at room temperature, washed again with PBS, and stained 10 min with Hoechst 33 258 (5 mg/L), washed, and observed under an Advanced Fluorescence Microscope (NIKON 80i, Tokyo, Japan) by an observer blind to the cell treatment.
Reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA was isolated from Huh7 cells with TRIzol reagent. First strand cDNA was synthesized using total RNA with the ReverTra Ace system (Toyobo, Tokyo, Japan) according to the manufacturer’s instructions. Transcripts of the gene for β-actin were used as an internal control. We used the following primers for PCR: Gli-1 (sense: 5'-ttcctaccagagtcccaagt-3', antisense: 5'-ccctatgtgaagccctattt-3', 185 bp), Shh (sense: 5'-CGCACGGGGACAGCTCGGAAGT-3', antisense: 5'-CTGCGCGGCCCTCGTAGTGC-3', 476 bp), Ptch (sense: 5'-GTTGGAAGAAAACAAACAGC-3', antisense: 5'-AGCCGTCAGGTAGATGTAGA-3', 277 bp), Smo (sense: 5'-TTACCTTCAGCTGCCACTTCTACG-3', antisense: 5'-GCCTTGGCAATCATCTTGCTCTTC-3', 321 bp); Bcl-2 (sense: 5'-CTGGTGGACAACATCGC-3', antisense: 5'-GGAGAAATCAAACAGAGGC-3', 135 bp), Bcl-xl (sense: 5'-GTAAACTGGGGTCGCATTGT-3', antisense: 5'-TGGATCCAAGGCTCTAGGTG-3', 146 bp), Bax (sense: 5'-CCAGCTGCCTTGGACTGT-3', antisense: 5'-ACCCCCTCAAGACCACTCTT-3', 135 bp), Bad (sense: 5'-AGGGCTGACCCAGATTCC-3', antisense: 5'-GTGACGCAACGGTTAAACCT-3', 178 bp), Bid (sense: 5'-GCTTCCAGTGTAGACGGAGC-3', antisense: 5'-GTGCAGATTCATGTGTGGATG-3', 116 bp)[30] and β-actin (sense: 5'-GTGGACATCCGCAAAGAC-3', antisense: 5'-GAAAGGGTGTAACGCAACT-3', 303 bp). The reactions were carried out in a DNA Thermal Cycler (Perkin-Elmer Corp. Norwalk, Connecticut, USA) with the following cycle conditions: denaturation at 94°C for 1 min, annealing at respective temperature for 30 s, and extension at 72°C for 1 min, for 35 cycles.
Western blot analysis
Western blotting was performed as described previously[31,32] with minor modification. Total cell extracts were prepared by lysing cells in RIPA lysis buffer. For gel electrophoresis, equivalent of total protein (40 μg) was loaded on each lane of a 10% sodium dodecyl sulfate-polyacrylamide gel.
Caspase-3 activity detection
After treatment, activation of caspase-3 was detected with 200 μg of cell lysate using the colorimetric protease assay kit (KeyGEN Biotech, Nanjing, China) according to the manufacturer’s protocol. The activity of caspase-3 was converted through measurement of absorbance (OD 405 values) using a microplate reader at 405 nm. Non-specific reactions were corrected by subtracting background absorbance readings from the combination of cell lysate and buffer.
Statistical analysis
Values were expressed as mean ± SD from at least three separate experiments. Differences between groups were assessed with one-way ANOVA or Student’s t-test using SPSS 11.5 for Windows (SPSS Inc, Chicago, IL, USA). Differences were considered significant if P < 0.05.
RESULTS
Expression of Hh pathway components in Huh7 cells
To begin to elucidate the effect of Gli-1 on Huh7 cells, we first examined the expressions of Hh pathway components in Huh7 cells and the normal hepatocyte cell line L02 cells. The expressions of Hh pathway components were determined by RT-PCR analyses. The expressions of Gli-1 were also assayed by immunocytochemistry detection. RT-PCR analyses showed that Shh, Smo and Gli-1 were expressed in Huh7 cells, but not in L02 cells (Figure 1A). Ptch was detected in both cells, however, the expressions were elevated in Huh7 cells. To further confirm these findings, we performed immunocytochemistry for Gli-1. Gli-1 protein was present in the cytoplasm of Huh7 cells, but not in L02 cells (Figure 1B). Because Gli-1 is one of the indicators of the Hh pathway activation, our data indicated that Hh pathway is active in Huh7 cells.
Figure 1.
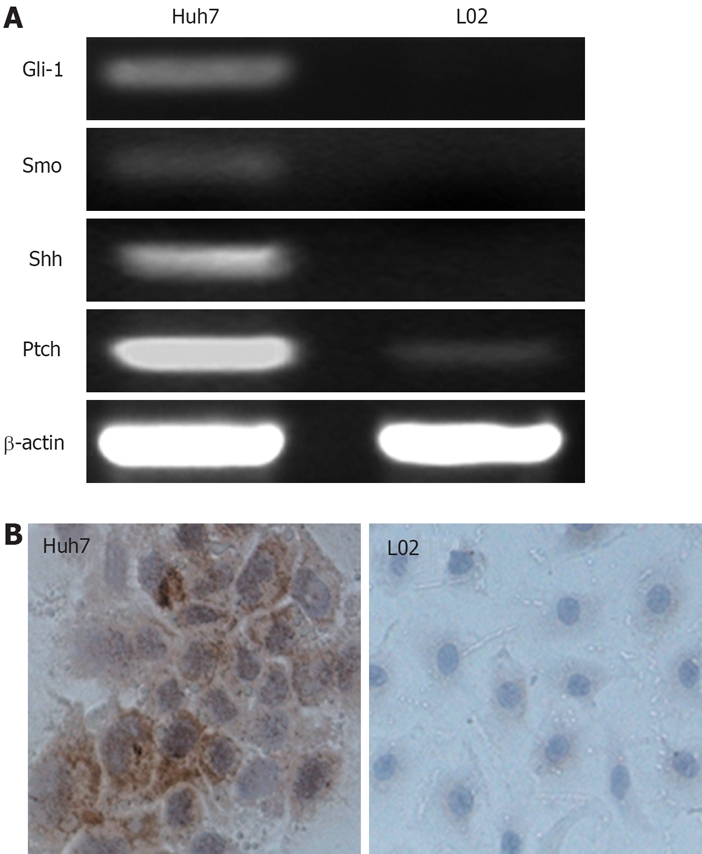
Expressions of Hh pathway components on Huh7 and L02 cells. A: The expressions of Gli-1, Smo and Shh mRNA was observed in Huh cells, but not in L02 cells; Ptch mRNA was detected in both cell lines, but the expressions was elevated in Huh7 cells; B: Immunocytochemical staining showed Gli-1 protein expression in the cytoplasm of Huh7 cells, but not L02 cells.
Gli-1 siRNA reduced Huh7 cell viability
Since Hh pathway is activated in Huh7 cells, we investigated whether Gli-1 siRNA could kill Huh7 cells. Firstly, we characterized the function of Gli-1 siRNA on Gli-1 mRNA expression. Huh7 cells were transfected with Gli-1 siRNA or nonspecific control siRNA at a concentration of 100 nmol/L. Cells treated with PBS were used as a mock control. Twenty-four hours and forty-eight hours after treatment, the expressions of Gli-1 mRNA were assayed by RT-PCR. The results indicated that Gli-1 siRNA completely blocked Gli-1 expressions in Huh7 cells at two time points (Figure 2A and B). Subsequently, the cell viability was assayed by MTT. The results showed that Gli-1 siRNA decreased cell viability to 88.3% ± 2.3% and 76.8% ± 3.4% after twenty-four hours and forty-eight hours treatment, respectively (Figure 2C).
Figure 2.
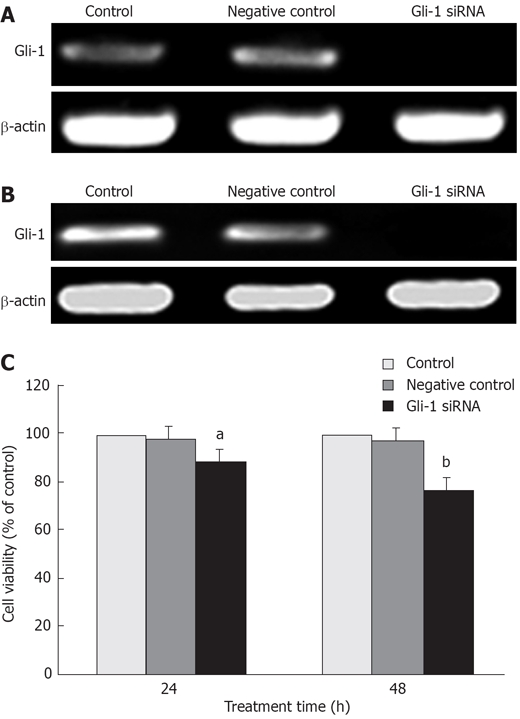
The effect of Gli-1 siRNA on cell viability in Huh7 cells. A: The expressions of Gli-1 mRNA 24 h after transfection with Gli-1 siRNA analyzed by RT-PCR. Gli-1 mRNA expressions were significantly down-regulated; B: The expressions of Gli-1 mRNA 48 h after transfection with Gli-1 siRNA analyzed by RT-PCR; C: The effect of Gli-1 siRNA on Huh7 cell viability by MTT assay. Cells were treated with Gli-1 siRNA for indicated time. Data are expressed as the mean ± SD of 3 independent experiments (aP < 0.05 , bP < 0.01 ; vs control).
Gli-1 siRNA induced apoptosis in Huh7 cells
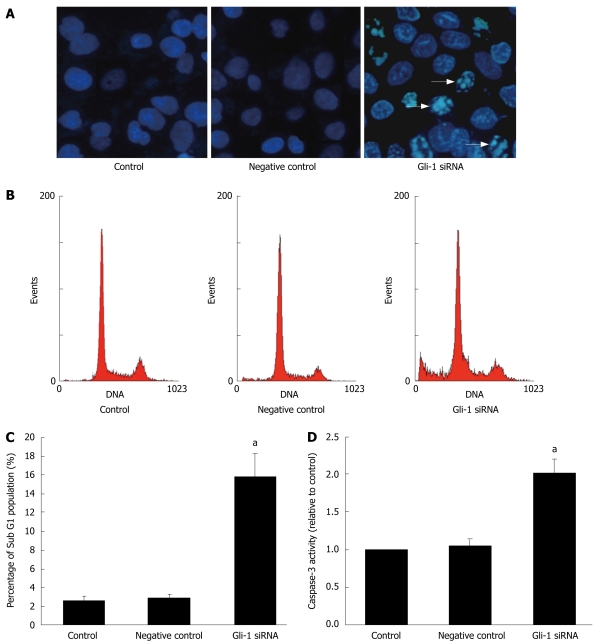
To test whether cell viability decreases by Gli-1 siRNA through apoptosis, we evaluated morphologic and biochemical changes typical of apoptosis. As illustrated in Figure 3A, chromatin condensation was seen by Hoechst 33 258 staining forty-eight hours after Huh7 cells treated with 100 nmol/L of Gli-1 siRNA. As apoptosis progresses into the late stage, apoptotic bodies with partially degraded DNA and DNA cleaved at internucleosomal sites appear. These apoptotic bodies were detected as the sub-G1 fraction on propidium iodide fluorescent flow cytometry analysis. Huh7 cells were treated for forty-eight hours with Gli-1 siRNA and subsequently stained with propidium iodide for flow cytometry. The cellular DNA content analysis revealed that Gli-1 siRNA induced more apoptosis than did either control or negative control (Figure 3B). The average percentages of the sub-G1 cells are plotted in Figure 3C. Gli-1 siRNA significantly increased the percentages of the sub-G1 cells by 15.8% ± 2.6%. Caspase-3 is classically regarded as executor. The caspase-3 activities in cell lysates were assayed using a colorimetric assay kit. In accordance with the increase of sub-G1 cells, Gli-1 siRNA increased 2.01-fold caspase-3 activity compared with the activity in untreated control cells (Figure 3D). These results indicated that Gli-1 siRNA reduced Huh7 cell viability through apoptosis induction.
Figure 3.
Gli-1 siRNA induced apoptosis in Huh7 cells. Huh7 cells were treated with Gli-1 siRNA for 48 h. A: Morphologic changes characteristic of apoptosis. The samples were stained with Hoechst 33 258 (5 mg/L) for 10 min and photographed using fluorescence microscopy; B: Gli-1 SiRNA induced apoptosis as measured by propidium iodide using a flow cytometry analysis. After treatment, the nuclei were stained with propidium iodide and analyzed by flow cytometry. Representative frequency distributions of DNA content of cells in the three treatment groups were shown; C: The bar chart showed the percentage of cells in the sub-G1 apoptotic fraction. The treatment groups were labeled on the horizontal axis. The error bars represented the standard deviation; D: Gli-1 siRNA induced activation of caspase-3. The activity of caspase-3 was assessed by caspase colorimetric assay. The data were showed as mean ± SD of 3 independent experiments. (aP < 0.05 vs control).
Gli-1 siRNA mediated apoptosis by decreased expressions of the Bcl-2 gene
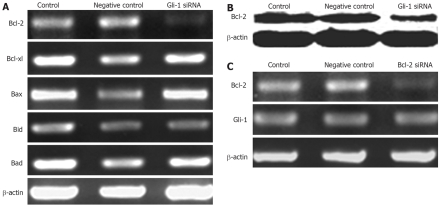
Several cytoplasmic proteins are critically involved in the regulation of apoptosis, in particular, members of the Bcl-2 family[33–35]. Gli-1 has been shown to up-regulate expressions of the key anti-apoptotic factor Bcl-2 in some malignancies[36,37]. To determine whether Gli-1 siRNA-induced apoptosis in Huh7 cells was associated with change of Bcl-2 family, Bcl-2 family was analyzed after treatment with Gli-1 siRNA. The expressions of Bcl-2 mRNA were remarkably decreased twenty-four hours after treatment with Gli-1 siRNA. In contrast, Bcl-xl, Bax, Bad, and Bid were unchanged (Figure 4A). Consequently, we used western blot to detect Bcl-2 protein. Treatment with Gli-1 siRNA for twenty-four hours resulted in an obvious decrease of Bcl-2 expression. To investigate whether Gli-1 is upstream of Bcl-2, Huh7 cells were treated with Bcl-2 siRNA. The findings showed that the expressions of Gli-1 were unchanged (Figure 4C). These results together with the data presented above indicated that Gli-1 was upstream of Bcl-2 in the hierarchical pathway of apoptosis regulation.
Figure 4.
Effects of Gli-1 siRNA on Bcl-2 family member expressions in Huh7 cells. A: The expressions of Bcl-2 family member mRNA 24 h after transfection with Gli-1 siRNA analyzed by RT-PCR. The expression of Bcl-2 was remarkably decreased, but no change for other members; B: The protein of Bcl-2 was detected by Western blot. The Bcl-2 protein was markedly decreased after 24 h treatment with Gli-1 siRNA; C: Bcl-2 siRNA effectively inhibited the expressions of Bcl-2, but the mRNA of Gli-1 was no change after transfection with Bcl-2 siRNA.
Gli-1 siRNA increased cytotoxic effects of 5-Fu in Huh7 cells
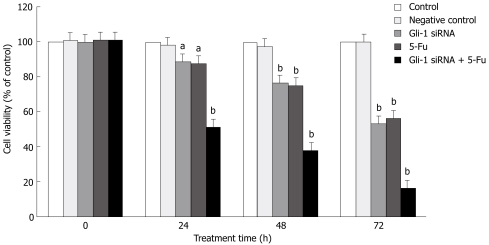
Hedgehog cascades have been proved to be involved in the resistance to clinical therapies and disease relapse in some cancers[38–40]. We further tested if Gli-1 siRNA could increase the cytotoxic effects of 5-Fu. The results showed that significant increased cytotoxicity effects were observed in Huh7 cells treated with Gli-1 siRNA plus 5-Fu (Figure 5). Moreover, Gli-1 siRNA enhanced 5-Fu-induced apoptosis in Huh7 cells (Table 1).
Figure 5.
Effect of Gli-1 siRNA combining with 5-Fu on Huh7 cells. The effect of Gli-1 siRNA combining with 5-Fu on Huh7 cell viability was measured by MTT assay. Cells were transfected with Gli-1 siRNA. After 6 h, cells were exposed to 5-Fu (25 μmol/L) or not for the indicated time periods. The bars are grouped according to the treatment time as labeled below the horizontal axis. The treatment groups were as labeled on the color key. Data are expressed as the mean ± SD of 3 independent experiments (aP < 0.05 ; bP < 0.01 vs control or negative control).
Table 1.
Combined effect of Gli-1 siRNA and 5-Fu on apoptosis in Huh7 cells
| Group | Percentage of sub G1 population (%) |
| Control | 2.63 ± 0.28 |
| Negative control | 2.78 ± 0.39 |
| Gli-1 siRNA | 15.8 ± 2.13a |
| 5-Fu | 27.29 ± 3.14a |
| 5-Fu + Gli-1 siRNA | 44.25 ± 4.59a,c |
Huh7 Cells were transfected with Gli-1 siRNA. After 6 h, cells were treated in the presence or absence of 25 μmol/L 5-Fu for 48 h. After treatment, the nuclei were stained with propidium iodide and analyzed by flow cytometry. The percentage of cells in the sub-G1 apoptotic fraction is shown in table. The data shown as mean ± SD were from three independent experiments. ANOVA analysis showed difference between groups was significant (aP < 0.05 vs control or negative control; cP < 0.05 vs other treatment groups).
DISCUSSION
Hedgehog signaling pathway has been found to play a critical role in regulating the growth, proliferation, and apoptosis of several cancers. Various types of cancers contain Hh activation. We have shown previously that hedgehog signaling is activated in HCC[22]. In this study, we found that Hh pathway components including Gli-1, the marker of Hh pathway activation, were expressed in the HCC cell line Huh7 but not in L02 normal liver cells. This provides a target to preferentially kill HCC cells.
Previous studies demonstrated that the naturally occurring plant extract cyclopamine or KAAD-cyclopamine that directly associates with the trans-membrane domains of SMO could inhibit proliferation, induce apoptosis in a subset of HCC cell lines[22,24]. These observations suggest a possibility for developing new therapeutic strategies using the Hh pathway inhibitor to kill cancer cells with Hh pathway activation. The Hh pathway can be blocked at several steps, such as blocking Smo with antagonist, e.g. cyclopamine or KAAD-cyclopamine[22–24,41], Shh antibody[17,20] and Gli-1 RNA interference[25]. Blocking Smo with an antagonist can generally inhibit the growth of tumor cells with activated Hh signaling, however, this drug may be less effective in tumors with genetic mutations downstream of SMO[42]. Mutation of Smo was found in human HCC[24]. Effective treatment of Huh7 cells will require identification of novel small molecular weight compounds acting downstream of Smo signaling. Thus, we hypothesized that Gli-1 siRNA that directly inhibit Gli-1 could be effective for Huh7 cells. The present study investigated the effects of Gli-1 siRNA on Huh7 cells. We demonstrated that Gli-1 siRNA down-regulated expression of Gli-1 and significantly decreased cell viabilities in Huh7 cells via inducing apoptosis. These findings are consistent and extended with the findings of Chen et al regarding induction of apoptosis and decrease of cell proliferation in ovarian carcinoma cells[43] and of Huang et al regarding Gli-1 which may be an important regulator in survival of HCC cells[22]. Furthermore, these observations are in agreement with the mechanism of the Hh pathway blockage reported previously. Nevertheless, our findings in this paper are unlike that of Gli-2 in playing a dominant role over Gli-1 in regulating the proliferation of HCC cells[44]. Thus, even in the same cell line, it seems that inhibition of Gli-1 has different effects on cell survival according to the type of knock down method. However, it is still possible that inhibition of Gli-2 may have more cytotoxic effect on HCC cells. It will be worthwhile to further evaluate. On the other hand, the cytotoxic effects of siRNA were moderate. This may be because of autocrine Shh expression in Huh7 cells that continuously drive the expression of Gli-1 to lower the effect of siRNA. Our unpublished data indicate that in low concentration of Gli-1 siRNA-treatment, Gli-1 almost recovers after twenty-four hours of treatment even when Gli-1 is silenced completely at the twelve hour time point. The findings suggest that autocrine Shh have impact on the effect of Gli-1 siRNA. It should be given the rational concentration to exert the best effect of Gli-1 siRNA on HCC.
The Bcl-2 family proteins are important regulators of apoptosis. Among the members of this family, Bcl-2 and Bcl-xl act as inhibitors of apoptosis, whereas Bax and Bak act as promoters of apoptosis. Mitochondrial apoptotic pathway is activated while Bax is over-expressed and transported to the mitochondria. Conversely, Bcl-2 inhibits Bax activity through binding to Bax. Thus, Bcl-2 was thought to be a very important anti-apoptotic protein in cells[34,35]. Several studies showed the increased expression of Bcl-2 in HCC contributed to cell death inhibition[45,46]. It is of interest to establish a direct link between Hh/Gli signaling and Bcl-2 expression. Bigelow et al showed that Gli1 was able to stimulate the Bcl-2 promotor[37]. Morton et al recently also demonstrated that activation of hedgehog signaling protected tumor cells from apoptosis through the stabilization of Bcl-2 protein[47]. We showed that Gli-1 siRNA induced apoptosis in Huh7 cells. On the basis of the findings reported here it is reasonable to assume that the effect of Gli-1 siRNA would, at least in part, be mediated by the regulation of Bcl-2 expression. In the present study, we showed that Gli-1 siRNA remarkably reduced the level of Bcl-2 mRNA expressions and Bcl-2 protein, but was not effective on other Bcl-2 family members. Moreover, the level of Gli-1 mRNA had no change after transfection with Bcl-2 siRNA in Huh7 cells. These findings suggest that over-expression of Bcl-2 is a consequence of the activation of Gli-1. Thus, silencing Gli-1 would induce apoptosis through down-regulation of Bcl-2 expressions in Huh7 cells. Nevertheless, the involved apoptotic pathway of Gli-1 siRNA-induced apoptosis in Huh7 cells through down-regulation of Bcl-2 remains to be investigated.
5-Fu is the most common chemotherapeutic agent in advanced HCC treatment. However, drug-resistance results in failure of chemotherapy. Loss of apoptosis pathway promoted by Hh signaling activation has been suggested to be the key mechanism by which HCC is resistant to 5-Fu. On the basis of the ability of Gli-1 siRNA to induce apoptosis in Huh7 cells, one would expect that Gli-1 siRNA should increase sensitivity of Huh7 cells to 5-Fu. In fact, the present study showed that Gli-1 siRNA increased chemosensitivity significantly. These results suggest that it is indeed possible to combine Gli-1 siRNA and 5-Fu to overcome drug resistance and enhance therapeutic activity. It would be important to further test the effect of other combinations of Gli-1 down-regulating agents with 5-Fu on a variety of HCC cells. Its potential therapeutic implications seem to justify future research efforts in this area.
In summary, our data provide a potential mechanism for Gli-1 siRNA-induced apoptosis. Our findings suggest that combination Gli-1 siRNA with chemotherapeutic drug could represent a more promising strategy against HCC. The effects of the strategies need further investigation in other HCC cell lines and in vivo.
COMMENTS
Background
Hedgehog pathway plays an important role in the development and maintenance of human hepatocellular carcinoma (HCC). Thus, there has been increasing interest in the application of specific Hh pathway inhibitors in cancer therapy for many years. Current strategies focus on the use of small molecule inhibitors of Smo. However, tumors with inactivating mutations in Smo or downstream components will most likely not respond to Smo antagonists. Gli-1 is the last and essential step of the pathway and would be the ideal target for HCC therapy. However, the effects of Gli-1 small interfering RNA (siRNA) on the human liver cancer cells remains unclear.
Research frontiers
We demonstrated that Gli-1 siRNA significantly decreased cell viabilities via inducing apoptosis through down-regulation of Bcl-2 expression in Huh7 cells. Furthermore, Gli-1 siRNA increased cytotoxic effects of 5-Fu in Huh7 cells.
Innovations and breakthroughs
Our data provide a potential mechanism for Gli-1 siRNA-induced apoptosis. Moreover, our findings show that Gli-1 siRNA significantly increased sensitivity of chemotherapy and suggest that combination Gli-1 siRNA with a chemotherapeutic drug could represent a more promising strategy against HCC. Overcome drug-resistance via Hh pathway inhibition will be a big breakthrough in HCC therapy. To our knowledge, this is the first discovery of Gli-1 siRNA causing apoptosis via down-regulation of bcl-2.
Applications
Our findings suggest that combination Gli-1 siRNA with chemotherapeutic drug could represent a more promising strategy against HCC. Furthermore, findings of Gli-1 siRNA significantly induced apoptosis through down-regulation of Bcl-2 expression are important in understanding regulation of apoptosis and a rational mechanism-based design of chemotherapy combinations for future clinical testing. Since about 75% of cancer cells of various origins and types are sensitive to drugs with a similar mechanism of action, the findings in this study are highly likely to be applicable to a wide range of cancers. Understanding these molecular mechanisms will allow us to identify novel therapeutic targets and design innovative combination chemotherapy regimens.
Terminology
Gli-1: It is a glioma-associated oncogene homologue 1 gene, Gli-1 is thought to be a direct transcriptional Hh target gene and the marker of Hh pathway activation. Bcl-2: is a family composed of various members, which are generated in mitochondria and key regulators of apoptosis. Bcl-2 is an anti-apoptotic protein and exhibits high expression in a wide range of human cancers including HCC. The increased expression of bcl-2 contributed to cell death inhibition. SiRNA: Small Interfering RNA (siRNA) is a 21-23-nt double-stranded RNA molecule. It guides the cleavage and degradation of its cognate RNA.
Peer review
This paper reports that Gli-1 siRNA induces apoptosis of human hepatocellular carcinoma cells and that this is mediated by down-regulation of Bcl-2. Furthermore, Gli-1 siRNA increased the cytotoxic effect of 5-Fu on Huh7 cells. These results extend the authors’ previous observations and in agreement with the mechanism of Hh pathway blockage reported previously. The authors tried to identify novel therapeutic targets and design a more promising strategy based on Hh pathway inhibitor against HCC.
Acknowledgments
The authors appreciate the Department of Surgical Laboratory, the First Affiliated Hospital of Sun Yat-Sen University, for providing experimental instruments and equipment.
Supported by The National Science Foundation of China, No. 30672053
Peer reviewer: Rashmi, Kaul, PhD, Assistant Professor, Department of Biochemistry and Microbiology, Oklahoma State University- Center for Health Sciences, 1111 W. 17th St. Tulsa, Oklahoma 74107, United States
S- Editor Liu Y L- Editor Alpini GD E- Editor Lu W
References
- 1.Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 2.Pasca di Magliano M, Hebrok M. Hedgehog signalling in cancer formation and maintenance. Nat Rev Cancer. 2003;3:903–911. doi: 10.1038/nrc1229. [DOI] [PubMed] [Google Scholar]
- 3.Ruel L, Rodriguez R, Gallet A, Lavenant-Staccini L, Therond PP. Stability and association of Smoothened, Costal2 and fused with Cubitus interruptus are regulated by Hedgehog. Nat Cell Biol. 2003;5:907–913. doi: 10.1038/ncb1052. [DOI] [PubMed] [Google Scholar]
- 4.Denef N, Neubuser D, Perez L, Cohen SM. Hedgehog induces opposite changes in turnover and subcellular localization of patched and smoothened. Cell. 2000;102:521–531. doi: 10.1016/s0092-8674(00)00056-8. [DOI] [PubMed] [Google Scholar]
- 5.Taipale J, Cooper MK, Maiti T, Beachy PA. Patched acts catalytically to suppress the activity of Smoothened. Nature. 2002;418:892–897. doi: 10.1038/nature00989. [DOI] [PubMed] [Google Scholar]
- 6.Ruiz i Altaba A, Palma V, Dahmane N. Hedgehog-Gli signalling and the growth of the brain. Nat Rev Neurosci. 2002;3:24–33. doi: 10.1038/nrn704. [DOI] [PubMed] [Google Scholar]
- 7.Cohen MM Jr. The hedgehog signaling network. Am J Med Genet A. 2003;123:5–28. doi: 10.1002/ajmg.a.20495. [DOI] [PubMed] [Google Scholar]
- 8.Wang B, Fallon JF, Beachy PA. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000;100:423–434. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- 9.Shin SH, Kogerman P, Lindstrom E, Toftgard R, Biesecker LG. GLI3 mutations in human disorders mimic Drosophila cubitus interruptus protein functions and localization. Proc Natl Acad Sci USA. 1999;96:2880–2884. doi: 10.1073/pnas.96.6.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bai CB, Stephen D, Joyner AL. All mouse ventral spinal cord patterning by hedgehog is Gli dependent and involves an activator function of Gli3. Dev Cell. 2004;6:103–115. doi: 10.1016/s1534-5807(03)00394-0. [DOI] [PubMed] [Google Scholar]
- 11.Buttitta L, Mo R, Hui CC, Fan CM. Interplays of Gli2 and Gli3 and their requirement in mediating Shh-dependent sclerotome induction. Development. 2003;130:6233–6243. doi: 10.1242/dev.00851. [DOI] [PubMed] [Google Scholar]
- 12.Motoyama J, Milenkovic L, Iwama M, Shikata Y, Scott MP, Hui CC. Differential requirement for Gli2 and Gli3 in ventral neural cell fate specification. Dev Biol. 2003;259:150–161. doi: 10.1016/s0012-1606(03)00159-3. [DOI] [PubMed] [Google Scholar]
- 13.Dai P, Akimaru H, Tanaka Y, Maekawa T, Nakafuku M, Ishii S. Sonic Hedgehog-induced activation of the Gli1 promoter is mediated by GLI3. J Biol Chem. 1999;274:8143–8152. doi: 10.1074/jbc.274.12.8143. [DOI] [PubMed] [Google Scholar]
- 14.Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 15.Athar M, Li C, Tang X, Chi S, Zhang X, Kim AL, Tyring SK, Kopelovich L, Hebert J, Epstein EH Jr, et al. Inhibition of smoothened signaling prevents ultraviolet B-induced basal cell carcinomas through regulation of Fas expression and apoptosis. Cancer Res. 2004;64:7545–7552. doi: 10.1158/0008-5472.CAN-04-1393. [DOI] [PubMed] [Google Scholar]
- 16.Berman DM, Karhadkar SS, Hallahan AR, Pritchard JI, Eberhart CG, Watkins DN, Chen JK, Cooper MK, Taipale J, Olson JM, et al. Medulloblastoma growth inhibition by hedgehog pathway blockade. Science. 2002;297:1559–1561. doi: 10.1126/science.1073733. [DOI] [PubMed] [Google Scholar]
- 17.Watkins DN, Berman DM, Burkholder SG, Wang B, Beachy PA, Baylin SB. Hedgehog signalling within airway epithelial progenitors and in small-cell lung cancer. Nature. 2003;422:313–317. doi: 10.1038/nature01493. [DOI] [PubMed] [Google Scholar]
- 18.Kubo M, Nakamura M, Tasaki A, Yamanaka N, Nakashima H, Nomura M, Kuroki S, Katano M. Hedgehog signaling pathway is a new therapeutic target for patients with breast cancer. Cancer Res. 2004;64:6071–6074. doi: 10.1158/0008-5472.CAN-04-0416. [DOI] [PubMed] [Google Scholar]
- 19.Karhadkar SS, Bova GS, Abdallah N, Dhara S, Gardner D, Maitra A, Isaacs JT, Berman DM, Beachy PA. Hedgehog signalling in prostate regeneration, neoplasia and metastasis. Nature. 2004;431:707–712. doi: 10.1038/nature02962. [DOI] [PubMed] [Google Scholar]
- 20.Berman DM, Karhadkar SS, Maitra A, Montes De Oca R, Gerstenblith MR, Briggs K, Parker AR, Shimada Y, Eshleman JR, Watkins DN, et al. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature. 2003;425:846–851. doi: 10.1038/nature01972. [DOI] [PubMed] [Google Scholar]
- 21.Thayer SP, di Magliano MP, Heiser PW, Nielsen CM, Roberts DJ, Lauwers GY, Qi YP, Gysin S, Fernandez-del Castillo C, Yajnik V, et al. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature. 2003;425:851–856. doi: 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang S, He J, Zhang X, Bian Y, Yang L, Xie G, Zhang K, Tang W, Stelter AA, Wang Q, et al. Activation of the hedgehog pathway in human hepatocellular carcinomas. Carcinogenesis. 2006;27:1334–1340. doi: 10.1093/carcin/bgi378. [DOI] [PubMed] [Google Scholar]
- 23.Sicklick JK, Li YX, Jayaraman A, Kannangai R, Qi Y, Vivekanandan P, Ludlow JW, Owzar K, Chen W, Torbenson MS, et al. Dysregulation of the Hedgehog pathway in human hepatocarcinogenesis. Carcinogenesis. 2006;27:748–757. doi: 10.1093/carcin/bgi292. [DOI] [PubMed] [Google Scholar]
- 24.Patil MA, Zhang J, Ho C, Cheung ST, Fan ST, Chen X. Hedgehog signaling in human hepatocellular carcinoma. Cancer Biol Ther. 2006;5:111–117. doi: 10.4161/cbt.5.1.2379. [DOI] [PubMed] [Google Scholar]
- 25.Sanchez P, Hernandez AM, Stecca B, Kahler AJ, DeGueme AM, Barrett A, Beyna M, Datta MW, Datta S, Ruiz i Altaba A. Inhibition of prostate cancer proliferation by interference with SONIC HEDGEHOG-GLI1 signaling. Proc Natl Acad Sci USA. 2004;101:12561–12566. doi: 10.1073/pnas.0404956101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanchez P, Ruiz i Altaba A. In vivo inhibition of endogenous brain tumors through systemic interference of Hedgehog signaling in mice. Mech Dev. 2005;122:223–230. doi: 10.1016/j.mod.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 27.Chen JK, Taipale J, Cooper MK, Beachy PA. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 2002;16:2743–2748. doi: 10.1101/gad.1025302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor MD, Liu L, Raffel C, Hui CC, Mainprize TG, Zhang X, Agatep R, Chiappa S, Gao L, Lowrance A, et al. Mutations in SUFU predispose to medulloblastoma. Nat Genet. 2002;31:306–310. doi: 10.1038/ng916. [DOI] [PubMed] [Google Scholar]
- 29.Cao LQ, Chen XL, Wang Q, Huang XH, Zhen MC, Zhang LJ, Li W, Bi J. Upregulation of PTEN involved in rosiglitazone-induced apoptosis in human hepatocellular carcinoma cells. Acta Pharmacol Sin. 2007;28:879–887. doi: 10.1111/j.1745-7254.2007.00571.x. [DOI] [PubMed] [Google Scholar]
- 30.Zhang XF, Li BX, Dong CY, Ren R. Apoptosis of human colon carcinoma HT-29 cells induced by ceramide. World J Gastroenterol. 2006;12:3581–3584. doi: 10.3748/wjg.v12.i22.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhen MC, Huang XH, Wang Q, Sun K, Liu YJ, Li W, Zhang LJ, Cao LQ, Chen XL. Green tea polyphenol epigallocatechin-3-gallate suppresses rat hepatic stellate cell invasion by inhibition of MMP-2 expression and its activation. Acta Pharmacol Sin. 2006;27:1600–1607. doi: 10.1111/j.1745-7254.2006.00439.x. [DOI] [PubMed] [Google Scholar]
- 32.She M, Yang H, Sun L, Yeung SC. Redox control of manumycin A-induced apoptosis in anaplastic thyroid cancer cells: involvement of the xenobiotic apoptotic pathway. Cancer Biol Ther. 2006;5:275–280. doi: 10.4161/cbt.5.3.2383. [DOI] [PubMed] [Google Scholar]
- 33.Swerdlow S, Distelhorst CW. Bcl-2-regulated calcium signals as common mediators of both apoptosis and autophagy. Dev Cell. 2007;12:178–179. doi: 10.1016/j.devcel.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 34.Marsden VS, Ekert PG, Van Delft M, Vaux DL, Adams JM, Strasser A. Bcl-2-regulated apoptosis and cytochrome c release can occur independently of both caspase-2 and caspase-9. J Cell Biol. 2004;165:775–780. doi: 10.1083/jcb.200312030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marsden VS, O'Connor L, O'Reilly LA, Silke J, Metcalf D, Ekert PG, Huang DC, Cecconi F, Kuida K, Tomaselli KJ, et al. Apoptosis initiated by Bcl-2-regulated caspase activation independently of the cytochrome c/Apaf-1/caspase-9 apoptosome. Nature. 2002;419:634–637. doi: 10.1038/nature01101. [DOI] [PubMed] [Google Scholar]
- 36.Regl G, Kasper M, Schnidar H, Eichberger T, Neill GW, Philpott MP, Esterbauer H, Hauser-Kronberger C, Frischauf AM, Aberger F. Activation of the BCL2 promoter in response to Hedgehog/GLI signal transduction is predominantly mediated by GLI2. Cancer Res. 2004;64:7724–7731. doi: 10.1158/0008-5472.CAN-04-1085. [DOI] [PubMed] [Google Scholar]
- 37.Bigelow RL, Chari NS, Unden AB, Spurgers KB, Lee S, Roop DR, Toftgard R, McDonnell TJ. Transcriptional regulation of bcl-2 mediated by the sonic hedgehog signaling pathway through gli-1. J Biol Chem. 2004;279:1197–1205. doi: 10.1074/jbc.M310589200. [DOI] [PubMed] [Google Scholar]
- 38.Sims-Mourtada J, Izzo JG, Ajani J, Chao KS. Sonic Hedgehog promotes multiple drug resistance by regulation of drug transport. Oncogene. 2007;26:5674–5679. doi: 10.1038/sj.onc.1210356. [DOI] [PubMed] [Google Scholar]
- 39.Mimeault M, Johansson SL, Vankatraman G, Moore E, Henichart JP, Depreux P, Lin MF, Batra SK. Combined targeting of epidermal growth factor receptor and hedgehog signaling by gefitinib and cyclopamine cooperatively improves the cytotoxic effects of docetaxel on metastatic prostate cancer cells. Mol Cancer Ther. 2007;6:967–978. doi: 10.1158/1535-7163.MCT-06-0648. [DOI] [PubMed] [Google Scholar]
- 40.Sims-Mourtada J, Izzo JG, Apisarnthanarax S, Wu TT, Malhotra U, Luthra R, Liao Z, Komaki R, van der Kogel A, Ajani J, et al. Hedgehog: an attribute to tumor regrowth after chemoradiotherapy and a target to improve radiation response. Clin Cancer Res. 2006;12:6565–6572. doi: 10.1158/1078-0432.CCR-06-0176. [DOI] [PubMed] [Google Scholar]
- 41.Williams JA, Guicherit OM, Zaharian BI, Xu Y, Chai L, Wichterle H, Kon C, Gatchalian C, Porter JA, Rubin LL, et al. Identification of a small molecule inhibitor of the hedgehog signaling pathway: effects on basal cell carcinoma-like lesions. Proc Natl Acad Sci USA. 2003;100:4616–4621. doi: 10.1073/pnas.0732813100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taipale J, Chen JK, Cooper MK, Wang B, Mann RK, Milenkovic L, Scott MP, Beachy PA. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature. 2000;406:1005–1009. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- 43.Chen X, Horiuchi A, Kikuchi N, Osada R, Yoshida J, Shiozawa T, Konishi I. Hedgehog signal pathway is activated in ovarian carcinomas, correlating with cell proliferation: it's inhibition leads to growth suppression and apoptosis. Cancer Sci. 2007;98:68–76. doi: 10.1111/j.1349-7006.2006.00353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim Y, Yoon JW, Xiao X, Dean NM, Monia BP, Marcusson EG. Selective down-regulation of glioma-associated oncogene 2 inhibits the proliferation of hepatocellular carcinoma cells. Cancer Res. 2007;67:3583–3593. doi: 10.1158/0008-5472.CAN-06-3040. [DOI] [PubMed] [Google Scholar]
- 45.Kanda T, Yokosuka O, Imazeki F, Arai M, Saisho H. Enhanced sensitivity of human hepatoma cells to 5-fluorouracil by small interfering RNA targeting Bcl-2. DNA Cell Biol. 2005;24:805–809. doi: 10.1089/dna.2005.24.805. [DOI] [PubMed] [Google Scholar]
- 46.Su SJ, Chow NH, Kung ML, Hung TC, Chang KL. Effects of soy isoflavones on apoptosis induction and G2-M arrest in human hepatoma cells involvement of caspase-3 activation, Bcl-2 and Bcl-XL downregulation, and Cdc2 kinase activity. Nutr Cancer. 2003;45:113–123. doi: 10.1207/S15327914NC4501_13. [DOI] [PubMed] [Google Scholar]
- 47.Morton JP, Mongeau ME, Klimstra DS, Morris JP, Lee YC, Kawaguchi Y, Wright CV, Hebrok M, Lewis BC. Sonic hedgehog acts at multiple stages during pancreatic tumorigenesis. Proc Natl Acad Sci USA. 2007;104:5103–5108. doi: 10.1073/pnas.0701158104. [DOI] [PMC free article] [PubMed] [Google Scholar]