Abstract
Objectives
There are currently no diagnostic indicators that are consistently reliable, obtainable, and conclusive for diagnosing and risk-stratifying pancreatic cysts. Proteomic analyses were performed to explore pancreatic cyst fluids to yield effective diagnostic biomarkers.
Methods
We have prospectively recruited 20 research participants and prepared their pancreatic cyst fluids specifically for proteomic analyses. Proteomic approaches applied were: 1) MALDI-TOF (matrix-assisted laser-desorption-ionization time-of-flight) mass spectrometry peptidomics with LC/MS/MS (HPLC-tandem mass spectrometry) protein identification. 2) 2D gel electrophoresis. 3) GeLC/MS/MS (tryptic digestion of proteins fractionated by SDS-PAGE and identified by LC/MS/MS).
Results
Sequencing of over 350 free peptides showed that exopeptidase activities rendered peptidomics of cyst fluids unreliable; Protein nicking by proteases in the cyst fluids produced hundreds of protein spots from the major proteins, making 2D gel proteomics unmanageable; GeLC/MS/MS revealed a panel of potential biomarker proteins that correlated with CEA (carcinoembryonic antigen).
Conclusions
Two homologs of amylase, solubilized molecules of four mucins, four solubilized CEACAMs (CEA-related cell adhesion molecules), and four S100 homologs, may be candidate biomarkers to facilitate future pancreatic cyst diagnosis and risk-stratification. This approach required less than 40 microliters of cyst fluid per sample, offering the possibility to analyze cysts smaller than 1 cm diameter.
Keywords: Pancreatic cyst fluid, biomarkers, proteomic, mucin, S100, CEACAM
Introduction
Increasing use of high resolution computerized tomography and magnetic resonance imaging in clinical practice has resulted in detection of a growing number of pancreatic cysts 1. As a result, clinicians are frequently asked to determine the biological nature of these cystic lesions and to make treatment recommendations accordingly. However, there are currently no diagnostic indicators that are consistently reliable, obtainable, and conclusive for diagnosing and risk-stratifying pancreatic cysts. The sensitivity of pancreatic cyst fluid cytology has been reported as only 27–64%. Several studies have suggested that a variety of tumor markers (e.g., CEA (carcinoembryonic antigen CEACAM5, a carcinoembryonic antigen-related cell adhesion molecule) 2, CA 19-9 (carbohydrate antigen 19-9), CA 15-3 (cancer antigen MUC1, mucin 1) may distinguish mucinous from non-mucinous cystic lesions, and also may predict whether a cyst harbors areas of malignant transformation 3–5, but no marker by itself is sufficiently reliable.
The biologic nature and histopathologic features of pancreatic cysts are varied 3, 6. Ten to twenty percent of pancreatic cysts are neoplastic, including neoplasms which grow as cystic structures (i.e., primary cystic neoplasms of the pancreas), and solid neoplasms that have undergone cystic degeneration. Serous cystadenomas (microcystic adenomas) account for approximately 32–39% of the primary cystic neoplasms and have very low malignant potential. Mucinous cystic neoplasms, which includes mucinous cystadenomas (MCAs) and intraductal papillary mucinous neoplasms (IPMNs), are a subgroup of primary cystic neoplasms that have malignant potential 7, 8, accounting for approximately 10–45% and 21–33% of primary cystic neoplasms, respectively 6, 9–11. Two subtypes of IPMN have been described 1, 12, a main duct variant and a branch duct variant; the latter may have a more indolent course. There are other less common forms of primary cystic neoplasms of the pancreas, such as solid pseudopapillary tumors.
In the absence of reliable methods of quantifying the malignant potential of a suspected pre-malignant cystic neoplasm of the pancreas, if existing clinical parameters suggest the presence of one such lesion in a person that is otherwise an acceptable surgical risk, partial or total pancreatomy may be recommended but can result in significant morbidity and mortality 13. Alternatively, a conservative "watch-and-wait” approach (i.e., serial imaging over time) is advocated for some patients, but this strategy may be suboptimal due to incremental costs accrued during surveillance, and the possibility that malignant transformation may occur between surveillance time points.
Our primary goal was to define a reliable and reproducible technique for analyzing pancreatic cyst fluids with proteomics. Our secondary goals were to ascertain whether mass spectrometry proteomics can be performed with much smaller volumes than is required for the clinical assays that are currently used to study pancreatic cyst fluid, and also to provide an assessment of whether proteomics can define potentially useful diagnostic and/or risk-stratifying pancreatic cyst biomarkers or biomarker profiles. We performed a meticulous proteomic study consistent with the guide line described in the recent review article in Pancreas 14.
Materials and Methods
Sample Acquisition
Aliquots of cyst fluid that were used for this project were obtained from materials that were aspirated for clinical purposes and prepared in a manner designed for proteomics analysis. The study was approved by the Institutional Review Board of the Fox Chase Cancer Center. Endoscopy ultrasound-fine needle aspiration (EUS-FNA) 15 was performed under conscious sedation using a linear echoendoscope. When a lesion was identified, EUS-FNA was performed with a 22 or 19-gauge needle through either a transduodenal or transgastric approach, depending on the location of the lesion within the pancreas. The highest priority was given to procuring a volume of fluid that was adequate to perform the necessary clinically indicated diagnostic assays [e.g., cytology, CEA (Mayo Medical Laboratories, code # 84074) and amylase (Mayo Medical Laboratories, code # 5079)]. As little as 40 μL of cyst fluid per patient was allocated for the proteomic study. For the purpose of this study, the cyst fluid cytology diagnoses were grouped into one of the following four categories: A- Benign: No evidence of benign mucinous epithelium, atypical cells or carcinoma; B- Benign mucinous epithelium; C- Atypical/suspicious; D- Malignant. Histopathology information was obtained for those cases that yielded surgical specimens.
Sample collection
Standard Operating Procedures were established and followed for all steps of cyst fluid collection and analysis. Each cyst fluid was photographed, diluted with three volumes of ice cold PBS, mixed, and centrifuged for 10 min at 13,000 x g at 4 °C to remove cells and any insoluble materials, photographed, and snap frozen in liquid nitrogen in aliquots and banked at −80 °C. To remove small peptides (Fraction A) bound to larger proteins, the cyst fluid was thawed on ice and denatured by three volumes of 0.1M glycine and 25% v/v final concentration of acetonitrile. The solution was filtered by ultrafiltration (pre-washed Amicon YM-30 Centricon #4208) at 4000 x g at 4 °C for about one hour to reach minimum retention volume designed for the unit. The proteins retained above the filter (Fraction B) were solubilized with 200 μL of 0.2% SDS solution and transferred to a 1.5 mL microcentrifuge tube. Three volumes of cold acetone was added to precipitate the proteins overnight at −20 °C and then centrifuged at 21,000 x g for 40 min. The pellet was washed once with 80 % cold acetone, centrifuged, and the pellet was air dried and resolubilized in 2D PAGE sample buffer (7 M urea, 2 M thiourea, 4% (w/v) CHAPS). Protein concentration was determined as previously described 16.
MALDI-TOF Peptidomics
Fraction A was dried in a SpeedVac and redissolved in 2% formic acid 5% acetonitrile and applied to a prewashed Vydac C18 Silica MicroSpin cartridge (#SEM SS18V, The Nest Group, Inc. Southborough, MA) according to manufacturer's instructions. The peptides, after column washing, were sequentially eluted by 50 μL each of 20%, 30%, 45%, and 80% acetonitrile step gradient in 0.2% formic acid. The eluted fractions were dried and then redissolved in 20 pL of 2% formic acid. One μL of each sample was applied to a gold-plated MALDI-TOF target plate of an Applied Biosystems QSTAR XL mass spectrometer equipped with an oMALDI II source, followed by 1 μL of matrix material composed of 8 mg/mL α-cyano-4-hydroxycinnamic acid in 50% acetonitrile 0.1% TFA. The spot was air dried. MALDI-TOF spectrum was collected by 5 min of laser irradiation in a random spiral at 20 shots per second, with the TOF analyzer scanning 300 to 3000 m/z. Typical resolution was higher than 14,000. A set of five synthetic peptide standards (Sigma cat. #MS-CAL2 ProteoMass Peptide MALDI-MS Calibration Kit) was used to calibrate the QSTAR at the beginning of each experiment.
LC/MS/MS sequencing of the peptides in Fraction A
Peptides in Fraction A were identified by combining above four step-gradient fractions of each cyst fluid and analyze by nanoLC/MS/MS as follows: The LC/MS/MS system consisted of above Applied Biosystems QSTAR XL hybrid quadruple TOF mass spectrometer supported by an Agilent 1100 nanoLC. The instrument was calibrated using above peptide standards in the nano-electrospray infusion mode. For 15 μg protein per gel lane, 10% of the digest of each gel slice was auto-injected onto a trap column (Agilent Zorbax 300SB-C18, 5 pm, 5x0.3 mm), washed, and eluted at 0.3 pL/min through an analytical column (Agilent Zorbax 300SB-C18, 3.5 pm, 150x0.1 mm) at room temperature for electrospray ionization from a New Objective fused-silica PicoTip (FS360-20-10-CE20) in a QSTAR Microion spray source at 0.3 μL/min. The elution gradient was in 0.2% formic acid with linear segments of 4.5%, 4.5%, 28%, 54%, 90% acetonitrile at 0, 4, 8, 80, 85 min, respectively. An IDA (Information Dependent Acquisition) LC/MS/MS protocol using MS periods of 2 s of TOF-MS and three cycles of 4 s of MS/MS each was used to obtain highly accurate spectra for protein identification for the three most intense peptide ions in each period. Each sample was analyzed at least in duplicate. Peptide identification was performed by MASCOT 2.2 software (Matrix Sciences) allowing variable methionine oxidation and no protease digestion, 150 ppm TOF-MS, and 0.5 Da MS/MS. Protein identification was accepted if: 1). at least two peptides in Fraction A identified the protein; 2). if more than one peptide of overlapping sequences in Fraction A identified the protein with MASCOT expectation score less than 0.5; 3). the protein was among the 462 proteins discovered below in Fraction B by GeLC/MS/MS analysis.
2-D Gel Electrophoresis Proteomics
100 μg protein of the Fraction B of six of the cyst samples was each analyzed by 2D gel as previously described in detail 16, 17. The first dimension was performed on pH 3-10 Immobiline DryStrip (0.5 x 3 x 180 mm, Amersham Pharmacia Biotech, Uppsala, Sweden). The protein spots were visualized by staining with Sypro Ruby fluorescent staining, robotic excised, digested with trypsin, and identified by peptide-mass-fingerprinting on a Bruker Reflex IV mass spectrometer as previously described in detail 16, 17.
SDS-PAGE
Three μg of proteins from Fraction B of each cyst were resolved in a pre-cast BioRad 4–12% gradient PAGE with 3 mm wide wells (BioRadTM, CA, USA). Electrophoresis was performed in MOPS buffer at 150V at room temperature. The gels were stained with SyproRuby fluorescence stain, visualized and recorded as previously described 16.
GeLC/MS/MS comparison of protein expression
15 μg of protein of Fraction B was reduced with dithiothreitol and alkylated by iodoacetamide at 25 °C for 1 hr 16 and then resolved in a pre-cast Novex 4–12% gradient PAGE with 3 mm wide wells (InvitrogenTM, CA, USA). Electrophoresis was performed in MOPS buffer at 150V at room temperature until the tracking dye was 1.5 cm from the top of the gel. The gel cassette was opened in a laminar flow hood. Each sample lane, two per gel, was cut into about 11 slices from the well to about 2 mm beyond the dye front. Each gel slice was again subjected to reduction and alkylation. Porcine trypsin (Sigma proteomic grade #T6567) was added as 63 ng in 7 μL 25 mM ammonium bicarbonate and incubated for 30 min. Unabsorbed trypsin of about 2 pL was removed and 20 μL of 25 mM ammonium bicarbonate was added and incubated at 37° C for about 16 hours. 10 μL of the peptide solution was mixed with 2.5 μL of 25% acetonitrile 1% formic acid, and 2 μL was injected into the LC/MS/MS system for protein identification as described above for Fraction A. For discovery of more proteins and peptides in cyst fluids and to overcome the possibility of false-negatives due to under-sampling of co-eluting peptides, after the first LC/MS/MS run of a sample, an exclusion list composed of the peptides sequenced in the first LC/MS/MS run was assembled and used to direct another LC/MS/MS run to sequence only new peptides in the same sample. The two peak lists were combined for database searching for protein identification and for relative quantitation of the proteins by emPAI score (exponentially modified protein abundance index) without isotope labeling 18. The emPAI score, [10^(number of observed peptides per protein/number of theoretical peptides per protein) – 1], is roughly proportional to the abundance of a protein in a complex mixture.
Validation of proteomic biomarkers by Western analysis
10 pg of protein of each sample was resolved on NuPAGE 4–12% Bis-Tris gel (InvitrogenTM, CA, USA). MOPS buffer was used to resolve proteins with molecular weight of 20k to 200k, and MES buffer to resolve smaller proteins. Blotting was performed at 200 mA for 4 h. Primary antibodies used were Anti-CEACAM6 (mouse monoclonal, 1:500, R&D Systems, MN, USA) and anti-S100A9 (mouse monoclonal, 1:500, Abcam Inc., MA, USA). Secondary antibody was goat anti-mouse IRDye 800CW (LI-COR Biosciences, NE, USA) with 1:10,000 dilution. The images were obtained with an Odyssey infrared imaging system (LI-COR Biosciences, NE, USA) at scan intensity of 5 and scan resolution of 169 μm.
Results
Clinical Information on the Cyst Fluids
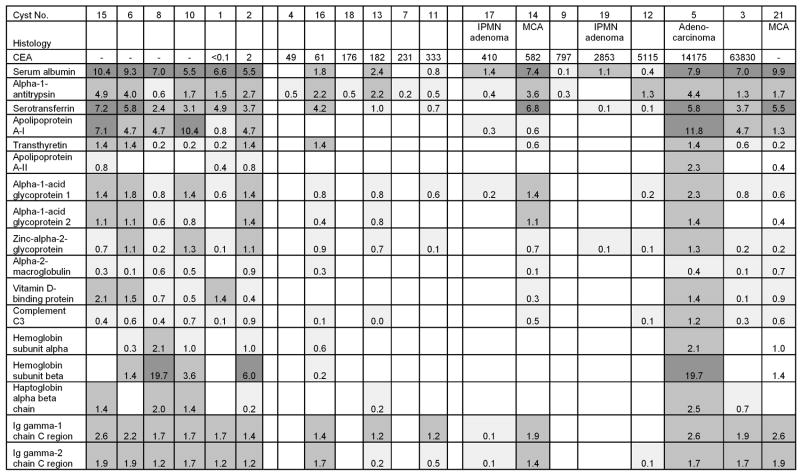
Demography of the patients, dimensions of the cysts, and the results of traditional clinical tests performed on the cyst fluids are shown in Table 1. All the patients were Caucasian. In Tables 1 and Figures 3–5, because CEA measurement by clinical immunoassays is believed to be the strongest indicator of mucinous versus non-mucinous cysts in the absence of direct measurement of the mucins, the cysts were presented in the order of increasing CEA. Two samples without CEA values were assigned to the right side because high CEA values would be anticipated based on the histopathology findings. For cysts 17, 14, 19, 5, and 21, subsequent surgical resection led to definitive histopathologic diagnosis as shown.
Table 1. Clinical information for the pancreatic cyst fluid samples.
CEA (ng/mL) and Amylase (units/mL) are results from clinical lab immuno assays. MCA=mucinous cystadenoma. Cytology categories: A- Benign: No evidence of benign mucinous epithelium, atypical cells or carcinoma. C- Atypical/suspicious cytology.
| Cyst No. | 15 | 6 | 8 | 10 | 1 | 2 | 4 | 16 | 18 | 13 |
| Histology | - | - | - | - | - | - | - | - | - | - |
| CEA | - | - | - | - | <0.1 | 1.8 | 49.4 | 60.5 | 176 | 182 |
| Amylase | - | - | - | - | - | 56 | - | - | - | 163400 |
| Cytology | A | A | A | C | A | A | C | A | A | A |
| Gender | F | F | F | M | F | F | F | F | F | M |
| Age | 33 | 53 | 83 | 50 | 51 | 74 | 73 | 56 | 76 | 67 |
| Cyst size | 15 mm x 12 mm | 14 mm x 17 mm | 22 mm x 14 mm | 18 mm x 28 mm | 16 mm x 13 mm | 20 mm x 24 mm | 31 mm x 11 mm | 34 mm x 32 mm | 22 mm x 14 mm | 24 mm x 17 mm |
| Location | pancreatic tail | pancreatic tail | pancreatic head | pancreatic body | pancreatic body | pancreatic tail | pancreatic neck | pancreatic body | pancreatic head | pancreatic head |
| Cyst No. | 7 | 11 | 17 | 14 | 9 | 19 | 12 | 5 | 3 | 21 |
| Histology | - | - | IPMN adenoma | MCA | - | IPMN adenoma | - | Adenocarcinoma | - | MCA |
| CEA | 231 | 333 | 410 | 582 | 797 | 2853 | 5115 | 14175 | 63,830 | - |
| Amylase | 12330 | 43152 | 25600 | 4036 | 75210 | - | - | 6 | - | - |
| Cytology | A | A | A | A | A | A | A | C | A | A |
| Gender | M | F | M | F | M | M | F | M | F | F |
| Age | 63 | 81 | 77 | 45 | 68 | 81 | 65 | 70 | 73 | 32 |
| Cyst size | 23 mm x 10 mm | 27 mm x 10 mm | 23 mm x 26 mm | 46 mm x 47 mm | 29 mm x 21 mm | 22 mm x 20 mm | 14 mm x 9 mm | 18 mm x 18 mm | 17 mm x 13 mm | - |
| Location | pancreatic body | pancreatic tail | uncinate process | - | pancreatic head | pancreatic tail | pancreatic head | pancreatic body/tail | pancreatic head | - |
Figure 3. Representatives of 137 plasma proteins distributed among the pancreatic cyst samples.
The numbers shown are emPAI roughly proportional to protein abundance. Dark (>1), medium (0.1 to 1), and light (<0.1) shading denotes relative protein abundance. No proteins were detected for the empty boxes. CEA = ng/mL. MCA=mucinous cystadenoma
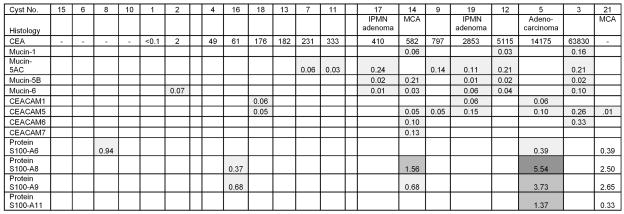
Figure 5. Proteomic potential biomarkers for mucins, CEACAMs, and S100s found in the pancreatic cyst samples.
Legends are the same as for Figure 3.
MALDI-TOF Peptidomics of pancreatic cyst fluids with identification by LC/MS/MS
For the analysis conditions described in this report, both Fraction A and Fraction B of pancreatic cyst fluids provided essentially unchanged mass spectrometry information after multiple freeze-thaw cycles, and unaffected by several hours of 10 °C incubation in 0.2% formic acid (data not shown). The MALDI-TOF the QSTAR XL mass spectrometer produced m/z of single-positive-charge peptides in Fraction A with a reproducible accuracy better than 20 ppm or <0.02 Da error for a 1000 Da peptide with a signal to noise ratio >10 (Supplemental Data 1, provided at our web site 1). Under the assumption that two peptides of greater than 0.3 Da difference in their centroid-mass in this system may be considered different in sequence, a total of 1045 non-redundant ions were accurately labeled by MALDI-TOF analysis of the cyst fluids 1–15 (Supplemental Data 2, provided at our web site 2). 243 of these ions were determined as mono-isotopic peptide ions. These weak +1 ions were not sequenced effectively by MALDI-TOF in this mass spectrometer, thus the Fraction A peptides were sequenced by LC/MS/MS in nano-electrospray mode as +2 and +3 charge ions. About 20% of the MALDI-TOF peaks can be matched to peptide sequenced by LC/MS/MS. A total of 317 peptides were identified in the SwissProt protein database as coming from 56 cyst fluid proteins (Table 2). 254 of these peptides of the highest confidence were subjected to Multiple Sequence Alignment by the ClustalW2 software 19. The peptides grouped into about 58 clusters of overlapping sequences (Supplemental Data 3, provided at our web site 3). About 77 peptides remained as singletons without overlapping sequences. Two examples of peptide clusters are shown in Figure 1.
Table 2. 56 proteins from which most degraded peptides appeared to originate.
A spreadsheet with detailed proteomic parameters for each cyst fluid is provided as Supplemental Data 4 at our Web site1.
| Alpha-1-antitrypsin |
| Alpha-2-HS-glycoprotein |
| Alpha-2-macroglobulin |
| Apolipoprotein A–I |
| Bile salt-activated lipase |
| Carboxypeptidase A1 |
| Carboxypeptidase A2 |
| Carboxypeptidase B |
| Cell adhesion molecule-related/down-regulated by oncogenes |
| Centrosomal protein of 72 kDa |
| Chymotrypsinogen B |
| Cleavage and polyadenylation specificity factor 6 |
| Collagen alpha-3 |
| Collagen alpha-6 |
| Complement C3 |
| Complement factor B |
| C-X-C chemokine receptor type 5 |
| Elastase-2A |
| Elastase-3A |
| Elastase-3B |
| Endothelial lipase |
| Epidermal growth factor receptor kinase substrate 8 |
| Epidermal growth factor receptor kinase substrate 8-like protein 1 |
| Fibrinogen alpha chain |
| Fibrinogen beta chain |
| Fibronectin |
| Gastricsin |
| Gastrokine-1 |
| Gelsolin |
| Haptoglobin |
| Haptoglobin-related protein |
| Hemoglobin subunit alpha |
| Hemoglobin subunit beta |
| Ig alpha-1 chain C |
| Kallikrein-1 |
| Lipase member H |
| Neuronal acetylcholine receptor protein subunit beta-2 |
| Obscurin |
| Osteopontin |
| Pancreatic alpha-amylase |
| Pancreatic lipase-related protein 1 |
| Pancreatic lipase-related protein 2 |
| Pancreatic secretory granule membrane major glycoprotein GP2 |
| Pancreatic triacylglycerol lipase |
| Pepsin A |
| Periostin |
| Polymeric-immunoglobulin receptor |
| Prominin-1 precursor |
| Pro-neuregulin-2, membrane-bound isoform |
| Retinoic acid-induced protein 3 |
| Serum albumin |
| Spectrin alpha chain |
| Thrombospondin-1 |
| Transthyretin |
| Trypsin-1 |
| Trypsin-2 |
| Trypsin-3 |
Figure 1. Alignment of some peptide clusters observed in cyst 13.
A: bile salt activated lipase, Apolipoprotein alpha.
2-D Gel Electrophoresis comparison of pancreatic cyst fluids
Six cyst fluids were analyzed by 2D gel electrophoresis (Figure 2). About 96 protein spots were identified by mass spectrometry for each cyst fluid. High resolution gel images with the protein identification annotated and their summary are provided as Supplemental Data 5–11 at our web site 4. Panel A of Figure 2 illustrated that serum albumin was split into at least 180 spots of different molecular weights and isoelectric points. Similarly, several other proteins were split into multiple spots of different molecular weights and isoelectric points.
Figure 2. 2D-gel electrophoresis proteomics of pancreatic cyst fluids.
Each panel of 2D gel from left to right is pH 3 to pH 10. A: Cyst 14 cystadenoma, B: Cyst 5 adenocarcinoma, C: Cyst 13 rich in pancreatic enzymes, D: Cyst 9 rich in pancreatic enzymes, E: Cyst 6 rich in plasma, F: Cyst 15 rich in plasma.
SDS-PAGE
The results of SDS-PAGE analysis of 20 cyst fluids are shown in Supplemental Data 12 at our web site 5.
The pancreatic cyst fluid GeLC/MS/MS proteome
The separation of Fraction A from Fraction B significantly increased the sensitivity of biomarker detection. Almost every protein identified in Figures 3–5 was abundant enough to be identified, and its relative abundance quantified for comparison, in its first LC/MS/MS run. The second run served to find more peptides for each protein.
Approximately 137 proteins normally found in plasma were observed among 13 of the pancreatic cyst fluids (Figure 3). The distribution of some of the 29 pancreatic enzymes identified within the cysts in this study is shown in Figure 4. These two classes of proteins are exclusive of each other in some cyst fluids.
Figure 4. Pancreatic enzyme proteins in the pancreatic cyst samples.
Legends are the same as for Figure 3.
Data Analysis
The mass spectrometry “wiff” data files were used to search the SwissProt protein database release 54.1 using MASCOT 2.2 (Matrix Sciences, London, U.K.), analyzing the MS/MS sequencing spectra of the +2 and +3 ions. Fixed modification of carbamidomethylcysteine, variable oxidation of methionine, and one trypsin miss were allowed for protein identification in GeLC/MS/MS, but the latter two were disallowed for calculating the emPAI scores. Peptide mass tolerance was +/−150 ppm and fragment mass tolerance was 0.5 Da. The protein identifications in Figure 5 required more than one peptide for each protein. False discovery rate due to coincidence in database was less than 3.5% for individual peptides as judged by hits at a decoy database containing randomized sequences in each entry.
The dominance of two major protein classes, blood proteins versus pancreatic enzymes, in each sample, do not allow effective classification of the potential biomarkers by typical statistical approaches that include unsupervised hierarchical clustering 20 and principal component analysis 21. Therefore, low abundance proteins with an emPAI score average for the expressing samples of less than 0.01 were first removed, leaving 462 proteins whose identification were with confidence (Supplemental Data 13 at our web site 6). This emPAI score represents about one peptide sequence identified in a protein of about 250,000 molecular weight, thus other lower score protein identifications not listed were within the approximately 3% false positive identification rate for this data. Next, 34 keratins, 137 blood proteins, and 29 pancreatic enzymes, were filtered from the proteome of each cyst fluid sample. The remaining 262 proteins were sorted by the average emPAI score calculated from the samples expressing each protein. Among the most abundant ones in this list of pancreatic cyst fluid proteins were three families of proteins some of whose homologs were previously proposed to be biomarkers of pancreatic cancer, namely mucins, CEACAM’s 22, and S100’s 23–25 (Figure 5).
Potential Proteomics Biomarkers
Ten of the cyst fluids contained one or more soluble mucin homologs, some of which have low amino acid sequence homology to each other. Seven of the cyst fluids revealed soluble CEACAM homologs. Five of the cyst fluids showed expression of S100 protein homologs (Supplemental Data 14, provided at our web site 7). The relative abundances of CEACAM5 (CEA) determined by emPAI score were in rough agreement with the clinical assays performed on the samples shown in Table 1, bearing in mind the differences in CEA measurement procedures. More specifically, the emPAI score for CEACAM5 was determined as score per unit protein used in mass spectrometry while the clinical immunoassay CEA unit was concentration (in ng per mL cyst fluid). In each case where the identification of a proteomic biomarker was at low abundance in a given cyst fluid, we ruled out the possibility of sample carry over contamination by verifying that the same biomarker was not detected in the cyst fluid loaded onto the HPLC column in the preceding sample.
Validation of the potential proteomic biomarkers by Western analyses
Figure 6 shows that CEACAM6 and S100A9 protein bands were observed by Western analysis in all the cyst fluid samples that were found by mass spectrometry to contain these proteins but not in cyst fluid samples in which they were absent.
Figure 6. SDS PAGE Western blotting validation of proteomic biomarkers in pancreatic cyst fluids.
A: CEACAM6, B: S100A9.
Discussion
Recent studies have indicated that proteomics is a suitable technology for biomarker discovery in a variety of bodily fluids. However, pancreatic cyst fluids have not been investigated in detail and may require special considerations. For example, serum protein composition is relatively constant compared with pancreatic cyst fluids. Some cyst fluids may be mucinous, or bloody, or dense, or light. Some cysts may drain into the ducts while others may have been in isolation for long periods of time. The challenges of pancreatic cyst fluid proteomics include the requirement to look past the proteins that are abundant. In this study, we applied three methods of proteomic investigation for pancreatic cyst fluids and discovered why both peptidomics and 2D gel electrophoresis are not practical approaches for this source of sample. We further show how GeLC/MS/MS can reveal useful biomarkers for advanced pancreatic neoplasm.
MALDI-TOF peptidomics of pancreatic cyst fluids with identification by LC/MS/MS
The high mass accuracy and stability of the mass spectrometer used in this study assures that the data can be reproducible by other comparable mass spectrometers so that the peptides identified by this study can be useful markers for other researchers.
It is interesting that about 56 of the most abundant proteins in the cyst fluids were the source of these degraded peptides (Table 2). Many of the peptide termini were not from tryptic digestion. Some were from chymotrypsin-like activities, and many appear to result from exopeptidase processing of one or both termini. This processing is evident in the two examples given in Figure 1 for cyst 13 where several peptides for each protein were detected and their alignment showed that they were derived from the same parent peptide. We believe this proteolytic processing occurred while the cyst fluids were inside the body because further processing was not observed after the samples were adjusted with glycine-acetonitrile. Protease inhibitors were not included in our sample collection procedures because they will prevent the study of cyst fluid proteases as biomarkers 26, and that the inhibitors would function for only an insignificant duration in the sample collection process. For a tissue with variable abundance of carboxypeptidases, aminopeptidases, matrix metalloproteases, and others, their combined effects on the constellation of peptides from the major proteins renders the peptidomics for pancreatic cyst fluids hopelessly complicated. The over-fitting of too many data points to too few patients can be misleading 27, 28.
2-D Gel Electrophoresis comparison of pancreatic cyst fluids
Differences among pancreatic cyst fluids were apparent (Figure 2). Plasma protein pattern was easily recognized in panels A, B, E, and F. However, image analysis was impractical for detecting biomarkers because most major proteins were split into numerous spots, some masking the low abundance biomarker proteins. For example, about half of the spots visible in Panel A were fragments of serum albumin (Supplemental Data 5 at our web site 8). Because cysts are known to sometimes undergo cellular degradation and necrosis, protein nicking in cyst fluids may not be preventable. Thus 2D gel proteomic approach is not appropriate for pancreatic cyst fluids.
SDS-PAGE Proteomics
SDS-PAGE of most cyst fluids distinguished two classes of major proteins; plasma infiltrate versus pancreatic enzymes (Supplemental Data 12 at our web site 9), but was unable to suggest useful biomarkers because of the dominance of the major protein bands.
GeLC/MS/MS proteomics of the pancreatic cyst fluids
The following discussion pertains to our findings in cyst fluids of amylases, mucins, CEA and S100 homologs which are believed to be important to pancreatic cancer:
Amylase biomarker
The possible indicator role of amylase differentiating mucinous from non-mucinous cysts, or determining the presence or absence of cancer within a cyst, has not been well studied. Elevated cyst fluid amylase and/or lipase suggest communication between the cyst and pancreatic duct, as for IPMNs and the majority of pancreatic pseudocysts, but cannot distinguish between these entities. Cyst 3 and cyst 14 in Figure 4 demonstrated that amylase by itself is not always a good indicator of the presence of pancreatic enzymes. Moreover, two amylase isozymes were identified (alpha amylase 2B and pancreatic alpha amylase encoded by two separate genes, AMY2B and AMY2A, respectively). The two isozymes have 98% sequence identity, but may differ in their regulation as shown in cysts 8, 10, and 14, which contained only one of the two isozymes. Current clinical assays which treat the two amylase activities as one do not capture the differential expression of the two amylase genes.
Mucin biomarkers
Abnormal expression of mucins and changes in their post-translational modification patterns were recognized as potential biomarkers of malignancy 3, 29. We showed that soluble mucins were detected even when cytology specimens did not reveal mucinous epithelial cells. Figure 5 shows that four soluble mucin homologs in cysts 2, 7, 11, 17, 14, 9, 19, 12, and 3 can be distinguished and conveniently measured via LC/MS/MS proteomics and may assist in future classification of cysts. Mucin 16 (Muc16) was also detected, with three or four peptides identified, in cysts 3 and 5, respectively. However, for a protein of 2,539,682 molecular weight, the emPAI score was smaller than 0.01 and thus not listed in Figure 5. Mucin 1 is MUC1, CA15-3, a known pancreatic cancer marker 30. Mucin 1 and mucin5AC are found in the stomach as well as in the pancreas 31 and thus can potentially appear as contaminant at low levels in pancreatic cyst fluids due to the FNA needle puncturing through the stomach wall. However, mucin 5B is found in the pancreas but not in the stomach 31. These two mucins are encoded by two different genes, MUC5AC (coding for both MUC5A and MUC5C) and MUC5B. Mucins 1, 5 and AC, but not mucin 5B have been demonstrated in pancreatic lesions by immunohistochemistry 32. None of the 11 unique peptides identifying mucin 5AC with confidence in this report are found among the 33 unique peptides that identified mucin 5B (Supplemental Data 14 at our web site 10). Importantly, this specificity of identification of soluble mucin 5B by mass spectrometry overcomes the concerns of potential gastric contamination.
CEACAM biomarkers
There are at least seven carcinoembryonic antigen homologs in humans 33, 34. CEACAM5 (CEA) is widely used in clinical tests for various cancers 35 and for distinguishing mucinous from non-mucinous pancreatic cysts. Pancreatic cyst fluid CEA levels of 192 ng/mL to 400 ng/mL appear to be specific for mucin-producing cystic neoplasms 36 most of the time 3, but not always. Our data demonstrated that additional CEACAM homologs can be easily identified and quantified using LC/MS/MS proteomics, some of which are present at higher concentrations than CEACAM5 and may increase the usefulness of CEA analysis. For example, cyst 14, a MCA, showed even higher levels of the CEACAM6 and CEACAM7 homologs than CEA (Figure 5). The validity of mass spectrometry identification of CEACAM6 proteins has been validated by Western blotting (Figure 6). Interestingly, there is little known about the functions of CEACAM homologs in pancreatic cysts and how they become solubilized in the cyst fluids. In fact, CEACAM 5, 6, 7, 8 are only found in primates and not in rodents 33, supporting our finding that CEACAM 6, and 7 should also be biomarkers of cancer in human pancreatic cyst fluids.
S100 biomarkers
The S100 protein family includes small Ca++ binding proteins that are soluble in 100% saturated ammonium sulfate solution, participate in many cellular functions 23, 37–39 including tumor promotion 40. S100A9 41, A6 and A11 24, 25 were elevated in pancreatic carcinoma tissue. S100A6 was elevated in the ductal epithelium of pancreatic cancer obtained by laser capture microdissection 42. Our study extended S100 detection to pancreatic cyst fluids. The confidence of distinguishing the four S100 homologs, A6, A8, A9, and A11 is very high. No peptides were identical among the 17 peptides sequenced for the four S100 homologs. S100A8 was detected with 6 peptides sequenced and 77% amino acid sequence coverage while homolog S100A9 had 6 peptides sequenced and 64% coverage. 34% sequence coverage was obtained for the 3 peptides sequenced for S100A11. Two peptides of excellent sequence quality and reproducibility were detected for S100A6. The coincidence of S100 homologs with the more advanced cyst neoplasia (Figure 5) requires further investigation. For example, S100 homologs with roles in inflammation may influence the etiology of pancreatic cysts40.
This study illustrated that LC/MS/MS mass spectrometry can provide comprehensive information on cancer biomarkers within pancreatic cysts using minimal volumes of fluid, correlating with increasing levels of CEA, and hence, the potential of a given cyst being a mucinous neoplasm. This sensitivity suggests that mass spectrometry can contribute diagnosis in situations which may result in small volume cyst fluid aspirates, including small cyst size and increased viscosity, where current clinical assays cannot be performed.
Although the experiments in this report required about eight months of continuous mass spectrometer time, knowing the protein names, an alternative expedient mass spectrometry method called Multiple Reaction Monitoring (MRM) can accurately quantify multiple known biomarkers against internal standards at the same time resulting in an even greater sensitivity than observed in this study 43–46. We believe proteomics will assist in the elucidation of the basic biological features and natural history of pancreatic cysts, provide a deeper understanding about the molecular profile within these cysts, and lead to better targets for early diagnosis and treatment of pre-malignant and malignant lesions of the pancreas.
Acknowledgments
The authors thank Dr. Kerry Campbell for critical reading of this manuscript. Support by JoEllen Weaver of the FCCC Biosample Repository is gratefully acknowledged.
Financial support:
Supported by the National Cancer Institute, CA119242 (A.Y) and the Ewing Trust (A.Y), Institutional Core Grant P30CA06927, the Fannie E. Rippel Foundation, the Shöller Foundation, Tobacco Settlement Funds from the Commonwealth of Pennsylvania, the Pew Charitable Trust, and the Kresge Foundation.
Footnotes
References
- 1.Yeo CJ, Sarr MG. Cystic and pseudocystic diseases of the pancreas. Curr Probl Surg. 1994;31(3):165–243. doi: 10.1016/0011-3840(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 2.Shami VM, Sundaram V, Stelow EB, et al. The level of carcinoembryonic antigen and the presence of mucin as predictors of cystic pancreatic mucinous neoplasia. Pancreas. 2007;34(4):466–469. doi: 10.1097/mpa.0b013e318033fa12. [DOI] [PubMed] [Google Scholar]
- 3.Brugge WR, Lewandrowski K, Lee-Lewandrowski E, et al. Diagnosis of pancreatic cystic neoplasms: a report of the cooperative pancreatic cyst study. Gastroenterology. 2004;126(5):1330–1336. doi: 10.1053/j.gastro.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Lewandrowski KB, Southern JF, Pins MR, et al. Cyst fluid analysis in the differential diagnosis of pancreatic cysts. A comparison of pseudocysts, serous cystadenomas, mucinous cystic neoplasms, and mucinous cystadenocarcinoma. Ann Surg. 1993;217(1):41–47. doi: 10.1097/00000658-199301000-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levy M, Levy P, Hammel P, et al. [Diagnosis of cystadenomas and cystadenocarcinomas of the pancreas. Study of 35 cases] Gastroenterol Clin Biol. 1995;19(2):189–196. [PubMed] [Google Scholar]
- 6.Brugge WR, Lauwers GY, Sahani D, et al. Cystic neoplasms of the pancreas. N Engl J Med. 2004;351(12):1218–1226. doi: 10.1056/NEJMra031623. [DOI] [PubMed] [Google Scholar]
- 7.Hamilton SR, Aaltonen LA. WHO Classification of Tumours. Pathology and Genetics of Tumours of Digestive System. Lyon, France: IARC Press; 2000. [Google Scholar]
- 8.Kloppel G, Solcia E, Longnecker DS, et al. Histological typing of tumours of the exocrine pancreas. Springer-Verlag; 1998. World Health Organization international histological classification of tumours. [Google Scholar]
- 9.Wilentz RE, Albores-Saavedra J, Zahurak M, et al. Pathologic examination accurately predicts prognosis in mucinous cystic neoplasms of the pancreas. Am J Surg Pathol. 1999;23(11):1320–1327. doi: 10.1097/00000478-199911000-00002. [DOI] [PubMed] [Google Scholar]
- 10.Kloppel G. Clinicopathologic view of intraductal papillary-mucinous tumor of the pancreas. Hepatogastroenterology. 1998;45(24):1981–1985. [PubMed] [Google Scholar]
- 11.Sarr MG, Carpenter HA, Prabhakar LP, et al. Clinical and pathologic correlation of 84 mucinous cystic neoplasms of the pancreas: can one reliably differentiate benign from malignant (or premalignant) neoplasms? Ann Surg. 2000;231(2):205–212. doi: 10.1097/00000658-200002000-00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yeo TP, Hruban RH, Leach SD, et al. Pancreatic cancer. Curr Probl Cancer. 2002;26(4):176–275. doi: 10.1067/mcn.2002.129579. [DOI] [PubMed] [Google Scholar]
- 13.Cullen JJ, Sarr MG, Ilstrup DM. Pancreatic anastomotic leak after pancreaticoduodenectomy: incidence, significance, and management. Am J Surg. 1994;168(4):295–298. doi: 10.1016/s0002-9610(05)80151-5. [DOI] [PubMed] [Google Scholar]
- 14.Gräntzdörffer I, Carl-McGrath S, Ebert MPRC. Proteomics of pancreatic cancer. Pancreas. 2008;36(4):329–336. doi: 10.1097/MPA.0b013e31815cc452. [DOI] [PubMed] [Google Scholar]
- 15.Jacobson BC, Adler DG, Davila RE, et al. ASGE guideline: complications of EUS. Gastrointest Endosc. 2005;61(1):8–12. doi: 10.1016/s0016-5107(04)02393-4. [DOI] [PubMed] [Google Scholar]
- 16.Li XM, Patel BB, Blagoi EL, et al. Analyzing alkaline proteins in human colon crypt proteome. J Proteome Res. 2004;3(4):821–833. doi: 10.1021/pr049942j. [DOI] [PubMed] [Google Scholar]
- 17.Patel BB, Li XM, Dixon MP, et al. Searchable high-resolution 2D gel proteome of the human colon crypt. J Proteome Res. 2007;6(6):2232–2238. doi: 10.1021/pr060641e. [DOI] [PubMed] [Google Scholar]
- 18.Ishihama Y, Oda Y, Tabata T, et al. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics. 2005;4(9):1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 20.Eisen MB, Spellman PT, Brown PO, et al. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95(25):14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peterson LE. Partitioning large-sample microarray-based gene expression profiles using principal components analysis. Comput Methods Programs Biomed. 2003;70(2):107–119. doi: 10.1016/s0169-2607(02)00009-3. [DOI] [PubMed] [Google Scholar]
- 22.Schmid RM. [Pancreatic cancer] Schweiz Rundsch Med Prax. 2006;95(44):1709–1712. doi: 10.1024/1661-8157.95.44.1709. [DOI] [PubMed] [Google Scholar]
- 23.Salama I, Malone PS, Mihaimeed F, et al. A review of the S100 proteins in cancer. Eur J Surg Oncol. 2008;34(4):357–364. doi: 10.1016/j.ejso.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 24.Ohuchida K, Mizumoto K, Ishikawa N, et al. The role of S100A6 in pancreatic cancer development and its clinical implication as a diagnostic marker and therapeutic target. Clin Cancer Res. 2005;11(21):7785–7793. doi: 10.1158/1078-0432.CCR-05-0714. [DOI] [PubMed] [Google Scholar]
- 25.Ohuchida K, Mizumoto K, Ohhashi S, et al. S100A11, a putative tumor suppressor gene, is overexpressed in pancreatic carcinogenesis. Clin Cancer Res. 2006;12(18):5417–5422. doi: 10.1158/1078-0432.CCR-06-0222. [DOI] [PubMed] [Google Scholar]
- 26.Villanueva J, Nazarian A, Lawlor K, et al. A sequence-specific exopeptidase activity test (SSEAT) for "functional" biomarker discovery. Mol Cell Proteomics. 2008;7(3):509–518. doi: 10.1074/mcp.M700397-MCP200. [DOI] [PubMed] [Google Scholar]
- 27.Koomen JM, Li D, Xiao LC, et al. Direct tandem mass spectrometry reveals limitations in protein profiling experiments for plasma biomarker discovery. J Proteome Res. 2005;4(3):972–981. doi: 10.1021/pr050046x. [DOI] [PubMed] [Google Scholar]
- 28.Baggerly KA, Morris JS, Edmonson SR, et al. Signal in noise: evaluating reported reproducibility of serum proteomic tests for ovarian cancer. J Natl Cancer Inst. 2005;97(4):307–309. doi: 10.1093/jnci/dji008. [DOI] [PubMed] [Google Scholar]
- 29.Hammel PR, Forgue-Lafitte ME, Levy P, et al. Detection of gastric mucins (M1 antigens) in cyst fluid for the diagnosis of cystic lesions of the pancreas. Int J Cancer. 1997;74(3):286–290. doi: 10.1002/(sici)1097-0215(19970620)74:3<286::aid-ijc9>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 30.Grote T, Logsdon CD. Progress on molecular markers of pancreatic cancer. Curr Opin Gastroenterol. 2007;23(5):508–514. doi: 10.1097/MOG.0b013e3282ba5724. [DOI] [PubMed] [Google Scholar]
- 31.Andrianifahanana M, Moniaux N, Batra SK. Regulation of mucin expression: mechanistic aspects and implications for cancer and inflammatory diseases. Biochim Biophys Acta. 2006;1765(2):189–222. doi: 10.1016/j.bbcan.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y, Gao J, Li Z, et al. Diagnostic value of mucins (MUC1, MUC2 and MUC5AC) expression profile in endoscopic ultrasound-guided fine-needle aspiration specimens of the pancreas. Int J Cancer. 2007;121(12):2716–2722. doi: 10.1002/ijc.22997. [DOI] [PubMed] [Google Scholar]
- 33.Kuespert K, Pils S, Hauck CR. CEACAMs: their role in physiology and pathophysiology. Curr Opin Cell Biol. 2006;18(5):565–571. doi: 10.1016/j.ceb.2006.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beauchemin N, Draber P, Dveksler G, et al. Redefined Nomenclature for Members of the Carcinoembryonic Antigen Family. Exp Cell Res. 1999;252(2):243–249. doi: 10.1006/excr.1999.4610. [DOI] [PubMed] [Google Scholar]
- 35.Gold P, Freedman SO. Specific carcinoembryonic antigens of the human digestive system. J Exp Med. 1965;122(3):467–481. doi: 10.1084/jem.122.3.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hammel P, Voitot H, Vilgrain V, et al. Diagnostic value of CA 72-4 and carcinoembryonic antigen determination in the fluid of pancreatic cystic lesions. Eur J Gastroenterol Hepatol. 1998;10(4):345–348. doi: 10.1097/00042737-199804000-00012. [DOI] [PubMed] [Google Scholar]
- 37.Roth J, Goebeler M, Sorg C. S100A8 and S100A9 in inflammatory diseases. Lancet. 2001;357(9261):1041. doi: 10.1016/S0140-6736(05)71610-X. [DOI] [PubMed] [Google Scholar]
- 38.Ryckman C, Vandal K, Rouleau P, et al. Proinflammatory activities of S100: proteins S100A8, S100A9, and S100A8/A9 induce neutrophil chemotaxis and adhesion. J Immunol. 2003;170(6):3233–3242. doi: 10.4049/jimmunol.170.6.3233. [DOI] [PubMed] [Google Scholar]
- 39.Leach ST, Yang Z, Messina I, et al. Serum and mucosal S100 proteins, calprotectin (S100A8/S100A9) and S100A12, are elevated at diagnosis in children with inflammatory bowel disease. Scand J Gastroenterol. 2007:1–11. doi: 10.1080/00365520701416709. [DOI] [PubMed] [Google Scholar]
- 40.Dougan M, Dranoff G. Inciting inflammation: the RAGE about tumor promotion. J Exp Med. 2008;205(2):267–270. doi: 10.1084/jem.20080136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu Z, Hu L, Evers S, et al. Differential expression profiling of human pancreatic adenocarcinoma and healthy pancreatic tissue. Proteomics. 2004;4(12):3975–3988. doi: 10.1002/pmic.200300863. [DOI] [PubMed] [Google Scholar]
- 42.Shekouh AR, Thompson CC, Prime W, et al. Application of laser capture microdissection combined with two-dimensional electrophoresis for the discovery of differentially regulated proteins in pancreatic ductal adenocarcinoma. Proteomics. 2003;3(10):1988–2001. doi: 10.1002/pmic.200300466. [DOI] [PubMed] [Google Scholar]
- 43.Zappacosta F, Collingwood TS, Huddleston MJ, et al. A quantitative results- driven approach to analyzing multisite protein phosphorylation: the phosphate- dependent phosphorylation profile of the transcription factor Pho4. Mol Cell Proteomics. 2006;5(11):2019–2030. doi: 10.1074/mcp.M600238-MCP200. [DOI] [PubMed] [Google Scholar]
- 44.Keshishian H, Addona T, Burgess M, et al. Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Proteomics. 2007;6(12):2212–2229. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolf-Yadlin A, Hautaniemi S, Lauffenburger DA, et al. Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc Natl Acad Sci U S A. 2007;104(14):5860–5865. doi: 10.1073/pnas.0608638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006;5(4):573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]