Abstract
There is emerging evidence that, beyond their cholesterol lowering properties, statins exhibit important antileukemic effects in vitro and in vivo, but the precise mechanisms by which they generate such responses remain to be determined. We have previously shown that statins promote differentiation of acute promyelocytic leukemia (APL) cells and enhance generation of all-trans-retinoic acid (ATRA)-dependent antileukemic responses. We now provide evidence that statin-dependent leukemic cell differentiation requires engagement and activation of the JNK kinase pathway. In addition, in experiments to define the molecular targets and mediators of statin-induced differentiation we found a remarkable effect of statins on ATRA-dependent gene transcription, evidenced by the selective induction of over 400 genes by the combination of atorvastatin and ATRA. Altogether, our studies identify novel statin molecular targets linked to differentiation, establish that statins modulate ATRA-dependent transcription, and suggest that combined use of statins with retinoids may provide a novel approach to enhance antileukemic responses in APL and possibly other leukemias.
Keywords: statins, atorvastatin, retinoic acid, leukemia
Introduction
Statins are agents with potent cholesterol reducing properties, resulting from their abilities to block HMG-CoA reductase activity and to inhibit mevalonate synthesis (1-3). As their administration results in rapid lowering of plasma LDL-cholesterol levels, statins are used extensively in the treatment of hypercholesterolemia and coronary artery disease (CAD) in humans (4-9). There is extensive evidence that the introduction of statins in the management and prevention of atherosclerosis had a dramatic impact in the current practice of clinical medicine and has changed the natural history of coronary artery disease in humans (4-9). Notably, extensive work over the years has established that these agents also exhibit important anti-inflammatory, pro-apoptotic and antineoplastic properties (10-14), raising the possibility that they may ultimately find a place in the prevention and/or treatment of patients with inflammatory or malignant disorders. However, so far most of the work to define the antitumor potential of statins has been focused on clinical epidemiological studies (reviewed in 14), while the therapeutic potential of these agents in combination with other chemotherapeutic and antineoplastic agents remains largely unexplored.
An area where the ability of statins to promote antitumor responses was recently directly examined is the treatment of acute myelogenous leukemia (AML), in adults (15). There has been extensive previous work documenting that statins exhibit antileukemic properties in vitro (16-19). Such findings are consistent with previously described observations establishing that synthesis and import contribute to protective cholesterol increments in acute myeloid leukemia cells (20). There are also consistent with the notion that a subset of acute myelogenous leukemias rely on increased LDL accumulation during treatment with particular drugs, indicating that combined use of statins with chemotherapy in certain cases of acute leukemia could be proven beneficial. Remarkably, it was also recently shown that Ikaros, a transcription factor that directs lymphoid lineage commitment and whose abnormal expression is implicated in the development of leukemias (21), modulates cholesterol uptake (22), further underscoring the relevance of cholesterol regulation in leukemogenesis.
Beyond the pro-apoptotic properties of statins, there is evidence that these drugs induce differentiation of cells of acute promyelocytic leukemia (APL) origin (23, 24). In addition, recent work from our laboratory has demonstrated that atorvastatin and fluvastatin enhance ATRA-induced differentiation and reverse ATRA-resistance of APL-derived cell lines or primary cells (24). These data have raised the possibility that statins can be used as differentiation enhancers in combination with ATRA. However, the mechanisms by which they promote such differentiation has been largely unknown. In the present study we provide direct evidence that engagement of the JNK pathway is essential for statin-dependent leukemic differentiation of cells of APL origin, underscoring the importance of JNK in the induction of statin-responses. We also performed experiments aimed to compare the effects of statin treatment on ATRA-dependent induction of expression of differentiation genes. In gene microarray studies, we found a dramatic atorvastatin-dependent enhancement of ATRA-inducible gene transcription in APL cells and were able to identify a group of JNK-linked genes selectively induced by the ATRA and atorvastatin combination. Altogether, these data provide insights on the molecular basis of the enhancing effects of statins on ATRA-induced antileukemic responses and support the hypothesis that combinations of statins with ATRA may lead to novel approaches for the treatment of APL and other leukemias.
Materials and Methods
Cells lines and reagents
The NB4 human APL cell line was grown in RPMI 1640 supplemented with 10% fetal bovine serum and antibiotics. The ATRA-resistant NB4.300/6 variant cell line (24, 25) was provided by Dr. Saverio Minucci (European Institute of Oncology, Milan, Italy) and was also grown in RPMI 1640 medium supplemented with 10% fetal bovine serum and antibiotics. Atorvastatin was purchased from 21CEC Pharmaceuticals Ltd. ATRA was purchased from Sigma. SP600125 was obtained from Calbiochem (La Jolla, CA). A cell-permeable JNK peptide inhibitor 1 (JNKI), was purchased from Axxora (San Diego, CA).
In vitro kinase assays
NB4 cells were pre-incubated for 60 min in the absence or presence of the JNK inhibitor SP600125 (20μM), and then treated with or without fluvastatin (10μM), or DMSO as solvent control for the indicated times. The cells were subsequently lysed in phosphorylation lysis buffer (PLB) (26, 27), and lysates were immunoprecipitated with an antibody against JNK1, purchased from Santa Cruz Biotechnology, Inc (Santa Cruz, CA), using protein G-Sepharose. The immunoprecipitated complexes were subsequently washed 3 times with PLB containing 0.1% Triton X-100 and 2 times with kinase buffer (25 mM Hepes, 25 mM MgCl2, 25 mM β-glycerophosphate, 2 mM dithiothreitol, and 0.1 mM Na3VO4, 20 μM ATP) and resuspended in 30 μl of kinase buffer containing 3 μg of c-Jun fusion protein purchased from Cell Signaling Technology, Inc. (Danvers, MA), used as an exogenous substrate. 10 μCi of [γ-32P] ATP was added to the mixture, and after 30 min of incubation at room temperature the reaction was terminated by the addition of SDS sample buffer. Proteins were analyzed by SDS-PAGE, and the phosphorylated form of c-Jun was detected by autoradiography.
Flow cytometric analysis
Granulocytic differentiation of APL cells was assessed by flow cytometric analysis, as previously described (28). Briefly, NB4 cells were pretreated for one hour with the indicated inhibitors, then treated with atorvastatin or fluvastatin for the indicated times, and cell differentiation was determined by flow cytometric analysis after staining with the anti-CD11b monoclonal antibody. The anti-CD11b monoclonal antibody and a matched isotype control were purchased from BD Biosciences (San Jose, CA).
Microarray analysis
Quality and quantity of total RNA from samples was determined using Agilent 2100 Bioanalyzer (Palo Alto, CA) and a Nanodrop spectrophotometer. cRNA was synthesized using Illumina RNA Amplification Kit Ambion (Austin, TX) starting from 500 ng of total RNA and following the procedure suggested by manufacturer. Sentrix® Human-6 Expression BeadChip hybridization, washing and staining were also done as suggested by the manufacturer. Arrays were scanned on Illumina BeadStation 500.
Microarray data analysis
BeadChip array data quality control was performed using Illumina BeadStudio software version 1.3.1.5. Probe average intensity signal was calculated with BeadStudio without background correction. Raw data were analyzed with Bioconductor (29). Average probe intensities were log2 transformed and normalized by loess method (30). To remove low quality signals two filters were applied sequentially to the 47293 reference sequences (RefSeq) available in the Sentrix® Human-6 Expression BeadChip: gene specific detection value should have been ≥0.6 in at least 50% of the samples (25845/47293 RefSeq passed this filter), gene specific interquantile range (IQR) should have been ≥0.2 (11287/25845 RefSeq passed this filter).
Statistical analysis for the time course experiments was performed using maSigPro Bioconductor library (31). MaSigPro follows a two steps regression strategy to find genes with significant temporal expression changes and significant differences between experimental groups. The method defines a general regression model for the data where the experimental groups are identified by dummy variables (i.e. ATRA, atorvastatin, ATRA + atorvastatin). The procedure first adjusts this global model by the least-squared technique to identify differentially expressed genes and selects significant genes applying false discovery rate control procedures (fdr ≤ 0.05). Secondly, backward stepwise regression is applied as a variable selection strategy to study differences between experimental groups and to find statistically significant different profiles (p-value ≤ 0.05). The final list of significant differentially expressed genes was defined using the R-squared values of this second regression model. RefSeq probes characterized by an R-squared regression model ≥ 0.6 were selected for further analysis. RefSeq probes identified by regression analysis were filtered to have at least one of the experimental point characterized by an |log2(fold change)| ≥ 1. Clustering analysis was performed using TMEV v3.1 suite (www.tigr.org). Functional analysis was performed using Ingenuity knowledge database (www.ingenuity.com).
Quantitative RT-PCR (TaqMan)
Cells were treated with ATRA (0.5μM) or atorvastatin (2 μM) or both agents for 8, 24 and 48 hrs, and RNA was isolated using the RNeasy kit from Qiagen (Valencia, CA). 1 μg of total cellular RNA was reverse transcribed into cDNA using the Omniscript RT kit and oligo(dT) primer purchased from Invitrogen (Carlsbad, CA). Real time reverse transcriptase PCR for CCL3, CCL4, IL1B, BTG-2 and NCF2 genes was carried out by an ABI7900 sequence detection system from Applied Biosystems (Foster City, CA) (32), using commercially available FAM-labeled probes and primers from Applied Biosystems. Relative quantitation of mRNA levels was plotted as -fold increase as compared with untreated samples. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used for normalization (33) ΔCt values (target gene Ct minus GAPDH Ct) for each triplicate sample were averaged, and ΔΔCt was calculated as previously described. mRNA amplification was determined by the formula 2-ΔΔCt(32).
IPA 4.0 analysis
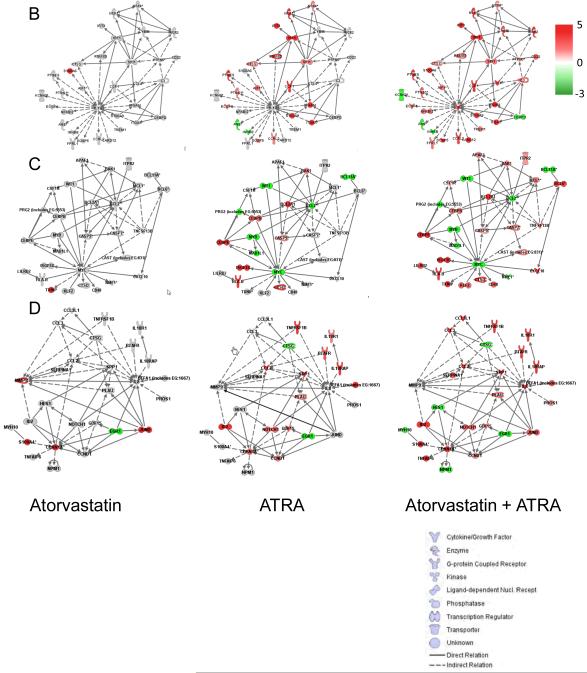
IPA 4.0 searches for the presence of relations existing between the differentially expressed genes and the genes annotated in the Ingenuity knowledge data base were analyzed using the data mining tool IPA 4.0 (www.ingenuity.com). For each treatment each time point was subjected to IPA 4.0 analysis and subsequently the results were subjected to a comparative analysis to identify functional classes associated with differentially expressed genes in response to different treatments. The data shown in Fig.3 B, C and D and in Fig.6 were generated with an updated IPA 7.0 version.
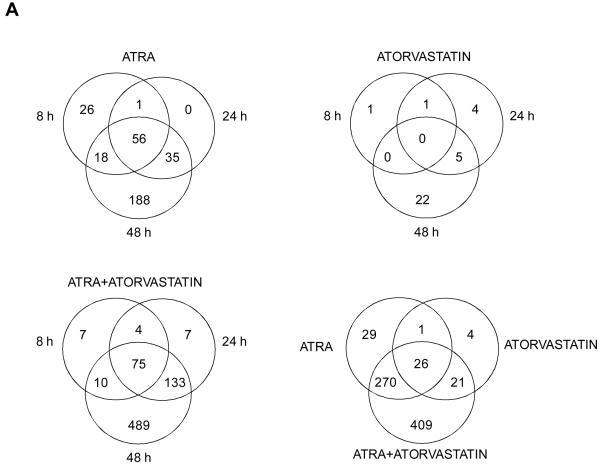
Figure 3.
Induction of gene expression by ATRA and/or atrovastatin in NB4 cells. A. Venn diagrams of differentially expressed RefSeqs characterized by an absolute log2 (fold change) of at least 1. Overlaps of the genes expressed at different time points for ATRA alone (left upper panel); Atrovastatin alone (right upper panel); and ATRA + Atorvastatin (left lower panel) are shown. Differentially expressed RefSeqs characterized by an absolute log2 (fold change) of at least 1 in at least one of the different time points of treatment is also shown (right lower panel). B-D. Relation found within differentially expressed genes, in at least one of the treatments and present in the enriched functional classes. Cluster 1, centered on IL1B gene at time point 48h (B), cluster 2, centered on MYC gene at time point 48h (C), cluster 3, centered on EGR1 gene at time point 48h (D).
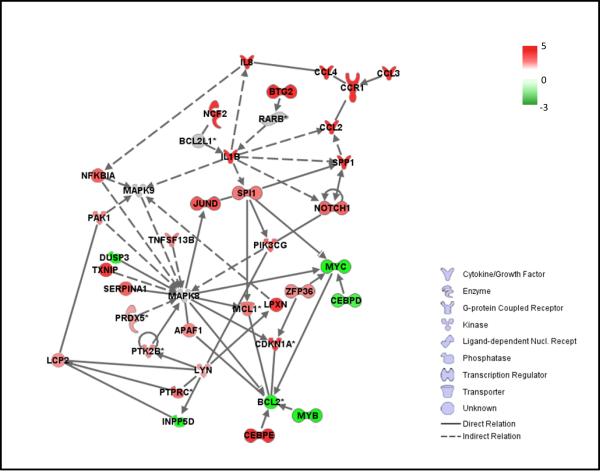
Figure 6.
Key connection links of the JNK kinase pathway to statin-induced genes associated with differentiation. IPA 7.0 analysis was performed as described in materials and methods. Functional relations were derived by connecting JNK to various genes selectively regulated after 48 hours treatment with the combination of atorvastatin and ATRA
Results
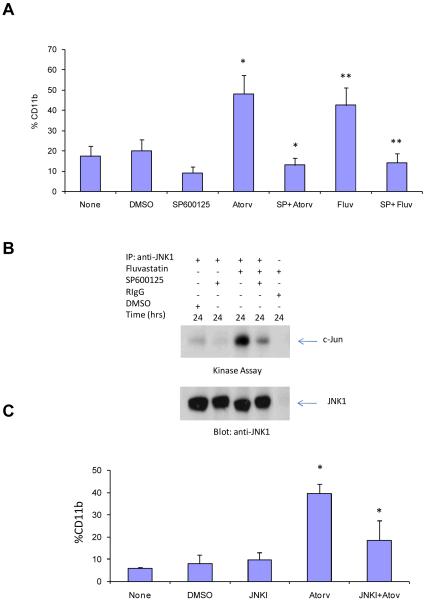
Our previous studies (24) had demonstrated that treatment of cells with statins results in activation of the JNK kinase pathway in leukemic cells and that such activation is required for statin-induced apoptosis. To determine whether engagement of the JNK kinase plays a role in the induction of differentiation of APL cells, experiments were performed in which JNK activity was blocked by either pharmacological or molecular means, and induction of differentiation was subsequently determined. Initially, the effects of a JNK pharmacological inhibitor, SP600125, were determined. When NB4 cells were pre-treated with SP600125, leukemic cell differentiation in response to either atorvastin or fluvastatin was blocked (Fig.1A), while as expected, SP600125 blocked statin-induction of JNK-kinase activity, detected by immune complex kinase assays (Fig. 1B). To definitively establish whether JNK activity is essential for statin-dependent leukemic cell differentiation, the effects of targeting JNK, using a peptide inhibitor (GRKKRRQRRR-PP-RPKRPTTLNLFPQVPRSQD-NH2) known to block JNK activation and downstream engagement of c-jun (34-36), were determined. As in the case of pharmacological inhibition, pre-treatment of cells with the peptide inhibitor abrogated the induction of differentiation of leukemic cells by atorvastatin (Fig. 1C), definitively establishing that such statin-regulated responses are JNK-dependent.
Figure 1.
Requirement of JNK for atorvastatin- or fluvastatin-induced differentiation of APL cells. A. NB4 cells were pre-incubated for 60 min in the absence or presence of SP600125 (20μM) and treated for 48 hrs in absence or presence of DMSO (control) or atorvastatin (2μM). The cells were subsequently stained with a phycoerythrin (PE)-conjugated anti-CD11b monoclonal antibody and analyzed by flow cytometry. The data represent means + S.E. of 4 independent experiments. Paired two-tailed t test analysis showed a two-tailed p=0.015 for atorvastatin alone versus the combination of SP600125 + atorvastatin and two-tailed p=0.03 for fluvastatin alone versus the combination of SP600125 + fluvastatin. B. (Upper panel) NB4 cells were pre-incubated for 60 min in the absence or presence of the JNK inhibitor SP600125 (20μM), and then treated with or without fluvastatin (10μM), or exposed to DMSO as solvent control for the indicated time. Cell lysates were immunoprecipitated (IP) with an antibody against JNK1 or control non-immune rabbit immunoglobulin (RIgG). The immunoprecipitates were then subjected to in vitro kinase assays, using c-Jun as an exogenous substrate. Proteins were analyzed by SDS-PAGE, and the phosphorylated form of c-Jun was detected by autoradiography. (Lower panel) The same blot was subsequently immunoblotted with an anti JNK1 antibody. C. NB4 cells were pre-incubated for 60 min in the absence or presence of JNKI peptide (5 μM) and treated for 48 hrs in absence or presence of DMSO (control) or atorvastatin (2μM). The cells were subsequently stained with a phycoerythrin (PE)-conjugated anti-CD11b monoclonal antibody and analyzed by flow cytometry. The data represent means + S.E. of three experiments. Paired two-tailed t test analysis showed a two-tailed p=0.0464 for atorvastatin versus JNKI + atorvastatin
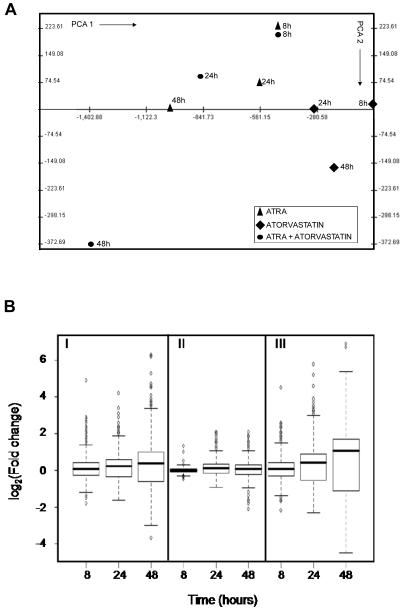
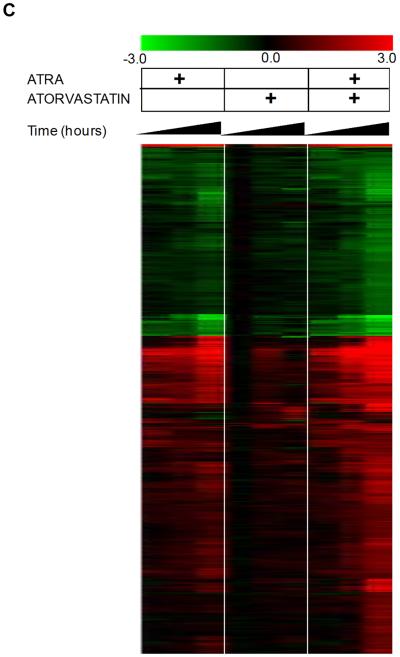
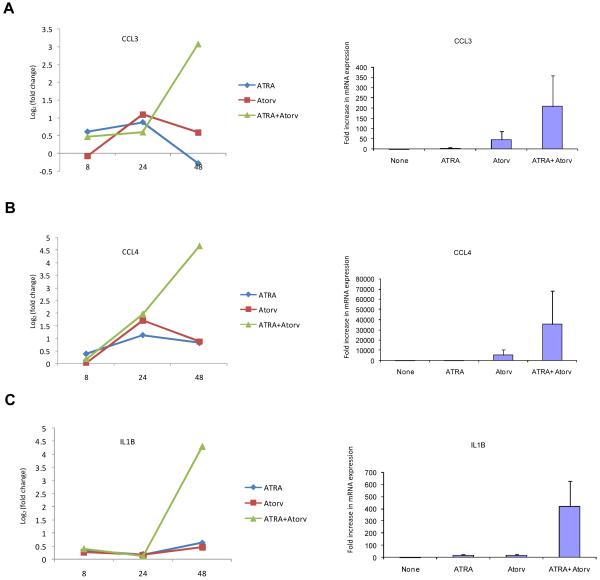
To determine whether the effects of statin-treatment on ATRA-induced leukemic cell differentiation reflect induction of specific genes, the patterns of gene expression induced by ATRA, atorvastatin, or by the combination of atorvastatin + ATRA were subsequently examined using DNA microarrays. We analyzed three prototypic situations (ATRA, atorvastatin, ATRA + atorvastatin) using Illumina Sentrix® Human-6 Expression BeadChips over three points time course (8, 24, 48 hours), and time course points were replicated in three independent experiments performed at different days. A total of 36 arrays were hybridized. After average probe intensity calculation, log2 transformation and normalization (29), probes characterized by low quality signal or invariant expression within the experimental conditions were discarded. A total of 11287 out of the 47293 RefSeq BeadChip probes were used to identify significantly differentially expressed genes. Statistical analysis was performed using a two step regression strategy (30). 1901 RefSeqs were found significantly differentially expressed in at least one condition over the time course treatments. After calculating the log2 (fold change) variation for all treatments with respect to the corresponding control time point, 782 out of 1901 RefSeq probes were characterized by a |log2(fold change)| ≥ 1 for at least one of the time points. Principal component analysis (PCA) (37), was performed on the three prototypic situations (Fig. 2A). PCA was used to attribute the overall variability in the data to a reduced set of variables (principal components). To each principal component a certain fraction of the overall variability of the data was attributed such that each successive component determined accounted for less of the variability than the previous one. From such analysis it became clear that the transcription effects induced by atorvastatin alone were clearly limited as compared to those induced by ATRA alone (Fig. 2A and B). Notably and most importantly, the combined addition of ATRA and atorvastatin produced a strong synergic effect. This was particularly evident by examining the fold change distribution width induced by the various treatments (Fig. 2B). The average effects of treatment versus no treatment at the different time points were also grouped by hierarchical agglomerative clustering (Fig. 2C). Two main groups can be observed in Fig. 2C. The set of RefSeqs mainly down-regulated (green) and the set of RefSeqs up-regulated (red). All three prototypic situations followed the same trend but with different levels of expression variation, with maximum effects seen in response to the combination of ATRA + atorvastatin (Fig. 2C). Moreover such expression was time-dependent with maximal gene expression occurring at 48 hours (Fig. 2C).
Figure 2.
Patterns of gene expression induced by atorvastatin or ATRA alone, or by the combination of ATRA + atorvastatin. A. Principal component analysis of treatments (PCA). PCA was performed using the average of the expression of the three replication for time point for the three prototypic situations. B. Distribution of fold changes existing between treatments versus untreated cells at the various time points in the set of 782 RefSeqs identified as significantly expressed differentially. I, ATRA; II, Atorvastatin; III, ATRA + Atorvastatin. C. Hierarchical clustering of 782 RefSeqs found differentially expressed by double step regression analysis.
To understand the functional relevance of the different groups of genes upregulated or downregulated by ATRA, atorvastatin, or the combination of ATRA+ atorvastatin, the 782 differentially expressed RefSeqs were divided in three groups (i.e. ATRA, atorvastatin and ATRA+ atorvastatin) on the basis of the presence of an absolute fold change variation of at least 1 with respect to the mock cell in at least one of the points of the time course-treatments. 705 RefSeqs were found directly associated to treatment with ATRA alone, while 250 RefSeq were found directly associated to treatment with atorvastatin alone. The combination of ATRA+atorvastatin could be associated with the highest number of RefSeq at 782. The overlaps existing between various time points for each treatment, as well as the overlaps between all treatments for RefSeq characterized by the presence of an absolute log2 (fold change) variation of at least 1 are summarized as Venn diagrams (Fig. 3A). As expected, the Venn diagrams clearly established that the transcriptional effects induced by atorvastatin alone are limited (Fig. 3A, right upper panel). In addition, they demonstrated a strong transcriptional synergic effect by the ATRA + atorvastatin combination (Fig. 3A, left lower panel).
The three treatment groups were then analyzed using the data mining tool IPA 4.0 (www.ingenuity.com). Using IPA 4.0, we searched for relationships between differentially expressed genes identified by us in these microarray studies, and genes annotated in the Ingenuity knowledge base, the largest manually gene annotation database based on functional information available in published studies. A comparative analysis was performed to identify the variation of expression of functional classes associated to differentially expressed genes with time. There were 15 top ranked enriched functional classes (Table I, supplementary data) and 7 of them were common to the 3 different treatments. Among the genes belonging to the 15 enriched classes, 332 Illumina Ids were associated to 254 Entrez Gene ids. Loading those genes in IPA 4.0, 13 networks (Table II, supplementary data) were obtained, and the top ranked 3 networks were all made of differentially expressed genes. It should be emphasized that networks were defined as groups of two or more genes linked by a functional association based on peer reviewed published data, while hub genes were those sharing a high number of relations with other components of networks.
In subsequent analysis, using an updated IPA 7.0 version, the effects of the different combination treatments at 48 hours on changes for the three top ranked networks were assessed. The 48 hours time point was selected for these studies as it represents the time where maximum transcriptional effects were seen. Clearly, marked synergistic effects were induced by the combination of ATRA with atorvastatin (Fig. 3 B, C and D). In cluster 1 (Fig. 3B), treatment of cells with atorvastatin alone regulated expression of very few genes in the network. This is consistent with the fact that this network is centered on IL1B. Upregulation of IL1B expression can be efficiently seen only within the combined ATRA + atorvastatin treatment (Fig. 3 B). In cluster 2 (Fig. 3C), a different example of a centered gene (MYC) is shown, which is downregulated by ATRA alone, but whose expression is not further modified by the addition of atorvastatin. In cluster 3 (Fig. 3D), a different pattern is shown, involving the EGR1 hub gene. In that case, the expression of EGR1 is decreased by either ATRA or atorvastatin alone, but it is further downregulated by the combination of ATRA + atorvastatin (Fig. 3D).
Altogether, our data established that atorvastatin treatment modulates ATRA-induced gene expression, raising the possibility that modulation of transcription may be a key mechanism by which statins enhance generation of antileukemic responses. This prompted us to classify genes selectively induced by the ATRA + atorvastatin combination, based on their functional relevance. Among the genes induced by the ATRA+ atorvastatin combination, there were several genes involved in cell differentiation (Table 3, supplementary data) and apoptosis (Table 4, supplementary data). In those tables, only genes whose expression was selectively modified/regulated by the combination of ATRA and atorvastatin at 48 hours are shown. Genes regulated synergistically by statins and ATRA, involved in apoptosis (Fig. 1, supplementary data) or differentiation (Fig. 2, supplementary data), were identified, suggesting the existence of cellular cascades and/or networks that regulate apoptosis or differentiation induced by the combination.
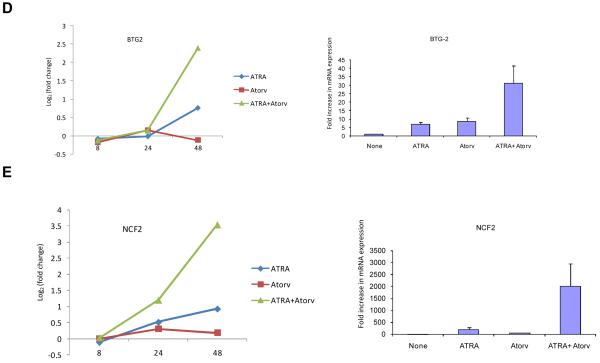
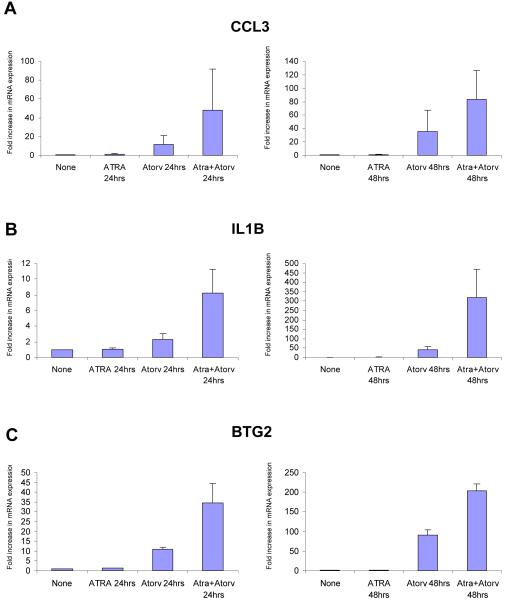
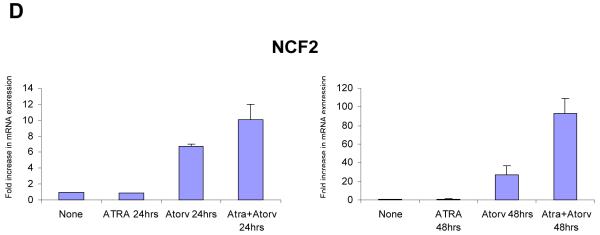
To further confirm the expression of key differentiation genes regulated by the ATRA + atorvastatin combination, the induction of expression of CCL3, CCL4, IL1B, BTG2 and NCF2 were assessed by real-time RT-PCR and compared to the patterns of expression seen in the microarray studies. There was strong induction of the expression of these genes after 48 hrs of treatment with the ATRA + atorvastatin combination (Fig. 4 A, B, C, D, and E, left panels), while similar results were obtained when real-time RT-PCR studies were performed (Fig. 4 A, B, C, D, and E, right panels).
Figure 4.
Synergistic effect of ATRA and atorvastatin on gene expression. NB4 cells were treated for 8, 24 and 48 hrs with ATRA (0.5μM), atorvastatin (2μM), or the combination of ATRA and atorvastatin, as indicated. Patterns of gene expression for CCL3 (A, left panel), CCL4 (B, left panel), IL1B (C, left panel), BTG2 (D, left panel), and NCF2 (E, left panel) were investigated using Illumina Sentrix Human-6 Expression BeadChips. Three independent microarray experiments were performed and 2 steps regression strategy was applied as statistical analysis. Expression of mRNA for CCL3 (A, right panel), CCL4 (B, right panel), IL1B (C, right panel), BTG2 (D, right panel), and NCF2 (E, right panel) were evaluated by quantitative real time RT-PCR (TaqMan). GAPDH was used for normalization. Data are expressed as fold increase over untreated samples and represent means ± S.E. of 3 independent experiments.
It is well established that resistance to ATRA develops in several cases of APL, despite initial sensitivity to the agent (36). In previous work, we had demonstrated that statins reverse ATRA-resistance and enhance leukemic differentiation of ATRA-resistant NB4 variants (24). As our data established that groups of genes associated with differentiation and/or apoptosis are selectively induced by the ATRA + atorvastatin combination, we sought to determine whether the expression of key differentiation genes by the combination also occurs in NB4.300/6 cells. NB4.300/6 cells were treated for either 24 hrs (Fig. 5, left panels) or 48 hrs (Fig. 5, right panels), with either ATRA or atorvastatin, or with the combination of ATRA and atorvastatin and the induction of expression of the CCL3, IL-1B, BTG2 and NCF2 genes was examined. As expected, there was no induction of expression of these genes in response to treatment with ATRA alone, at either 24 or 48 hours of treatment, while induction of some genes was seen in response to treatment with atorvastatin alone (Fig. 5). However, the combination of ATRA and atorvastatin resulted in significant enhancement of the expression of all genes in these cells (Fig. 5), consistent with reversal of ATRA-dependent refractoriness in the presence of atorvastatin.
Figure 5.
Expression of genes associated with differentiation in ATRA resistant APL cells. NB4.306 cells were treated for 24 hrs (left panels) or 48 hrs (right panels) with ATRA (0.5μM), atorvastatin (2μM), or the combination of ATRA and atorvastatin, as indicated. Expression of mRNAs for CCL3 (A), IL1B (B), BTG2 (C) or NCF2 (D) was evaluated by quantitative real time RT-PCR (TaqMan), using GAPDH for normalization. Data are expressed as fold increase over untreated samples and represent means ± S.E. of 2-independent experiments.
As our data established that JNK plays an important role in statin dependent differentiation (current study) and apoptosis (24), we used IPA 4.0 analysis centered on JNK (MAPK8) to identify networks of genes specifically induced by ATRA + atorvastatin in our microarray assays, with known functional linkages to differentiation and/or apoptosis. For that purpose the 48 hour time point of combined treatment was used, as at that time maximal regulation of transcriptional effects had been observed. Several pathways and signaling elements linked to differentiation or apoptosis were found to be directly or indirectly linked to JNK (Fig. 6), further suggesting the existence of a mechanism by which induction of JNK kinase activity by statins promotes differentiation and apoptosis of leukemic cells.
Discussion
Acute promyelocytic leukemia is a subtype of acute myelogenous leukemia (AML) characterized by the presence of the t(15;17) chromosomal translocation, resulting in the abnormal fusion of the retinoic acid receptor α (RARα) gene with the PML gene and the abnormal PML-RARα fusion protein (39-41). It is well established that all-trans-retinoic acid (ATRA) is a potent inducer of differentiation of acute promyelocytic leukemia cells in vitro and in vivo (reviewed in 38, 42, 43), and after its introduction in the management of the disease in the 1980s it has become a key component of standard combination regimens for the treatment of this leukemia. However, it is now also well known that leukemic cell resistance to ATRA develops with time, and this has led to the introduction and development of another agent with potent anti-APL properties, arsenic trioxide, for the management of ATRA-resistant APL (reviewed in 44). The development of retinoic acid resistance in a subset of patients with APL underscores the need for the development of novel therapeutic approaches to overcome such resistance. Moreover, beyond their potential usefulness in APL, development of similar novel approaches may prove to be of value in other AML subtypes that are normally resistant the effects of ATRA.
We have recently shown that atorvastatin and fluvastatin enhance the induction of differentiation and apoptosis of APL cells by ATRA (24). In addition, we have established that these statins reverse the development of ATRA resistance, both in NB4 variant APL cell lines and in primary resistant acute promyelocytic leukemia cells (24). Such results have raised the possibility that combined use of ATRA with late generation statins could provide a novel approach to overcome leukemic cell resistance. Importantly, our previous studies have also shown that statins enhance the suppressive effects of ATRA on bone marrow- or peripheral blood-derived primary leukemic progenitors from patients with non-APL forms of AML, raising the possibility that statins may sensitize other resistant AML subtypes to the antileukemic effects of ATRA in vivo (24). Although the clinical testing of such a hypothesis is pending, understanding the mechanisms by which statins promote induction of ATRA-dependent antileukemic effects is highly relevant and potentially important. Identifying such mechanisms could result in the identification of novel cellular elements that could be targeted in a more selective and specific manner for the treatment of acute leukemias.
The potent effects of new generation statins as inducers of APL cell differentiation and their ability to reverse ATRA-resistance, prompted us to perform studies to define the cellular mechanisms involved in the generation of such responses. In initial studies, we examined the effects of pharmacological or molecular inhibition of the JNK kinase pathway in the induction of differentiation of the NB4 APL cell line that express the t(15;17) translocation. These studies demonstrated that JNK activity is essential for the atorvastatin- or fluvastatin-dependent leukemic cell differentiation, as evidenced by the reversal of such differentiation by either the pharmacological JNK inhibitor SP600125 or by a peptide JNK inhibitor. Thus, activation of JNK by statins is essential for generation of both pro-apoptotic responses (24) and leukemic cell differentiation in APL. It should be noted that JNK is the only major pathway activated by statins in APL cells, and as we have shown in previous work, the p38 Map kinase pathway is not modulated by statins, while the Erk Map kinase is inhibited (24).
The selective engagement of JNK by statins and its important role in leukemic cell differentiation, prompted us to perform further work aimed to identify statin-dependent induction of genes that participate in leukemic cell differentiation or apoptosis and may be linked to JNK regulated pathways. For that purpose, we performed extensive microarray studies using Illumina BeadChips, to define the gene expression profiles seen in NB4 cells in response to statin treatment alone or the combination of statin treatment and ATRA. A number of genes that appear to be regulated by ATRA and atorvastatin, which have relevance to apoptosis and differentiation were identified by using such approach. Although atorvastatin alone induced expression of a minimal number of genes, it exhibited a remarkable effect in promoting ATRA-inducible gene expression. A large number of genes associated with apoptosis or differentiation were found to be selectively induced by the combination of atorvastatin and ATRA, a finding consistent with the potent enhancing effects of statins on ATRA-induced differentiation and pro-apoptotic responses in leukemic cells (24). Among the genes selectively expressed by the combination were a number of chemokines, including macrophage inflammatory protein MIP-1alpha (CCL3) and MIP-1beta (CCL4), which are well known to play important regulatory effects on hematopoietic progenitor growth and differentiation (45, 46). Similarly, IL1B, a key regulator of hematopoiesis (47), was a prominently induced gene by the ATRA + atorvastatin combination. Our microarray data analysis also demonstrated that the expression of a number of other genes was drastically augmented in response to the treatment with ATRA plus atorvastatin. Among those genes are BTG2 and NCF2. BTG2 is involved in growth inhibition and cell cycle regulation (48), and its overexpression increases RARalpha transcriptional activity and cell differentiation in response to ATRA in myeloid leukemia cells and CD34+ hematopoietic progenitors (49). NCF2 expression has been also linked to myelopoiesis and differentiation (50), underscoring the functional relevance of gene induction by the ATRA and statin combination.
Importantly, in experiments in which the expression of gene targets was examined using real-time RT-PCR, we were able to directly demonstrate upregulation of CCL3, IL1B, BTG2 and NCF2 in the variant cell line NB4.300/6 that is completely refractory to the effects of ATRA (25), but undergoes differentiation and apoptosis in response to treatment by combinations of ATRA and statins (24). Thus, statins enhance ATRA-dependent expression of key genes involved in differentiation and apoptosis both in ATRA-sensitive and resistant APL cells. Although the precise contributions of distinct elements in the generation of the antileukemic effects of ATRA remain to be defined in future studies, our data establish a unique mechanism by which statins reverse ATRA-antileukemic responses via regulation of the JNK pathway. Further work in that direction is warranted and may lead to the definition of new target proteins for the development of novel approaches for the treatment of ATRA-resistant APL and other AML subtypes.
Supplementary Material
Acknowledgements
We thank Dr. Saverio Minucci (European Institute of Oncology, Milan, Italy) for providing us the NB4.300/6 cell line. We also thanks Dr. Daniela Cantarella at the Oncogenomics microarray facility (IRCC, Candiolo, Italy) for providing us microarray data acquisition. This work was supported by NIH grants CA121192 and CA77816, a Merit Review Grant from the Department of Veterans Affairs, and by the Hairy Cell Leukemia Research Foundation.
References
- 1.Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990;343:425–30. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 2.Istvan ES, Deisenhofer J. Structural mechanism for statin inhibition of HMG-CoA reductase. Science. 2001;292:1160–64. doi: 10.1126/science.1059344. [DOI] [PubMed] [Google Scholar]
- 3.Ness GC, Zhao Z, Lopez D. Inhibitors of cholesterol biosynthesis increase hepatic low-density lipoprotein receptor protein degradation. Arch Biochem Biophys. 1996;325:242–48. doi: 10.1006/abbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 4.Sacks FM, Pfeffer MA, Moye LA, et al. Cholesterol and recurrent Events Trial investigators. The effect of pravastatin on coronary events after myocardial infarction in patients with average cholesterol levels. N Engl J Med. 1996;335:1001–9. doi: 10.1056/NEJM199610033351401. [DOI] [PubMed] [Google Scholar]
- 5.The long-term intervention with pravastatin in ischaemic disease (LIPID) study group Prevention of cardiovascular events and death with pravastatin in patients with coronary heart disease and a broad range of initial cholesterol levels. N Engl J Med. 1998;339:1349–57. doi: 10.1056/NEJM199811053391902. No Authors Listed. [DOI] [PubMed] [Google Scholar]
- 6.Nissen SE, Tuzcu EM, Schoenhagen P, et al. Effect of intensive compared with moderate lipid-lowering therapy on progression of coronary atherosclerosis: a randomized controlled trial. JAMA. 2004;291:1071–80. doi: 10.1001/jama.291.9.1071. [DOI] [PubMed] [Google Scholar]
- 7.LaRosa JC, Grundy SM, Waters DD, et al. Intensive lipid lowering with atorvastatin in patients with stable coronary disease. N Engl J Med. 2005;352:1425–35. doi: 10.1056/NEJMoa050461. [DOI] [PubMed] [Google Scholar]
- 8.Ballantyne CM, Bertolami M, Hernandez Garcia HR, et al. Achieving LDL cholesterol, non-HDL cholesterol, and apolipoprotein B target levels in high-risk patients: Measuring effective reductions in Cholesterol using rosuvastatin therapy (MERCURY) II. Am Heart J. 2006;151(975):e1–9. doi: 10.1016/j.ahj.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 9.Rackley CE. Monotherapy with HMG-CoA reductase inhibitors and secondary prevention in coronary artery disease. Clin Cardiol. 1996;19:683–9. doi: 10.1002/clc.4960190903. [DOI] [PubMed] [Google Scholar]
- 10.Chan KK, Oza AM, Siu LL. The statins as anticancer agents. Clin Cancer Res. 2003;9:10–9. Review. [PubMed] [Google Scholar]
- 11.Andela VB, Pirri M, Schwarz EM, et al. The mevalonate synthesis pathway as a therapeutic target in cancer. Clin Orthop Relat Res. 2003;(415 Suppl):S59–66. doi: 10.1097/01.blo.0000093846.72468.66. Review. [DOI] [PubMed] [Google Scholar]
- 12.Brower V. Of cancer and cholesterol: studies elucidate anticancer mechanisms of statins. J Natl Cancer Inst. 2003;95:844–6. doi: 10.1093/jnci/95.12.844. [DOI] [PubMed] [Google Scholar]
- 13.Demierre MF, Higgins PD, Gruber SB, Hawk E, Lippman SM. Statins and cancer prevention. Nat Rev Cancer. 2005;5:930–42. doi: 10.1038/nrc1751. [DOI] [PubMed] [Google Scholar]
- 14.Sassano A, Platanias LC. Statins in tumor suppression. Cancer Lett. 2008;260:11–9. doi: 10.1016/j.canlet.2007.11.036. [DOI] [PubMed] [Google Scholar]
- 15.Kornblau SM, Banker DE, Stirewalt D, et al. Blockade of adaptive defensive changes in cholesterol uptake and synthesis in AML by the addition of pravastatin to idarubicin + high-dose Ara-C: a phase 1 study. Blood. 2007;109:2999–06. doi: 10.1182/blood-2006-08-044446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu J, Wong WW, Khosravi F, Minden MD, Penn LZ. Blocking the Raf/MEK/ERK pathway sensitizes acute myelogenous leukemia cells to lovastatin-induced apoptosis. Cancer Res. 2004;64:6461–68. doi: 10.1158/0008-5472.CAN-04-0866. [DOI] [PubMed] [Google Scholar]
- 17.Dimitroulakos J, Nohynek D, Backway KL, et al. Increased sensitivity of acute myeloid leukemias to lovastatin-induced apoptosis: a potential therapeutic approach. Blood. 1999;93:1308–18. [PubMed] [Google Scholar]
- 18.Li HY, Appelbaum FR, Willman CL, Zager RA, Banker DE. Cholesterol-modulating agents kill acute myeloid leukemia cells and sensitize them to therapeutics by blocking adaptive cholesterol responses. Blood. 2003;101:3628–34. doi: 10.1182/blood-2002-07-2283. [DOI] [PubMed] [Google Scholar]
- 19.Xia Z, Tan MM, Wong WW, Dimitroulakos J, Minden MD, Penn LZ. Blocking protein geranylgeranylation is essential for lovastatin-induced apoptosis of human acute myeloid leukemia cells. Leukemia. 2001;15:1398–407. doi: 10.1038/sj.leu.2402196. [DOI] [PubMed] [Google Scholar]
- 20.Banker DE, Mayer SJ, Li HY, William CL, Appelbaum FR, Zager RA. Cholesterol synthesis and import contribute to protective cholesterol increments in acute myeloid leukemia cells. Blood. 2004;104:1816–24. doi: 10.1182/blood-2004-01-0395. [DOI] [PubMed] [Google Scholar]
- 21.Winandy S, Wu P, Georgopoulos K. A dominant mutation in the Ikaros gene leads to rapid development of leukemia and lymphoma. Cell. 1995;83:289–99. doi: 10.1016/0092-8674(95)90170-1. [DOI] [PubMed] [Google Scholar]
- 22.Loeper S, Asa SL, Ezzat S. Ikaros Modulates Cholesterol Uptake: A link between tumor suppression and differentiation. Cancer Res. 2008;68:3715–23. doi: 10.1158/0008-5472.CAN-08-0103. [DOI] [PubMed] [Google Scholar]
- 23.Dimitroulakos J, Thai S, Wasfy GH, Hedley DW, Minden MD, Penn LZ. Lovastatin induces a pronounced differentiation response in acute myeloid leukemias. Leuk Lymphoma. 2000;40:167–78. doi: 10.3109/10428190009054894. [DOI] [PubMed] [Google Scholar]
- 24.Sassano A, Katsoulidis E, Antico G, et al. Suppressive effects of statins on acute promyelocytic leukemia cells. Cancer Res. 2007;67:4524–32. doi: 10.1158/0008-5472.CAN-06-3686. [DOI] [PubMed] [Google Scholar]
- 25.Dermime S, Grignani F, Clerici M, et al. Occurrence of resistance to retinoic acid in the acute promyelocytic leukemia cell line NB4 is associated with altered expression of the PML/RAR alpha protein. Blood. 1993;82:1573–77. [PubMed] [Google Scholar]
- 26.Uddin S, Yenush L, Sun XJ, et al. Interferon-alpha engages the insulin receptor substrate-1 to associate with the phosphatidylinositol 3′-kinase. J. Biol. Chem. 1995;270:15938–15941. doi: 10.1074/jbc.270.27.15938. [DOI] [PubMed] [Google Scholar]
- 27.Verma A, Mohindru M, Deb DK, et al. Activation of Rac1 and the p38 mitogen-activated protein kinase pathway in response to arsenic trioxide. J Biol Chem. 2002;277:44988–95. doi: 10.1074/jbc.M207176200. [DOI] [PubMed] [Google Scholar]
- 28.Alsayed Y, Uddin S, Mahmud N, et al. Activation of Rac1 and the p38 mitogen-activated protein kinase pathway in response to all-trans-retinoic acid. J Biol Chem. 2001;276:4012–19. doi: 10.1074/jbc.M007431200. [DOI] [PubMed] [Google Scholar]
- 29.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 31.Conesa A, Nueda MJ, Ferrer A, Talon M. maSigPro: a method to identify significantly differential expression profiles in time-course microarray experiments. Bioinformatics. 2006;22:1096–102. doi: 10.1093/bioinformatics/btl056. [DOI] [PubMed] [Google Scholar]
- 32.Haslett JN, Sanoudou D, Kho AT, et al. Gene expression comparison of biopsies from Duchenne muscular dystrophy (DMD) and normal skeletal muscle. Proc Natl Acad Sci U S A. 2002;99:15000–5. doi: 10.1073/pnas.192571199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–08. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 34.Dickens M, Rogers JS, Cavanagh J. A cytoplasmic inhibitor of the JNK signal transduction pathway. Science. 1997;277:693–6. doi: 10.1126/science.277.5326.693. [DOI] [PubMed] [Google Scholar]
- 35.Bonny C, Oberson A, Negri S, Sauser C, Schorderet DF. Cell-permeable peptide inhibitors of JNK: novel blockers of beta-cell death. Diabetes. 2001;50:77–82. doi: 10.2337/diabetes.50.1.77. [DOI] [PubMed] [Google Scholar]
- 36.Barr RK, Kendrick TS, Bogoyevitch MA. Identification of the critical features of a small peptide inhibitor of JNK activity. J Biol Chem. 2002;277:10987–97. doi: 10.1074/jbc.M107565200. [DOI] [PubMed] [Google Scholar]
- 37.Raychaudhuri S, Stuart JM, Altman RB. Principal components analysis to summarize microarray experiments: application to sporulation time series. Pacific Symposium on Biocomputing. 2000:452–63. doi: 10.1142/9789814447331_0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tallman MS, Nabhan C, Feusner JH, Rowe JM. Acute promyelocytic leukemia: evolving therapeutic strategies. Blood. 2002:99759–67. doi: 10.1182/blood.v99.3.759. [DOI] [PubMed] [Google Scholar]
- 39.Warrell RP, Jr, de The H, Wang ZY, Degos L. Acute promyelocytic leukemia. N Engl J Med. 1993;329:177–189. doi: 10.1056/NEJM199307153290307. [DOI] [PubMed] [Google Scholar]
- 40.Borrow J, Goddard AD, Sheer D, Solomon E. Molecular analysis of acute promyelocytic leukemia breakpoint cluster region on chromosome 17. Science. 1990;249:1577–1580. doi: 10.1126/science.2218500. [DOI] [PubMed] [Google Scholar]
- 41.Melnick A, Licht JD. Deconstructing a disease: RARα, its fusion partners, and their roles in the pathogenesis of acute promyelocytic leukemia. Blood. 1999;93:3167–3215. [PubMed] [Google Scholar]
- 42.Miller WH, Jr, Waxman S. Differentiation induction as a treatment for hematologic malignancies. Oncogene. 2002;21:3496–3506. doi: 10.1038/sj.onc.1205328. [DOI] [PubMed] [Google Scholar]
- 43.Kambhampati S, Verma A, Li Y, Parmar S, Sassano A, Platanias LC. Signaling pathways activated by all-trans-retinoic acid in acute promyelocytic leukemia cells. Leuk Lymphoma. 2004;45:2175–2185. doi: 10.1080/10428190410001722053. [DOI] [PubMed] [Google Scholar]
- 44.Douer D, Tallman MS. Arsenic trioxide: new clinical experience with an old medication in hematologic malignancies. J Clin Oncol. 2005 Apr 1;23(10):2396–410. doi: 10.1200/JCO.2005.10.217. [DOI] [PubMed] [Google Scholar]
- 45.Liesveld JL, Harbol AW, Belanger T, Rosell KE, Abboud CN. MIP-1alpha and TGF-beta production in CD34+ progenitor-stromal cell coculture systems: effects of progenitor isolation method and cell-cell contact. Blood Cells Mol Dis. 2000 Aug;26(4):261–75. doi: 10.1006/bcmd.2000.0305. [DOI] [PubMed] [Google Scholar]
- 46.Jing H, Yen JH, Ganea D. A novel signaling pathway mediates the inhibition of CCL3/4 expression by prostaglandin E2. J Biol Chem. 2004 Dec 31;279(53):55176–86. doi: 10.1074/jbc.M409816200. [DOI] [PubMed] [Google Scholar]
- 47.Dinarello CA. Biologic basis for interleukin-1 in disease. Blood. 1996 Mar 15;87(6):2095–147. [PubMed] [Google Scholar]
- 48.Tirone F. The gene PC3 (TIS21/BTG2), prototype member of the PC3/BTG/TOB family: regulator in control of cell growth, differentiation, and DNA repair? J Cell Physiol. 2001 May;187(2):155–65. doi: 10.1002/jcp.1062. [DOI] [PubMed] [Google Scholar]
- 49.Passeri D, Marcucci A, Rizzo G, Billi M, Panigada M, Leonardi L, Tirone F, Grignani F. Btg2 enhances retinoic acid-induced differentiation by modulating histone H4 methylation and acetylation. Mol Cell Biol. 2006 Jul;26(13):5023–32. doi: 10.1128/MCB.01360-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lindsey S, Huang W, Wang H, Horvath E, Zhu C, Eklund EA. Activation of SHP2 protein-tyrosine phosphatase increases HoxA10-induced repression of the genes encoding gp91(PHOX) and p67(PHOX) J Biol Chem. 2007 Jan 26;282(4):2237–49. doi: 10.1074/jbc.M608642200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.