Abstract
Extracellular matrix (ECM) molecules modify gene expression through attachment-dependent (i.e., focal adhesion related) integrin receptor signalling. It was previously unknown whether the same molecules acting as soluble peptides could generate signal cascades without the associated mechanical anchoring, a condition that may be encountered during matrix remodelling, degradation and relevant to invasion and metastatic processes. In the current study the role of ECM ligand regulated gene expression through this attachment independent process was examined.
It was observed that fibronectin, laminin, collagens type I and II induce Smad2 activation in MCF-10A and MCF-7 cells. This activation is not caused by TGFβ ligand contamination or autocrine TGF involvement and is 3–5 fold less robust than the TGFβ1 ligand. The resulting nuclear translocation of Smad4 in response to ECM ligand indicates downstream transcriptional responses occurring. Co-immunoprecipitation experiments determined that type II collagen and laminin act through interaction with integrin α2β1 receptor complex. The ECM ligand induced Smad activation (termed signalling crosstalk) resulted cell type and ligand specific transcriptional changes which are distinct from the TGFβ ligand induced responses.
These findings demonstrate that cell-matrix communication is more complex than previously thought. Soluble ECM peptides drive transcriptional regulation through corresponding adhesion and non-attachment related processes. The resultant gene expressional patterns correlate with pathway activity and not by the extent of Smad activation. These results extend the complexity and the existing paradigms of ECM-cell communication to ECM ligand regulation without the necessity of mechanical coupling.
Keywords: Extracellular matrix, TGFβ signalling, Breast Cancer, Gene expression
Introduction
In breast cancer development ECM regulates gene expression and phenotype through adhesion mediated signalling (1, 2). A strong body of evidence indicates the importance of this process in many aspects of tissue homeostasis regulation from stromal fibroblast activation (3) to epithelial to mesenchymal transformation in tumorigenesis (4). Previous studies have focused on the role of ECM as a signal initiator in the context of an adhesion-related process. Tissue remodelling and protease degradation generates neoepitopes from ECM components that potentially act as “soluble” peptides in the peri-cellular microenvironment (5–8). These neoepitopes have been reported to induce changes in migration and cell behaviour in some experimental systems (9–16). While matrix effects are recognized, the contribution these peptides make to cellular phenotype in breast cancer is unknown. This potentially complements cytokines which are liberated and activated during tissue remodelling such as Transforming Growth Factor β (TGFβ) which is involved in epithelial mesenchymal transformation (17).
Transforming Growth Factor β isoforms are produced and deposited into the ECM as inactive complexes by many cell types (18). Ligand activation can be achieved by several mechanisms, including through integrin αvβ6 and αvβ8 receptor interactions that liberate them for receptor binding (19, 20). Signalling occurs when TGFβ isoforms bind and activates the TGFβ receptor complex (type I and II) which is subsequently endocytosed and phosphorylate Smad2 and 3 proteins at their C-terminal SSXS amino acid sequence (21–25). Only ALK4, 5, 7 receptors propagate signalling through Smad2 and 3 recognizing inputs from Activin A, GDF1, GDF11, Nodal, and TGFβ’s ligands (26–28). These kinases are sensitive to SB431542 inhibition with IC50 values 140, 94 nM (ALK4, 5), and ~1 μM (ALK7) respectively (29). All previously reported Smad dependent TGFβ signalling events are require TGFβ ligands for receptor activation. Novel type II collagen and angiotensin II related Smad activation mechanisms have been reported (30, 31). It remains unknown whether the collagen mediated process i) is active in epithelial cells; ii) exists for other ECM molecules; iii) is independent of TGFβ ligand; iv) has specific transcriptional consequences. The current manuscript reports the consequences of soluble ECM induced Smad2 activation. It characterizes type II collagen (CII) and laminin (LAM) effects on the TGFβ/BMP signalling, and pathway specific transcriptional responses in MCF-10A “normal” and MCF-7 (ER+) invasive human breast cancer cell lines. The results indicate that (a) soluble fibronectin, laminin, and collagens type I and II induce Smad2 phosphorylation, which is limited in magnitude if compared to native signalling, (b) this activation induces Smad4 nuclear translocation, (c) resultant Smad activation modulates gene expression in a ligand and cell type specific manner, which is distinct from TGFβ1 induced responses, and (d) this activity cannot be attributed to TGFβ contamination of ECM preparations.
Results
Smad2 is activated by ECM treatments
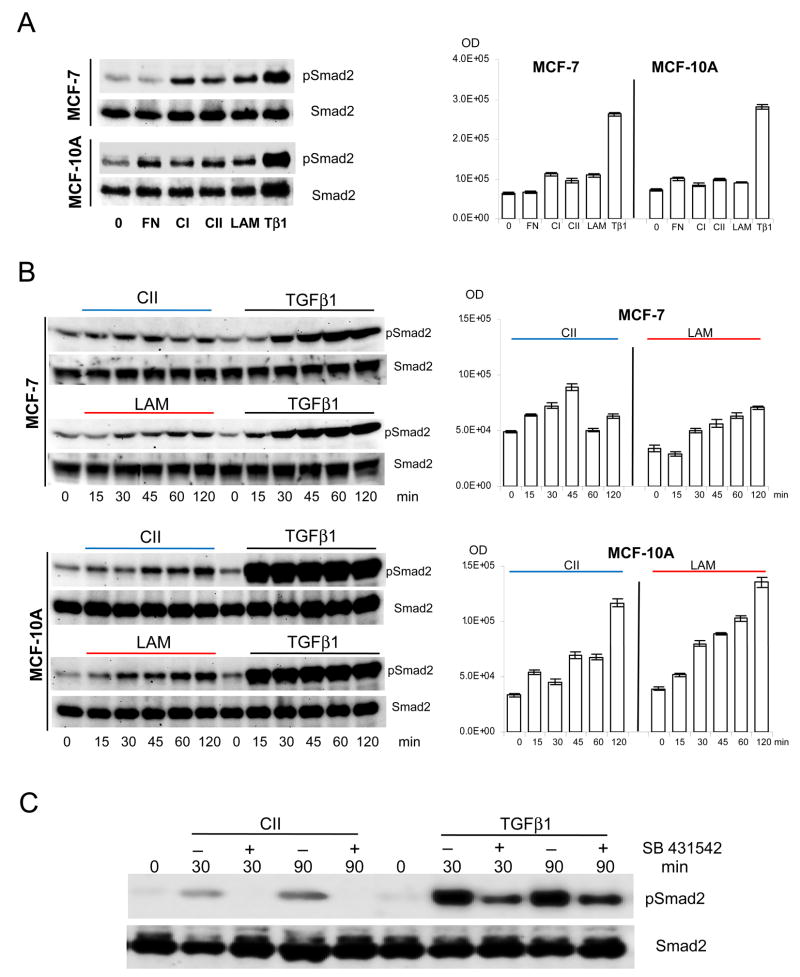
Laminin is a major component of basal membrane surrounding the acinus and breast epithelial cells. To mimic the effect of matrix degradation and cellular remodelling, we investigated how protease digested laminin (LAM), fibronectin (FN), type I and II collagen (CI, CII), peptides can effect Smad2 phosphorylation (Fig. 1A–B). While FN does not induce Smad2 phosphorylation in MCF-7 cells, MCF-10A cells display a 38% increase in Smad2 activation when compared to untreated cells (Fig. 1A). Both cell lines respond to type I collagen (CI) with Smad activation, which is 33% greater in MCF-7 than in MCF-10A. The CI, CII, and LAM peptides induced responses in MCF-7 are 76%, 50%, 72%, and in MCF-10A are 15%, 34%, and 24.5% increased respectively from the un-stimulated controls. In comparison, TGFβ1 results in a 400% Smad2 activation in both cell types. We chose to investigate CII and LAM effects further because α2β1 integrin receptors are the major binding complex for both peptides. Smad activation kinetics was compared at time periods between 0–120 minutes following CII and LAM exposure (Fig. 1B). MCF-10A responses to soluble ECM peptides are greater in magnitude (LAM=347% and CII=350%) than MCF-7 (LAM=207% and CII=29.3% compare densitometry at 120 minutes, right). Phospho-Smad2 levels gradually increased as a result of exposure, with the exception of MCF-7 CII, which peaks earlier at 45 minutes. All ECM Smad2 activations are significantly lower in magnitude than in TGFβ1 initiated responses. Similar responses were documented with JJ012, 105KC chondrosarcoma, C28 chondrocyte, Mv1Lu mink lung epithelial, and WM35 melanoma cell lines with Smad2 and Smad3 activation kinetics (data not shown). To confirm further that Smad2 activation is dependent of ALK4, 5, and/or 7 kinase, sensitivity to TGFβ type I receptor inhibitor SB-431542 was determined (Fig. 1C). As shown, the inhibitor completely abolishes the ECM induced Smad2 phosphorylation while significantly down-regulates the native TGFβ1 ligand induced responses, indicating that both pathways require this kinase activity.
Figure 1.
The MCF cells were handled, plated, synchronized, treated, and harvested as described under Material and Methods section. A. The Smad activation (pSmad2) was tested without ligand (0), with fibronectin (FN), type I and II collagen (CI, CII), and laminin (LAM) (all at 50 μg/ml) and TGFβ1 (at 10 ng/ml) at 45 minutes. B. The time curve for Smad activation is comparable to the known activation kinetics with the observation of limited effectiveness in accumulation of generated pSmad2 signal by ECM treatments. Compare MCF-7 CII and LAM treatments (pSmad2 and Smad2 lanes), vs. MCF-10A CII and LAM and their representative densitometry results at right. The significance of ECM treatments were analyzed on raw images acquired by UVP Bio-Imager (supplied by the software) in triplicates, and then subjected to one-way ANOVA analysis (MATLAB 7.5.0) to establish the probability values (p). The difference in densitometry results of (A) and (B) were significant, p<0.001. C. The p100 plates were pre-incubated with SB-431542 for 30 minutes at 5.0 μM final concentration at 37°C in presence of 5% CO2 before pathway induction with CII and TGFβ1 exposures as above. The parallel samples were then harvested at indicated time points, pelleted, snap frozen in liquid nitrogen, and stored at −80°C until use. 125.0 μg standardized total protein was subjected to SDS-PAGE analysis and western blotting.
Crosstalk signalling is independent from TGFβ peptides, mobilizes full pathway activation and involves integrin α 2β 1 complex
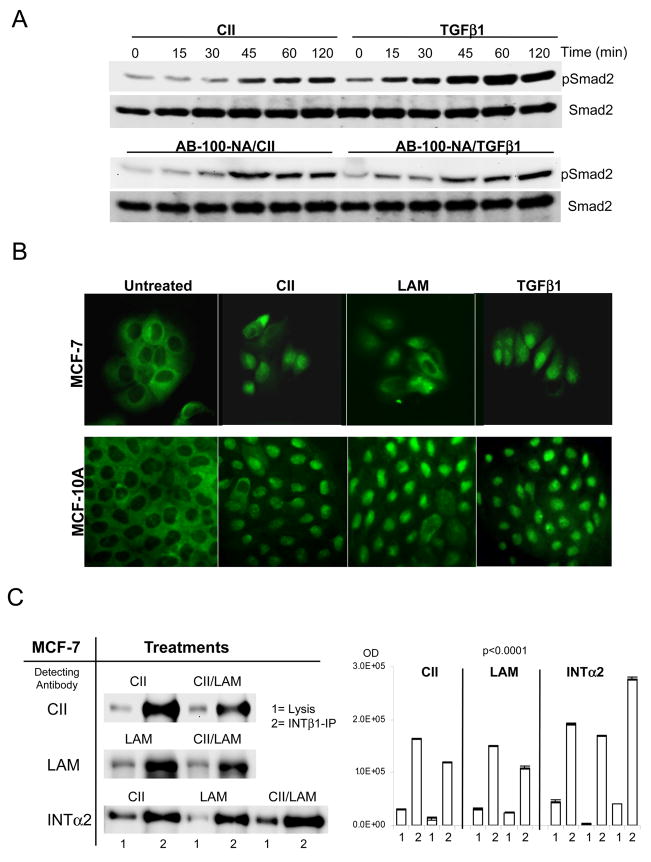
The Smad2 activation by ECM samples raised the question whether the result was caused either by TGFβ contamination, or/with the contribution of endogenously produced cellular inactive TGFβ. To address this concern the Smad activation dynamics were re-examined with CII treatment in the presence of pan-specific TGFβ neutralizing antibody (Fig. 2A, ND50 against hTGFβ1, pTGFβ1.2, pTGFβ2, rcTGFβ3 and raTGFβ5 as 5.0, 1.0, 15.0, 4.0, 1.0 μg/ml respectively). Only TGFβ1 induction was down-regulated by the antibody (compare pSmad2 bands in AB-100-NA/TGFβ1 with TGFβ1 exposure). Contrary to this, the ECM treatment induced activation was not affected (AB-100-NA/CII vs. CII), indicating that endogenously produced TGFβ isoforms did not contribute to the ECM induced Smad activation. The possible TGFβ contamination of ECM peptides was analyzed by MALDI-TOF Mass Spectrometry. No TGFβ1 contamination was detectable in the ECM peptide samples (4.0–14.0 μM ECM sample loaded, 0.4 μM TGFβ1 was the reference control, with instrumental sensitivity in femto/atto-M range, data not shown).
Figure 2.
The cells were treated as described in the materials and methods section. A. Parallel plates were pre-treated with pan-specific AB-100-NA TGFβ neutralizing antibody (R&D with ND50 against hTGFβ1, pTGFβ1.2, pTGFβ2, rcTGFβ3 and raTGFβ5 as 5.0, 1.0, 15.0, 4.0, 1.0 μg/ml respectively). Used 25 μg/ml concentrations for 2 hours (RT) to neutralize exogenously added and endogenous cellular production, then standard activation time curve was established with 50 μg/ml CII, and 2.5 ng/ml TGFβ1 as control. For Smad4 nuclear translocation, MCF-10A and MCF-7 cells were plated at 3×105 cell/well concentration in 6 well plates and synchronised overnight in serum free media then treated as above. B. Following one hour incubation the cells were washed, fixed and processed with Smad4 primary antibody overnight, followed with Alexa-488 secondary antibody for two hours. The images were acquired on Zeiss AxioII microscope with GFP/FITC filter set. C. The CII and LAM are the major peptides for integrin α2β1 receptor complex. After lysis and quantization, 2.0 mg total protein were subjected to integrinβ1 (INTβ1-IP bands 2) for 2 hours then precipitated overnight at 4°C. The ECM treatments are competing (signal down-regulation is less than 50%) for the same receptor population by combined CII/LAM treatments (detecting antibodies [left] pre-treatments [right] with CII detection upper, and LAM detection lanes at centre. Corresponding densitometry, left, and centre). For validation of the right receptor complex pull-down, integrinα2 detection was used (INTα2 lane at bottom, IP bands 2, treatments CII, LAM, CII/LAM, and INTα2 densitometry right). The respective Smad2 bands are generated by stripping and re-probing the membranes. Western blots were repeated in duplicates and corresponding densitometry (right panels) analysis of raw acquired images (UVP imager software) was normalized to Smad2 signals. Statistical analysis was performed as under Figure 1, p=0.0001 was considered highly significant.
The ECM peptide induced Smad activation capacity prompted the analysis of Smad4 nuclear translocation (Fig. 2B) in order to verify that the peptide induced Smad activation is capable of initiating downstream events. As shown, in untreated cells Smad4 is not present in the nuclei, while ECM ligand and TGFβ1 exposure cause nuclear translocation of the signal.
The binding of collagen type II and laminin to the α2β1 integrin receptor complex was confirmed by co-immunoprecipitation experiments in MCF cells (Fig. 2C). Integrin β1 (INTβ1) antibody precipitates integrin α2 (INTα2) receptor independently from ligand exposure (bottom lane, related densitometry right). Its presence increased with combined peptides by 53.84% (CII/LAM response IP bands 2 far right) indicating that additional α2 β1 receptor populations were accessible for complex formation and binding. This result is complemented by ligand competition, in which the detected CII and LAM decreased 27.1 and 27.79% respectively (CII/LAM upper, middle lanes band 2). These results indicate that CII and LAM are in competition for α2β1 integrin receptor binding. The lysis (bands=1) shows the controls of appropriate targets in the total cell lysate. Immunoprecipitation experiments in MCF-10A cells duplicated the results observed in MCF-7 cells (data not shown).
Comparison of gene regulation differences in MCF-10A and MCF-7 cells
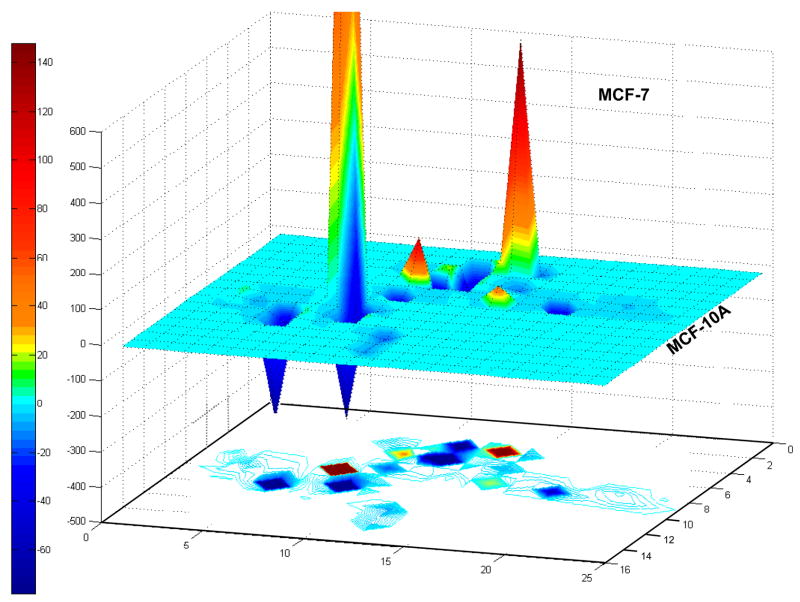
Since Smad2 activation was documented with the phospho-specific antibody recognizing only the double phosphorylated (S465/S467) molecule and this induced Smad4 nuclear translocation, there was an expectation that the pathway activity will cause Smad related gene expressional changes, which can be analyzed by the TGFβ/BMP pathway specific expressional QPCR array. It is also equally important, how these pathway specific genes are regulated differentially between MCF-10A normal and MCF-7 (ER+) invasive untreated human breast cancer cells. Specific TGFβ/BMP signalling arrays were used to characterize cellular responsiveness to each of the ECM peptides. A 4-hour time point was chosen to include still the stable early gene activation events together with the lasting mid-time and late regulations but exclude the transient fluctuations. The cellular expression of 84 genes in MCF-7 was compared to MCF-10A cells (Fig. 3). The MCF-7 expressional profile changes show fundamental up-regulation in 5 genes (BMP7, >CDKN2B, >PDGFB, >GSC), and major down-regulation in 6 genes (INHA, >TGFBR3, >TGFBI, >PLAU, >NOGGIN, >ENG, details are provided in the Supplemental Figure and Table 1). The comparison shows that 63% of genes (53 out of 84) regulated differentially in MCF-7. From these, 13 genes (25%) are up-, and 40 genes (75.5%) are down regulated. Particularly, the Adhesion and Extracellular Molecules group are affected strongly, where 83% of genes are down-regulated from the affected 18, suggesting that MCF-7 is less dependent on adhesion related functions than MCF-10A.
Figure 3.
Experiments were performed in triplicates, and 5.0 μg total RNA was used to generate cDNA. for each plate. The analysis templates for each assay are provided by the SA Biosciences Co. web page (www.superarray.com) which uses 2−ΔΔCt method to calculate fold differences from the QPCR Crossing points (Ct) with confidence analysis T-Test data for each gene investigated. The fold differences then were analyzed for functional Venn groups (Supplemental Material) and the generated gene distribution representative matrix (missing or empty data set to 0) was visualized in MATLAB to give the 3D representation of expressional values and the underlying contours representing the intensity and topological distribution of these changes.
ECM treatments generate distinct expressional responses
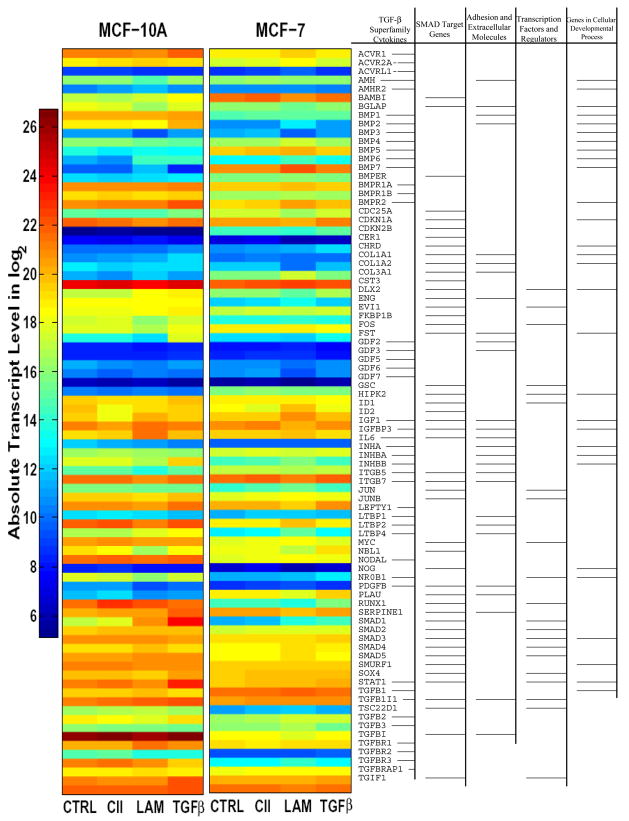
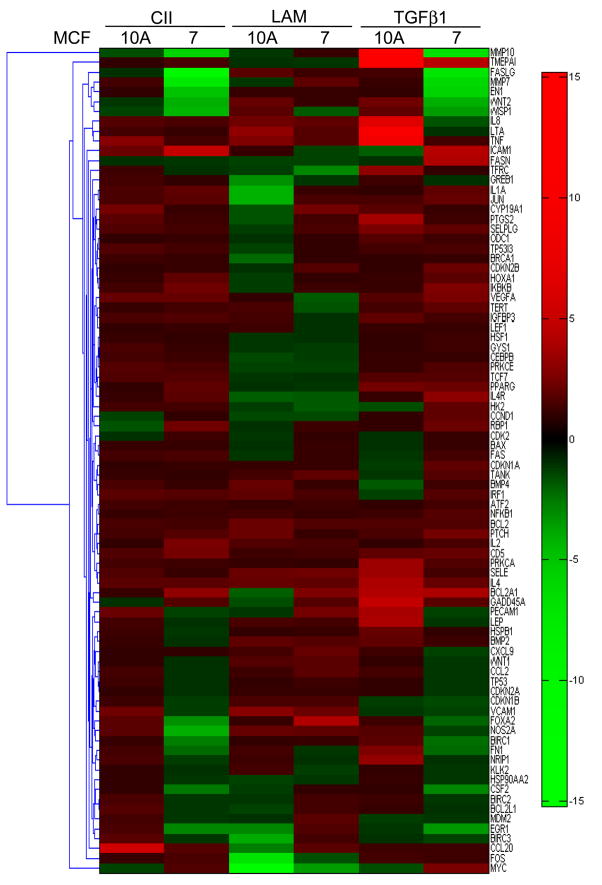
The heat-maps display absolute transcript levels of untreated controls (CTRL), CII, LAM, and TGFβ1 (TGFβ) treated plates (Fig. 4). The results demonstrate that CII, LAM, and TGFβ, regulate different sets of genes, depending on ligand exposure and cell types. The LAM induces more dynamic alternation in gene expression than CII or even TGFβ1 (CTRL vs. LAM, CII, and TGFβ columns in MCF-10A/MCF-7 cells). Specifically, the BMP7, CDC25A, and COL3A1 genes show fundamental up-regulation in MCF-7 and remain responsive to LAM treatment only. The 84 genes are functionally grouped into five major areas (right) according to the assay description (PAHS-035, SA Bioscience Frederick MD). The comparison indicates that overall responsiveness to TGFβ signalling is down-regulated in MCF-7 cells, TGFβ isoforms 1, 2, 3, ACVR1, 2A, receptors, as well as Smad3, 4 transmitters are all down-regulated, and the pathway inhibitor BAMBI is up-regulated.
Figure 4.
Real-time polymerase chain reaction with SYBR Green master mix were used to quantify the expression levels of 84 genes ontologically related and regulated by TGFβ/BMP Signalling pathway, or the 84 genes of Signal Transduction Pathway Finder specific arrays (SA Biosciences, Frederick, MD). The heat-map shows absolute mRNA copy numbers which were calculated from PCR cycle thresholds (Cts, Fig. 4). For example, on the color coded log2 scale, a value of 10 represents 210 or 1024 transcripts. Two endogenous controls, GAPDH and ACTB, were used for normalization. Functional gene clustering (with major groups according to the array manual) indicated at right. In the Signal Transduction Pathway Finder Array (Fig. 5), the fold expression differences were analyzed through the SA Biosciences webpage, then transferred into MATLAB and visualized with the Bioinformatics Toolbox Clustergram function. All experiments were run in triplicates.
While the TGFβ/BMP Signalling specific pathway array focuses on expressional changes related only to Smad signalling activity, and CII, LAM engagement with their integrin receptors are also inducing parallel signalling pathways. How the ECM peptides affected overall signalling activity in MCF-10A and MCF-7 cells was determined with a Signal Transduction Pathway finder array (PAHS-014, Fig. 5). Overall, in normal MCF10A cells, CII, LAM, TGFβ1 response patterns are minimally overlapping (comparison of 10A columns) indicating that the treatments differentially effects specific signalling pathways in these cells. Interestingly, the invasive MCF-7 cell line responds to CII and TGFβ1 in similar but not identical ways (note that there is no contamination or endogenous TGFβ ligand involvement in the ECM induced Smad activation). The LAM affects different pathways and to a different extent (comparison of MCF-7 columns). All three treatments activated the CREB pathway similarly.
Figure 5.
Real-time polymerase chain reaction with SYBR Green master mix were used to quantify the expression levels of 84 genes ontologically related and regulated by TGFβ/BMP Signalling pathway, or the 84 genes of Signal Transduction Pathway Finder specific arrays (SA Biosciences, Frederick, MD). The heat-map shows absolute mRNA copy numbers which were calculated from PCR cycle thresholds (Cts, Fig. 4). For example, on the color coded log2 scale, a value of 10 represents 210 or 1024 transcripts. Two endogenous controls, GAPDH and ACTB, were used for normalization. Functional gene clustering (with major groups according to the array manual) indicated at right. In the Signal Transduction Pathway Finder Array (Fig. 5), the fold expression differences were analyzed through the SA Biosciences webpage, then transferred into MATLAB and visualized with the Bioinformatics Toolbox Clustergram function. All experiments were run in triplicates.
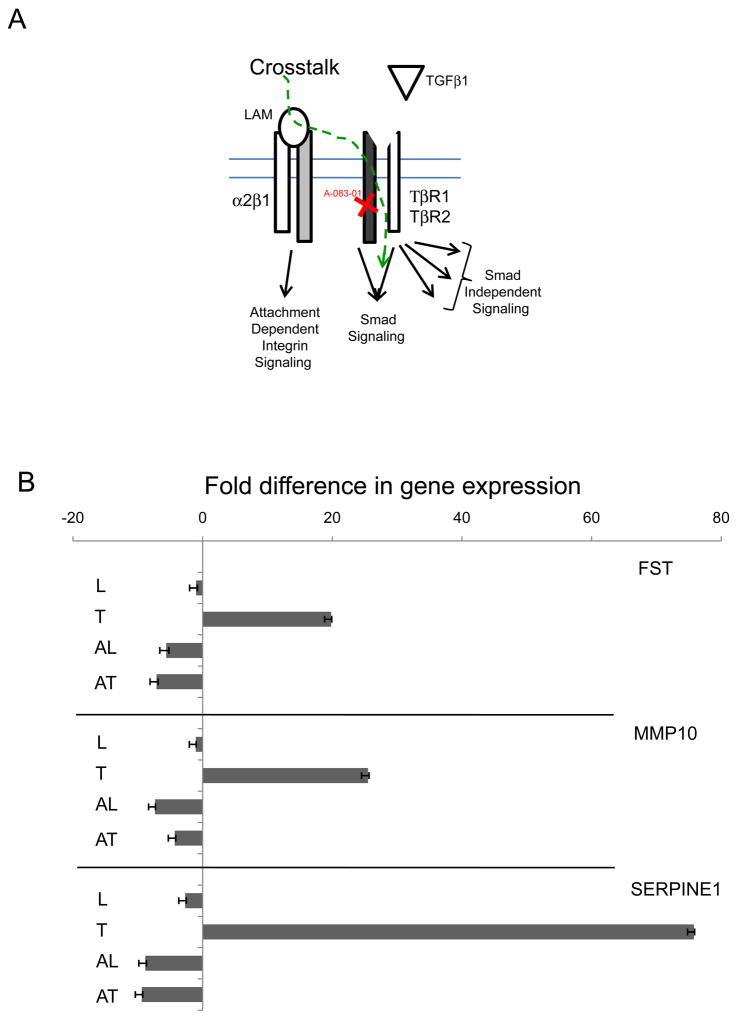
Validation of crosstalk sensitive genes
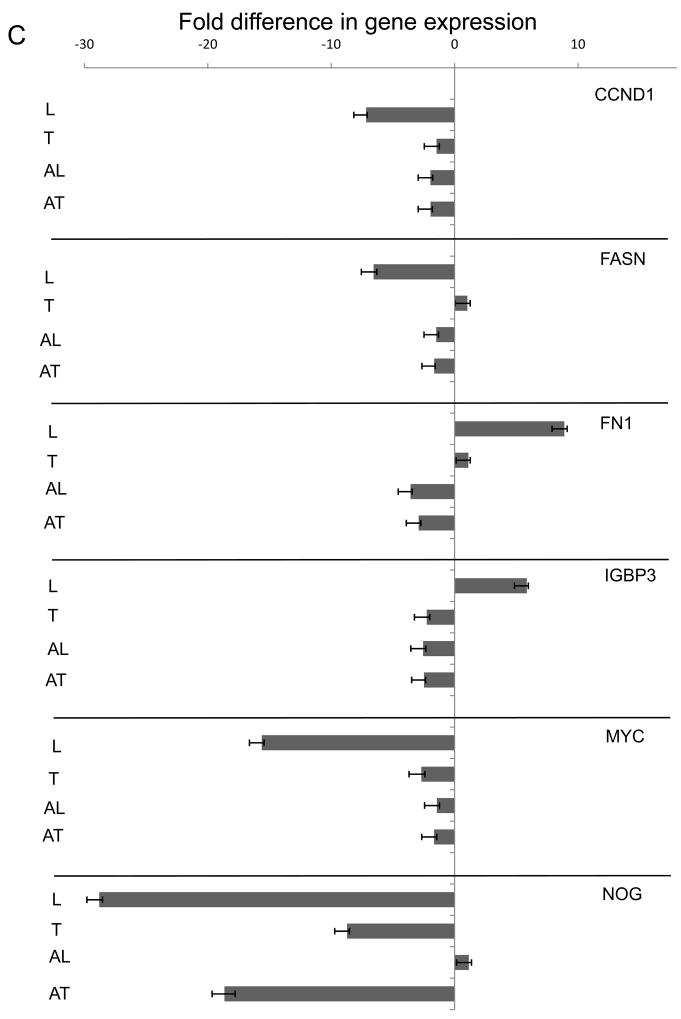
To demonstrate that individual gene expression change depends on crosstalk and/or TGFβ1 ligand induction and pathway activity, selected genes from the TGF/BMP Signalling specific and Signal Transduction Pathway Finder arrays were chosen for further expressional analysis in present of TGFβ type I receptor inhibitor (A-083-01, Fig. 6). ECM induced and inhibitor sensitive genes (Fig. 6A) represent the crosstalk modulated segment of expressional patterns. The classical examples of TGFβ1 regulation is shown on FST(+19.810), MMP10(+25.503) and SERPINE1(+75.790, Fig. 6B). They are not sensitive to laminin (L), up-regulated by TGFβ1 (T) and the inhibitor TGFβ1 + A-083-01 (AT) reverses the effect. The LAM + A-083-01 (AL) combination shows down-regulation similar to AT demonstrating that LAM indeed does not affect these genes. Selected genes responding to LAM (L) induction (Fig. 6C) are CCND1(−7.17), FASN(−6.51), FN1(+8.895), IGBP3(+5.848), MYC(−15.63), NOG(−28.797) which then reversed by the inhibitor pre-treatment (AL), while neither TGFβ1 and its combination with A-083-01does not affect them. The exception is NOG where LAM (L) induced down-regulation reversed by inhibitor (AL) but still negative (−8.717) while TGFβ1 (T) does not affect NOG(1.149) and the inhibitor combination (AT) down-regulates it (−18.65).
Figure 6.
Parallel triplicate experiments were plated and synchronized as described in the Material and Methods section. A-083-01 (5μM) pre-treatment were used (30 minutes) on selected samples followed by LAM and TGFβ1 exposure alone and in combination with the inhibitor. Samples were harvested after 4-hour incubation to enhance the stable expressional profile changes. Selected genes were assayed on cDNA library generated (Materials and Methods) by ABI TaqMan probes (Supplemental Table 2) on ABI 7900 HT Fast Real Time QPCR instrument. The results were transferred to Excel (Microsoft) and graphed with error bars generated by standard deviation of Ct values from the three independent experiments.
Supplemental material
The functional comparison of untreated MCF-10A normal and MCF-7 cells was generated by Venn diagram (4 groups) presenting fold up- and down-regulation of genes in MCF-7 relative to MCF-10A normal control (Supplemental Fig. 1). The Venn groups, the fold differences, and related Student’s T-test, p values are listed in the Supplemental Table 1. The results presented in Figures 3, 4, 5 and 6 were validated with independent TaqMan QPCR assays designed against selected targets (Supplemental Table 2). Overall 31 genes were validated in independent assays. The differences between the TaqMan and SuperArray assays (SA) are greater when the transcript levels change dramatically (for example in BMP7 and CDKN2B), because the TaqMan assays are more target sensitive than the SYBR Green detection. Some of the genes are present in both arrays as CDKN2B, CDKN2A, JUN, IL2, FOS, and others. The validation proves that the array gene expression data are legitimate and accurate.
Discussion
The progression of breast cancer is associated with an epithelial to mesenchymal transition and involves components of TGFβ signalling (32, 33) and signalling from the extracellular matrix (34–37). MCF-10A and MCF-7 (ER+) cell lines are well established model systems for the study of tumor microenvironment in breast cancer progression (38). In addition to the influence of the ECM, the capacity of neoepitopes created by protease digestion of matrix molecules to modulate cell migration has also been reported (15). The signalling induced by proteolytic fragments of ECM proteins is largely unknown. Using this model system we demonstrate that soluble ECM peptides induce Smad2 activation in human breast cancer cells. The role of integrin mediated signalling is well recognized (39) and in conjunction with this study suggests that a signalling outcome depends on the modulation of a signalling network rather than effecting a single pathway. The perceived complexity of cell-extracellular matrix communication and matrix originated signal transduction was previously based on the assumption that these processes are all related to adhesion with an immobile matrix (40). To extend these original paradigms, this study demonstrates that soluble ECM molecules activate Smad signalling by binding to their representative integrin receptors and parallel, indirectly activating the TGFβ signalling pathway.
The pSmad2 specific antibody that was utilized recognizes the dual phosphorylation of C-terminal SSXS motif. This activation of Smad2 and 3 is described only by the ALK4, 5, 7 receptors (22, 27, 41). The demonstrated TGFβ type I (TβRI receptor) receptor inhibitor SB-431542 sensitivity of both pathways verifies ALK5 involvement without excluding the possible participation of ALK4 and 7. The detailed molecular mechanisms by which the ECM and native TGFβ ligands induced Smad activation overlaps and differ is beyond the scope and focus of this current manuscript. The fact that this ECM induced signalling can be observed within 15 minutes after ligand exposure is in good agreement with the known kinetics for Smad activation by TGFβ ligand (21, 42).
The α2β1 integrin complex is a major receptor for both LAM and CII ligand (43). It is also implicated in mediating the malignant transformation in pancreatic cancer cell lines (44). Moreover, the laminin is particularly important in MCF cells for acinus development as a scaffolding matrix (matrigel) as well as media component (45). The co-immunoprecipitation experiments clarified that CII and LAM are competing ligands for the α2β1 integrin and binding increases the association of the heteromeric receptor complex. The soluble ECM induced Smad activation (termed as crosstalk signalling) verified in multiple cell lines, indicating that this process can represents a general mechanism for ECM molecules when acting as soluble peptides.
The TGFβ signalling is tightly controlled by the extracellular matrix through regulating the availability of free peptides for TGFβ receptor activation (46). It can be hypothesized that cell binding to ECM could synergistically enhance the binding of latent TGFβ binding protein (LTBP) and latency associated protein (LAP) complexes to their integrin αvβ6 and αvβ8 receptors and release the peptides by the extra ECM stimuli to initiate Smad signalling. This scenario can be excluded for three reasons: 1) The observed ECM-Smad activation kinetics makes it unlikely, would require an interaction between the α2β1, αvβ6, αvβ8, TβR1 and TβR2 receptors plus LAP and LTBP complexes, and the ECM induced Smad signals can be detected within 15 minutes. 2) We show that the AB-100-NA neutralizing antibody blocks only the TGFβ1 induced signalling but not affecting the crosstalk itself, indicating that the two process are separate. 3) No TGFβ isoforms contamination was found by MALDI-TOF-MS. Moreover, if Smad activation by ECM is a result of TGFβ1 ligand participation, it would be expected that ECM exposure activate the same genes with perhaps lower amplitude depending on the signalling thresholds as TGFβ1 does.
Contrary to this, each ECM ligand elicits a different set of expressional responses when compared with each other and to TGFβ1. MCF cells are regulated by type IV collagen and laminin but normally do not encounter type II collagen in their in vivo environment (38). Therefore the CII is an ideal control to show that just because it binds to the same α2β1 integrin complex, activating the same crosstalk and integrin signalling pathways, the expressional responses are still separated and selective for the representative treatments. For example: the CII induced Smad signalling in the MCF-7 cells peaks at 45 minutes (Fig. 1, 2), and with greater magnitude than the corresponding LAM signal. However, this is not capable of generating any significant expressional responses (Fig. 4, MCF7 panel CTRL vs. CII) whereas in MCF-10A, the same level and time the CII related Smad signal regulates ~11% of genes from the total of 84 (MCF-10A panel CTRL vs. CII). This suggest that gene expressional responses are ligand and cell type dependent, therefore are unlikely to be the result of TGFβ ligand i.e., both LAM and CII induce similar pSmad2 levels but elicits different separate transcriptional responses.
The data indicate that the MCF-7 cells are also less responsive to TGFβ signalling than MCF-10A. The comparison of untreated MCF-10A and MCF-7 cell lines shows that 63% of genes (53 out of 84) differentially regulated in MCF-7. From these 13 genes (25%) are up- and 40 genes (75.5%) are down-regulated. Notably, the adhesion and extracellular molecules cluster are affected strongly, where 83% of genes are down-regulated from the affected 18, indicating that MCF-7 is less dependent on adhesion related functions than MCF-10A (Supplemental Figure). The regulation of this selective target gene population reflects the increase of invasive capacity of MCF-7 cells when compared to MCF-10A line.
The laminin induced expressional changes were validated on selected genes displaying crosstalk sensitivity (Fig. 6, i.e. genes respond to LAM induction which then reversed by A-083-01 TGFβ type I receptor inhibitor), by ABI TaqMan probes. As shown, the inhibitor selectively blocks TGFβ1 induced FST, MMP10, and SERPINE1. These are the classical responsive genes of TGFβ pathway activity. Furthermore, the inhibitor also reverses genes regulated by LAM, (not TGFβ1) verifying that the fold expressional change of these genes was indeed induced by laminin (crosstalk) throughout the TGFβ pathway, and not by alternative branch of attachment dependent integrin signalling.
This manuscript demonstrates that ECM molecules induce transcriptional responses through a non-attachment related signalling process that parallels classical integrin signalling. This process affects cellular pathways on a ligand and cell type dependent manner in human breast cancer and other cells that were investigated. The invasive MCF-7 cells show similar but not identical responses to type II collagen as to TGFβ1, while the laminin response patterns are distinct from both. Crosstalk process modulate signalling capacity of the invasive MCF-7 cells which has been shown be fundamental in epithelial to mesenchymal transition and breast cancer progression (17, 47, 48).
Consequently the ECM-cell communication affects transcriptional regulation in a way what is more complex than previously thought. Furthermore, when matrix components acting on attachment independent way (during matrix remodelling and degradation) this process can affect gene expression, and contribute to ECM originated signalling controlling cellular activity and phenotype.
Materials and Methods
Cell Culture
MCF-10A cells (human normal mammary epithelial cells) were propagated in MEGM media (Clonetics CC-3150 with supplied SingleQuots growth factors CC-4136) with 100 ng/ml Cholera toxin (Calbiochem 227035). MCF-7 cells were cultured in IMEM media (Gibco 10373-017, + gentamycine) containing 10% FBS, 10 mg/L Phenol Red (Sigma P02990) + 10 μg/mL insulin (Sigma I-0516).
Chemicals
Fibronectin (Sigma, Saint Louis MO., F4759), type I Collagen (Sigma C9301), type II collagen (Chondrex 20022) was dissolved in 0.05 M CH3COOH as 1.0, 2.0, and 2.0 mg/ml stock solutions respectively. The laminin (Sigma L2020) was supplied as 1.0 mg/ml stock in 50 mM Tris-HCl, pH 7.5, with 150 mM NaCl. TGFβ1 (R&D 240-B) stock was 10 ng/μl in 4.0 mM HCl with 1.0 mg/ml BSA carrier protein. The SB-431542 and A-083-01 were purchased from Tocris Bioscience (Ellisville MO).
Antibodies
The primary antibodies were purchased from Cell Signaling, Beverly, MA (anti-phospho-Smad2 #3101, and anti-Smad2 #3122), and Santa Cruz Biotechnology Inc., Santa Cruz CA (anti-Smad4 sc-7966 collagen type II sc-7764, laminin β1 sc-5583, Integrin α2 sc-9089). Secondary HRP conjugated antibodies are from Santa Cruz (sc-2020) and Amersham/GE Piscataway NJ (#NA93AV) and anti-mouse Alexa-488 is from Molecular Probes/Invitrogen (A21202).
ECM Treatments
Cells were plated in p100 plates at density of 6.0×106 (MCF-10A) to 8.0×106 (MCF-7) cell/p100 to give a confluent culture after overnight incubation. The cultures were synchronized by serum free DMEM:F12 for 24 hours to maximize the signal to noise ratio. ECM peptides were applied at 50 μg/ml concentration in 4 ml serum free DMEM:F12 for the specified time. TGFβ1 positive control was used at 10 ng/ml concentration. To harvest, cells were washed twice on ice with ice cold PBS, scraped in 1 ml PBS, pelleted at 8000 rpm for 2 minutes, and the pellet was snap frozen in liquid nitrogen and stored at −80°C until processing.
Western Blotting and Image Acquisition
Cells were lysed in buffer containing 50 mM Tris-HCl pH 7.4, 1% NP-40, 0.25% Na-deoxycholate, 150 mM NaCl, 1 mM EDTA, 1 mM Na-vanadate, and 1 mM Phenylmethylsulfonyl fluoride, completed with protease inhibitor cocktail from Roche. The samples were normalized for protein with ND-1000 (NanoDrop Technologies Inc., Wilmington DE). For western blotting, 125 μg protein per lane were analyzed with primary antibodies incubated overnight at 4°C, followed by secondary HRP conjugated antibodies for 2 hours at room temperature. Bands were detected with SuperSignal West Pico ECL detection kit (Pierce, Rockford IL) on UVP Biospectrum Digital Imaging system (UVP Inc. Upland CA). The raw images were quantized by optical density through the supplied densitometry analysis software and normalized to total Smad levels.
Co-Immunoprecipitation
Confluent synchronized p100 plates were pre-treated for two hours at 4°C with collagen type II and laminin and in combinations. Then the cells were harvested, samples stored as described above. Samples were processed in 800 μl lysis buffer for 30 minutes on ice. Supernatants were separated, and the residual pellets were processed again with a fresh aliquot (200 μl) and sonicated (converter V2391, Virsonic 100 unit VirTis (SP industries Co. Gardiner NY). The combined supernatants were quantized and 2.0 mg total protein samples were incubated with mouse anti-human VLA-2 (α2β1 Chemicon, MAB1998) against functional collagen receptor, or by the mouse anti-human Integrinβ1 (Santa Cruz sc-9970) for 2 hours then precipitated with 45 μl protein A agarose (Sigma P-7786), anti-mouse IgG (Sigma A-6531), or Protein G agarose for overnight at 4°C. Samples were washed extensively, then solubilized in 80 μl 2x Laemli buffer, and analyzed on 8% SDS-PAGE.
Gene Expression Analysis
Human TGFβ/BMP Signalling (PAHS-035) and Signal Transduction Pathway Finder (PAHS-014) specific RT2 profiler PCR arrays were obtained from SA Bioscience Co. (Frederick MD US). Cells were plated in triplicates, exposed to peptides, then harvested at four hours later and stored as described above. The RNA was purified with RNeasy Mini Kit with on column DNase treatment (Quiagen Corp. US 74104) according to the manufacturer protocol. For the cDNA synthesis, 5 μg total RNA was used with High Capacity cDNA Reverse Transcription Kit (Applied Biosystems 4322171) as described by the kit manual. The array analysis was carried out following the manufacturer protocol with SYBR Green PCR (ROX) Master Mix (Applied Biosystems Foster City CA # 4309155) and 1.0–5.0 μg cDNA/plate on ABI 7900HT Fast QPCR system. Gene expression differences were determined using the 2−ΔΔCt method according to AB and SA Biosciences protocols. The expressional pattern differences between the MCF-10A and MCF-7 cells and the heat maps were generated in MATLAB 7.5.0 software using the expressional fold differences analyzed through the SA Biosciences web page, and with absolute values of the transcripts measured by QPCR.
Statistical Analysis
The one-way ANOVA subroutine of MATLAB 7.5.0 was used to verify the significance of western blotting results (quantized through the UVP Bio-Imager densitometry software in triplicates). The array analysis related p values were generated through the representative web page links supported software. Values presented in the supplemental materials.
Supplementary Material
Acknowledgments
We thank to Drs Karoline Briegel for the MCF cell lines, Victor Asirvatham for help with the MALDI-TOF MS, and Dr. Muhammad Umar Jawad and Ms Sara Garamszegi for their helpful comments. This work was supported by grants from National Institutes of Health CA096796-01 (SPS), a Ruth L. Kirschstein National Research Service Award NIH 1 F32 HL083673-01 (LAS), Woman’s Cancer Association Madelon Ravlin Memorial Award WCA 66461Y (NG and SPS), American Cancer Society ACS 66105 to (NG). Other supports were provided by the University of Miami, Department of Orthopaedics, and the Sylvester Comprehensive Cancer Center.
References
- 1.Bissell MJ, Radisky DC, Rizki A, Weaver VM, Petersen OW. The organizing principle: microenvironmental influences in the normal and malignant breast. Differentiation. 2002;70(9–10):537–46. doi: 10.1046/j.1432-0436.2002.700907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fata JE, Werb Z, Bissell MJ. Regulation of mammary gland branching morphogenesis by the extracellular matrix and its remodeling enzymes. Breast Cancer Res. 2004;6(1):1–11. doi: 10.1186/bcr634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6(5):392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 4.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2(6):442–54. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 5.Atley LM, Mort JS, Lalumiere M, Eyre DR. Proteolysis of human bone collagen by cathepsin K: characterization of the cleavage sites generating by cross-linked N-telopeptide neoepitope. Bone. 2000;26(3):241–7. doi: 10.1016/s8756-3282(99)00270-7. [DOI] [PubMed] [Google Scholar]
- 6.Downs JT, Lane CL, Nestor NB, et al. Analysis of collagenase-cleavage of type II collagen using a neoepitope ELISA. J Immunol Methods. 2001;247(1–2):25–34. doi: 10.1016/s0022-1759(00)00302-1. [DOI] [PubMed] [Google Scholar]
- 7.Li WW, Nemirovskiy O, Fountain S, Rodney Mathews W, Szekely-Klepser G. Clinical validation of an immunoaffinity LC-MS/MS assay for the quantification of a collagen type II neoepitope peptide: A biomarker of matrix metalloproteinase activity and osteoarthritis in human urine. Anal Biochem. 2007;369(1):41–53. doi: 10.1016/j.ab.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 8.Otterness IG, Downs JT, Lane C, et al. Detection of collagenase-induced damage of collagen by 9A4, a monoclonal C-terminal neoepitope antibody. Matrix Biol. 1999;18(4):331–41. doi: 10.1016/s0945-053x(99)00026-8. [DOI] [PubMed] [Google Scholar]
- 9.Buzza MS, Zamurs L, Sun J, et al. Extracellular matrix remodeling by human granzyme B via cleavage of vitronectin, fibronectin, and laminin. J Biol Chem. 2005;280(25):23549–58. doi: 10.1074/jbc.M412001200. [DOI] [PubMed] [Google Scholar]
- 10.Chang C, Lauffenburger DA, Morales TI. Motile chondrocytes from newborn calf: migration properties and synthesis of collagen II. Osteoarthritis Cartilage. 2003;11(8):603–12. doi: 10.1016/s1063-4584(03)00087-6. [DOI] [PubMed] [Google Scholar]
- 11.Kenny HA, Kaur S, Coussens LM, Lengyel E. The initial steps of ovarian cancer cell metastasis are mediated by MMP-2 cleavage of vitronectin and fibronectin. J Clin Invest. 2008;118(4):1367–79. doi: 10.1172/JCI33775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koshikawa N, Schenk S, Moeckel G, et al. Proteolytic processing of laminin-5 by MT1-MMP in tissues and its effects on epithelial cell morphology. FASEB J. 2004;18(2):364–6. doi: 10.1096/fj.03-0584fje. [DOI] [PubMed] [Google Scholar]
- 13.Midwood KS, Mao Y, Hsia HC, Valenick LV, Schwarzbauer JE. Modulation of cell-fibronectin matrix interactions during tissue repair. J Investig Dermatol Symp Proc. 2006;11(1):73–8. doi: 10.1038/sj.jidsymp.5650005. [DOI] [PubMed] [Google Scholar]
- 14.Pilcher BK, Dumin JA, Sudbeck BD, Krane SM, Welgus HG, Parks WC. The activity of collagenase-1 is required for keratinocyte migration on a type I collagen matrix. J Cell Biol. 1997;137(6):1445–57. doi: 10.1083/jcb.137.6.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Remy L, Trespeuch C, Bachy S, Scoazec JY, Rousselle P. Matrilysin 1 influences colon carcinoma cell migration by cleavage of the laminin-5 beta3 chain. Cancer Res. 2006;66(23):11228–37. doi: 10.1158/0008-5472.CAN-06-1187. [DOI] [PubMed] [Google Scholar]
- 16.Steadman R, Irwin MH, St John PL, Blackburn WD, Heck LW, Abrahamson DR. Laminin cleavage by activated human neutrophils yields proteolytic fragments with selective migratory properties. J Leukoc Biol. 1993;53(4):354–65. doi: 10.1002/jlb.53.4.354. [DOI] [PubMed] [Google Scholar]
- 17.Lamouille S, Derynck R. Cell size and invasion in TGF-beta-induced epithelial to mesenchymal transition is regulated by activation of the mTOR pathway. J Cell Biol. 2007;178(3):437–51. doi: 10.1083/jcb.200611146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munger JS, Harpel JG, Giancotti FG, Rifkin DB. Interactions between growth factors and integrins: latent forms of transforming growth factor-beta are ligands for the integrin alphavbeta1. Mol Biol Cell. 1998;9(9):2627–38. doi: 10.1091/mbc.9.9.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Munger JS, Huang X, Kawakatsu H, et al. The integrin alpha v beta 6 binds and activates latent TGF beta 1: a mechanism for regulating pulmonary inflammation and fibrosis. Cell. 1999;96(3):319–28. doi: 10.1016/s0092-8674(00)80545-0. [DOI] [PubMed] [Google Scholar]
- 20.Sheppard D. Integrin-mediated activation of transforming growth factor-beta(1) in pulmonary fibrosis. Chest. 2001;120(1 Suppl):49S–53S. doi: 10.1378/chest.120.1_suppl.s49. [DOI] [PubMed] [Google Scholar]
- 21.Di Guglielmo GM, Le Roy C, Goodfellow AF, Wrana JL. Distinct endocytic pathways regulate TGF-beta receptor signalling and turnover. Nat Cell Biol. 2003;5(5):410–21. doi: 10.1038/ncb975. [DOI] [PubMed] [Google Scholar]
- 22.Attisano L, Wrana JL. Signal transduction by the TGF-beta superfamily. Science. 2002;296(5573):1646–7. doi: 10.1126/science.1071809. [DOI] [PubMed] [Google Scholar]
- 23.Dore JJ, Jr, Yao D, Edens M, Garamszegi N, Sholl EL, Leof EB. Mechanisms of transforming growth factor-beta receptor endocytosis and intracellular sorting differ between fibroblasts and epithelial cells. Mol Biol Cell. 2001;12(3):675–84. doi: 10.1091/mbc.12.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garamszegi N, Dore JJ, Jr, Penheiter SG, Edens M, Yao D, Leof EB. Transforming growth factor beta receptor signaling and endocytosis are linked through a COOH terminal activation motif in the type I receptor. Mol Biol Cell. 2001;12(9):2881–93. doi: 10.1091/mbc.12.9.2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Penheiter SG, Mitchell H, Garamszegi N, Edens M, Dore JJ, Jr, Leof EB. Internalization-dependent and -independent requirements for transforming growth factor beta receptor signaling via the Smad pathway. Mol Cell Biol. 2002;22(13):4750–9. doi: 10.1128/MCB.22.13.4750-4759.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Massague J. TGFbeta in Cancer. Cell. 2008;134(2):215–30. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425(6958):577–84. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 28.Massague J, Chen YG. Controlling TGF-beta signaling. Genes & Development. 2000;14(6):627–44. [PubMed] [Google Scholar]
- 29.Inman GJ, Nicolas FJ, Callahan JF, et al. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Molecular Pharmacology. 2002;62(1):65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- 30.Schneiderbauer MM, Dutton CM, Scully SP. Signaling “cross-talk” between TGF-beta1 and ECM signals in chondrocytic cells. Cell Signal. 2004;16(10):1133–40. doi: 10.1016/j.cellsig.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 31.Wang W, Huang XR, Canlas E, et al. Essential role of Smad3 in angiotensin II-induced vascular fibrosis. Circulation Research. 2006;98(8):1032–9. doi: 10.1161/01.RES.0000218782.52610.dc. [see comment] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moustakas A, Pardali K, Gaal A, Heldin CH. Mechanisms of TGF-beta signaling in regulation of cell growth and differentiation. Immunol Lett. 2002;82(1–2):85–91. doi: 10.1016/s0165-2478(02)00023-8. [DOI] [PubMed] [Google Scholar]
- 33.Muraoka RS, Dumont N, Ritter CA, et al. Blockade of TGF-beta inhibits mammary tumor cell viability, migration, and metastases. J Clin Invest. 2002;109(12):1551–9. doi: 10.1172/JCI15234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bissell MJ, Kenny PA, Radisky DC. Microenvironmental regulators of tissue structure and function also regulate tumor induction and progression: the role of extracellular matrix and its degrading enzymes. Cold Spring Harb Symp Quant Biol. 2005;70:343–56. doi: 10.1101/sqb.2005.70.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kenny PA, Lee GY, Bissell MJ. Targeting the tumor microenvironment. Front Biosci. 2007;12:3468–74. doi: 10.2741/2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.LaBarge MA, Petersen OW, Bissell MJ. Of microenvironments and mammary stem cells. Stem Cell Rev. 2007;3(2):137–46. doi: 10.1007/s12015-007-0024-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee GY, Kenny PA, Lee EH, Bissell MJ. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat Methods. 2007;4(4):359–65. doi: 10.1038/nmeth1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30(3):256–68. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 39.Comoglio PM, Boccaccio C, Trusolino L. Interactions between growth factor receptors and adhesion molecules: breaking the rules. Curr Opin Cell Biol. 2003;15(5):565–71. doi: 10.1016/s0955-0674(03)00096-6. [DOI] [PubMed] [Google Scholar]
- 40.Geiger B, Bershadsky A. Assembly and mechanosensory function of focal contacts. Curr Opin Cell Biol. 2001;13(5):584–92. doi: 10.1016/s0955-0674(00)00255-6. [DOI] [PubMed] [Google Scholar]
- 41.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113(6):685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 42.Vilar JM, Jansen R, Sander C. Signal processing in the TGF-beta superfamily ligand-receptor network. PLoS Comput Biol. 2006;2(1):e3. doi: 10.1371/journal.pcbi.0020003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plow EF, Haas TA, Zhang L, Loftus J, Smith JW. Ligand binding to integrins. J Biol Chem. 2000;275(29):21785–8. doi: 10.1074/jbc.R000003200. [DOI] [PubMed] [Google Scholar]
- 44.Grzesiak JJ, Bouvet M. The alpha2beta1 integrin mediates the malignant phenotype on type I collagen in pancreatic cancer cell lines. Br J Cancer. 2006;94(9):1311–9. doi: 10.1038/sj.bjc.6603088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer. 2005;5(9):675–88. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 46.ten Dijke P, Arthur HM. Extracellular control of TGFbeta signalling in vascular development and disease. Nat Rev Mol Cell Biol. 2007;8(11):857–69. doi: 10.1038/nrm2262. [DOI] [PubMed] [Google Scholar]
- 47.Alcorn JF, Guala AS, van der Velden J, et al. Jun N-terminal kinase 1 regulates epithelial-to-mesenchymal transition induced by TGF-{beta}1. J Cell Sci. 2008;121(Pt 7):1036–45. doi: 10.1242/jcs.019455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song J. EMT or apoptosis: a decision for TGF-beta. Cell Res. 2007;17(4):289–90. doi: 10.1038/cr.2007.25. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.