Abstract
The interaction between brome mosaic virus (BMV) coat protein (CP) and viral RNA is a carefully orchestrated process resulting in the formation of homogeneous population of infectious virions with T=3 symmetry. Expression in vivo of either wild type or mutant BMV CP through homologous replication never results in the assembly of aberrant particles. In this study, we report that deletion of amino acid residues 41–47 from the N-proximal region of BMV CP resulted in the assembly of polymorphic virions in vivo. Purified virions from symptomatic leaves remain non-infectious and Northern blot analysis of virion RNA displayed packaging defects. Biochemical of variant CP by circular dichroism and MALDI-TOF, respectively, revealed that the engineered deletion affected the protein structure and capsid dynamics. Most significantly, CP subunits dissociated from polymorphic virions are incompetent for in vitro reassembly. Based on these observations, we propose a chaperon mediated mechanism for the assembly of variant CP in vivo and also hypothesize that 41KAIKAIA47 N-proximal peptide functions as a molecular switch in regulating T= 3 virion symmetry.
Keywords: Virion polymorphism, genome packaging, virus assembly, brome mosaic virus, cowpea chlorotic mottle virus
Introduction
Eukaryotic RNA viruses with icosahedral symmetry exhibit remarkable physical homogeneity in vivo. During the assembly process, coat protein (CP) subunits are known to adopt several different conformations. An optimal conformation must be selected for the maintenance of physical homogeneity among assembled virions. Factors regulating the selection of optimal CP subunit conformations remain unknown. Nevertheless high resolution studies reveal that protein conformation involves alteration between flexible regions located near N and C termini that are commonly referred to as “molecular switches”. Their elimination often results in the appearance of polymorphic structures (Dong et al., 1998; Lokesh et al., 2002).
Brome mosaic virus (BMV) is a small RNA plant virus that belongs to the genus Bromovirus in the family Bromoviridae (Rao, 2001). The genome of BMV is divided among three single-stranded, positive sense RNA molecules (Rao, 2001). Viral replication is dependent on efficient interaction between non structural proteins 1a and 2a encoded respectively by genomic RNA1 (gB1) and 2 (gB2) (Kao et al., 1992). Genomic RNA3 (gB3) is dicistronic. Its 5′ half encodes another non-structural movement protein (MP) that promotes cell-to-cell spread while the capsid protein gene (CP) encoded in the 3′ half is translationally silent, but is expressed from a subgenomic RNA (sgB4) that is synthesized from progeny (−) gB3 by internal initiation mechanism (Miller et al., 1985). Therefore synthesis of sgB4 is contingent on replication of gB3. Physical and biochemical characterization of BMV virions suggested that gB1 and gB2 are packaged independently into two separate virions whereas gB3 and sgB4 are co-packaged into a third virion (Rao, 2006).
The structure of BMV virion has been determined to a 3.4Å resolution revealing a T=3 icosahedron and composed of 180 identical subunits of a single 19.4 kDa protein (Ding et al., 1995). In the BMV virion, amino acid residues 41–189 form the pentameric capsid A subunits, and residues 25–189 and 1–189 for the B and C subunits, respectively, compose the hexameric capsomeres (Ding et al., 1995). The first 25 N-terminal amino acids of BMV CP contain an arginine rich motif (N-ARM) and are not visible in the electron density map (Ding et al., 1995; Speir et al., 1995). The N-terminal residues may not be visible due to the basic residues in the N-ARM being internal and interacting with RNA, while the remainder of the CP is highly structured. The ease with which bromovirus preparations can be dissociated into CP subunits and nucleic acid and reassembled in vitro into virions resembling native forms (Choi et al., 2002; Zhao et al., 1995) allow the identification of RNA sequences required for efficient assembly of RNA 3 into virions (Choi and Rao, 2003). Similar approaches have delineated regions of the N-ARM of BMV CP required for directing assembly of infectious virions (Choi and Rao, 2000a; Rao and Grantham, 1995; Rao and Grantham, 1996)
Interaction between amino and carboxyl termini is essential for the formation of CP dimers, the building blocks for bromovirus assembly (Zlotnick et al., 2000). Consequently removal of amino acid residues 1-49 and/or 177-189 from amino and carboxyl termini eliminate contact between the two termini and abolish virion assembly (Choi and Rao, 2000a; Rao and Grantham, 1996; Sacher and Ahlquist, 1989; Zhao et al., 1995). Because bromovirus virions are predominately stabilized by RNA-protein interactions, RNA is a prerequisite component for the formation of icosahedral capsids in vivo without which no empty virions are found. BMV CP has been shown to form capsids with T=1 symmetry in vitro due to the loss of either 35 (Ding et al., 1995) or 63 N-terminal amino acids (Cuillel et al., 1981), and in vivo when the CP mRNA was expressed either autonomously in a yeast system (Krol et al., 1999) or via tobacco mosaic virus-based expression vector (Choi and Rao, 2000b). To date, virion polymorphism in BMV has not been observed in vivo when its CP was expressed in the presence of homologous replication. In this study, while analyzing BMV CP regions involved in RNA packaging, we identified a seven amino acid peptide with the sequence of KAIKAIA, corresponding to N-terminal 41–47 residues that function as a molecular switch. BMV CP harboring the deletion of 41KAIKAIA47 (Δ7aa) resulted in the formation of polymorphic virions. These virions and CP subunits respectively exhibited unique properties with respect to RNA packaging and assembly characteristics in vivo and in vitro. Based on the incompetence of Δ7aa CP subunits to assemble in vitro but not in vivo, we hypothesize that, in vivo, a chaperon-mediate assembly mechanism promotes virion formation.
Results
Rationale and Characteristics of Δ7aa variant
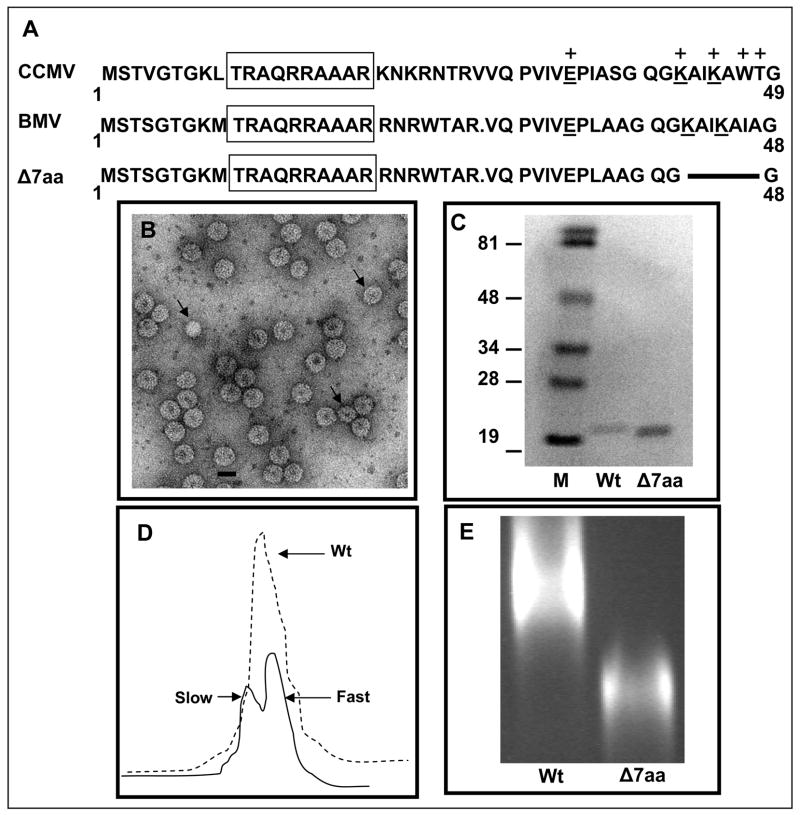
Previous mutational analysis of the N-terminal 25 amino acid region of BMV CP revealed that determinants specific for packaging CP subgenomic RNA4 were localized in the N-terminal 10-20 amino acids constituting the arginine rich motif (N-ARM) and the same amino acids however were not specific for the genomic RNA (Choi and Rao, 2000a). This data suggested that determinants specific for packaging BMV genomic RNA likely resided elsewhere in the CP ORF. Although the crystallographic structure of BMV has been determined, no recognizable RNA is visible in the electron density maps (Ding et al., 1995). By contrast, ordered RNA was observed in cowpea chlorotic mottle virus (CCMV) and depending on the local capsid environment, ordered electron density for residues 27 to 190 or for residues 42 to 190 was observed (Speir et al., 1995). These observations allowed Speir et al to predict the RNA binding amino acid residues encompassing the entire CP ORF. Since BMV and CCMV are structurally very similar (Ding et al., 1995; Speir et al., 1995) and their CP sequences are 70% identical and highly homologous over the remainder, we have extrapolated RNA binding amino acid residues of CCMV CP to that of BMV CP. A region of amino acids 30-49 immediately downstream to the basic N-terminal ARM of CCMV CP has been envisioned to contain five RNA binding amino acid residues (one Glu34, two Lys42/45, one Trp47 and one Thr48; Fig. 1A). Among these three residues (one Glu33, two Lys41/44) are conserved in sequence and position in BMV CP (Fig. 1A). To verify the functionality of this N-proximal RNA binding core sequence, a deletion of seven amino acids encompassing residues 41-47 was engineered into a biologically active clone of BMV RNA3 (pT7B3). This variant is referred to as Δ7aa (Fig. 1A).
Figure 1.
Characteristic features of BMV CP variant Δ7aa. (A) Sequence alignment of the N-terminal amino acids of CCMV and BMV CP. Within this region, the sequences of BMV and CCMV are highly homologous. BMV CP is one amino acid residue shorter than that of CCMV due to a deletion at position 27. The boxed region indicates the N-terminal arginine rich motif (N-ARM). Residues of CCMV CP involved in RNA binding are indicated by (+). The conserved residues involved in RNA binding are underlined. The extent of the engineered deletion in variant Δ7aa is shown by the solid line. (B) Electron micrograph of purified virions of Δ7aa. Purified virions were subjected negative staining prior to viewing under EM. The smaller size particles are indicated by the arrow head. Bar = 20 nm. (C) Western blot analysis of wt and Δ7aa CP. Purified virions of wt and Δ7aa were suspended in SDS-PAGE sample buffer, denatured by boiling the sample and subjected to 16% SDS-PAGE. After transferring the fractionated proteins to a nitrocellulose membrane, the blot was probed with antibodies specific for BMV. Note the difference in mobility of CP for wt and Δ7aa. M, marker lane. Sizes of protein markers in kDa are shown to the left. (D) Sucrose density gradient sedimentation analysis of partially purified virions of Δ7aa. Partially purified virions were subjected to centrifugation through 10%–40% linear sucrose density gradient. Wt BMV virions sedimenting as a single peak is indicated by the dotted line while those of Δ7aa sedimenting as slow and fast peaks by the solid line. (E) Virion mobility profiles of wt and Δ7aa. Purified virions of wt and Δ7aa were electrophoresed in a 1% agarose/TAE gel containing EtBr. Notice the mobility of Δ7aa virions is different from that of wt.
Biological activity and packaging phenotypes of variant B3/Δ7aa
In vitro synthesized capped transcripts of B3/Δ7aa variant were co-inoculated with wt B1 and B2 to Chenopodium quinoa. Control plants inoculated with all three wt transcripts displayed characteristic chlorotic local lesions followed by systemic mottling phenotype by 7–10 day post inoculation (Table 1). Whereas, inoculation with B3/Δ7aa variant resulted in the induction of chlorotic local lesions whose onset was delayed by 2 days. The infection, however, failed to spread systemically even 3 weeks post inoculation (Table 1).
Table 1.
Biological activity and virion properties of wild type BMV and its CP variant Δ7aa
| Mutant | Sequence alterations | Symptoms on C. quinoac | Virion Properties |
||
|---|---|---|---|---|---|
| Yield (mg/g) | Size (nm) | Electophoretic Mobilityd | |||
| Wild Type | Wild Type | CLL; SM | 5 | 28 | Normal |
| Δ7aaa | N-41KAIKAIA47 deleted | CLL; no systemic infection | 0.4 | 28/26 | Faster |
| Δl7aa-agrob | N-41KAIKAIA47 deleted | CLL; no systemic infection | 1.5 | 28/26 | Faster |
Δ7aa infection in plants was mediated through in vitro synthesized transcript inoculation.
Δ7aa infection in plants was mediated through agroinfiltration.
CLL, chlorotic local lesions; SM, systemic mottling
Electrophoretic mobility was carried out in 1% agarose gels prepared and electrphoresed in TAE buffer prior to staining with ethidium bromide.
In BMV, induction of local lesions requires virion assembly (Rao and Grantham, 1995). Therefore, the infection phenotypes elicited by B3/Δ7aa suggested that the variant CP is competent for virion assembly promoting local lesion induction. To verify this, virions were purified from symptomatic inoculated leaves of B3/Δ7aa variant. Compared to wt, the virus yield for variant B3/Δ7aa variant was reduced by 90% (Table 1). When examined under the electron microscope, by contrast to the purified virion preparation of wt, which appeared to be homogeneous with respect to size and morphology, virions of B3/Δ7aa variant exhibited polymorphism containing at least two distinct populations. An area measuring 7.5 μm2 on each electron micrograph was used to visually count the differently–sized virions. Based on three independent experiments, 90% of the virion population measured at about 28 nm while the remaining 10% measured at about 26 nm (Fig. 1B). The observed effect may be dynamic and only affect a fraction of the assembly events. This type of behavior is obvious in CCMV reassembly reactions where various particle populations form without any mutation in the coat protein(Bancroft, 1970; Bancroft et al., 1969). The presence of the engineered deletion was confirmed by the faster migration pattern of CP in Western blots probed with anti-BMV CP (Fig. 1C). Additional sucrose-gradient sedimentation of partially purified virions of B3/Δ7aa variant segregated the two virion populations into a faster (identical to that of wt BMV virions) and a slower sedimenting peaks (Fig. 1D). Furthermore, agarose gel electrophoresis of purified virions clearly distinguished 7aa from wt by its faster migration profile (Fig. 1E).
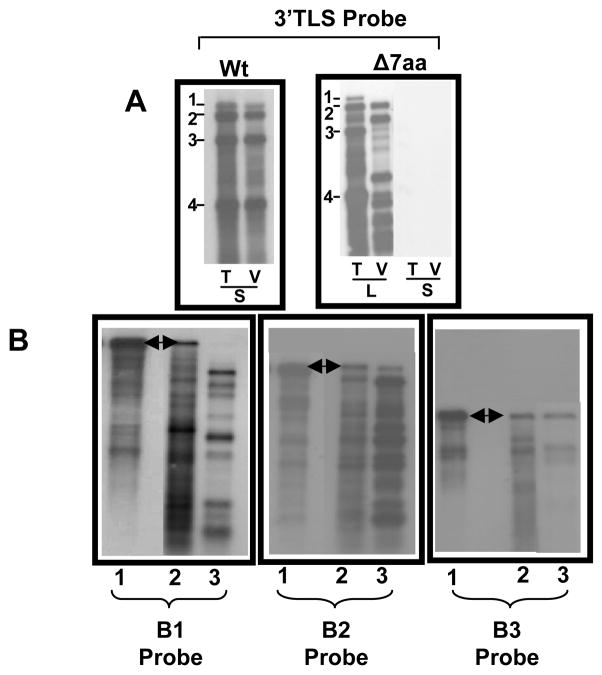
To verify the packaging competence of B3/Δ7aa variant, total and virion RNA profiles were comparatively analyzed by Northern blot hybridization using riboprobe complementary to either the highly conserved 3′ TLS sequence present on all four BMV RNAs (Fig. 2A) or specific for each BMV genomic RNA (Fig. 2B). By contrast to wt, total and virion RNA recovered from B3/Δ7aa variant progeny revealed a strikingly different packaging profile (Fig. 2A and B). Hybridization of total RNA preparations with riboprobes complementary either to a 3′ TLS region or specific for BMV genomic RNAs, detected the presence of all four RNAs (Fig. 2A). However, similar hybridization analysis of multiple blots containing virion RNA failed to detect B1 but not B2 or B3 (Fig. 2B). Prolonged exposure of blots also failed to reveal the presence of any detectable B1 (data not shown). The non-infectious nature of purified virions of Δ7aa confirmed the deficiency in B1 packaging. In addition to genomic RNAs, several truncated RNA species with faster migrating pattern was consistently detected with each hybridization probe, indicating that fragmentation of genomic RNA had occurred. This fragmentation profile was consistently detected in several repeated experiments.
Figure 2.
Progeny analysis of wt and Δ7aa. (A) Northern blot analysis of total (T) and virion RNA (V) recovered from either local (L) and systemic (S) infections of wild type (Wt) and Δ7aa. Approximately 5 μg of total nucleic acid or 0.5 μg of virion RNA was denatured with formamide/formaldehyde and subjected to 1.2% agarose electrophoresis prior to vacuum blotting to a nylon membrane. The blot was then hybridized with 32P-labeled riboprobes complementary to homologous 3′ TLS present on all four BMV RNAs. The position of all four BMV RNAs is shown to the left of each panel. (B) Northern blot analysis of virion RNAs recovered from variant Δ7aa. Multiple Northern blots containing total (lane 2) and virion RNA (lane 3) of the variant Δ7aa were hybridized with riboprobes specific for B1 or B2 or B3. Lane 1 contained virion of wt BMV RNA. Conditions of hybridization are as described above. The arrow head indicates the position of respective full length genomic RNAs.
Analysis of RNA content in polymorphic forms of Δ7aa
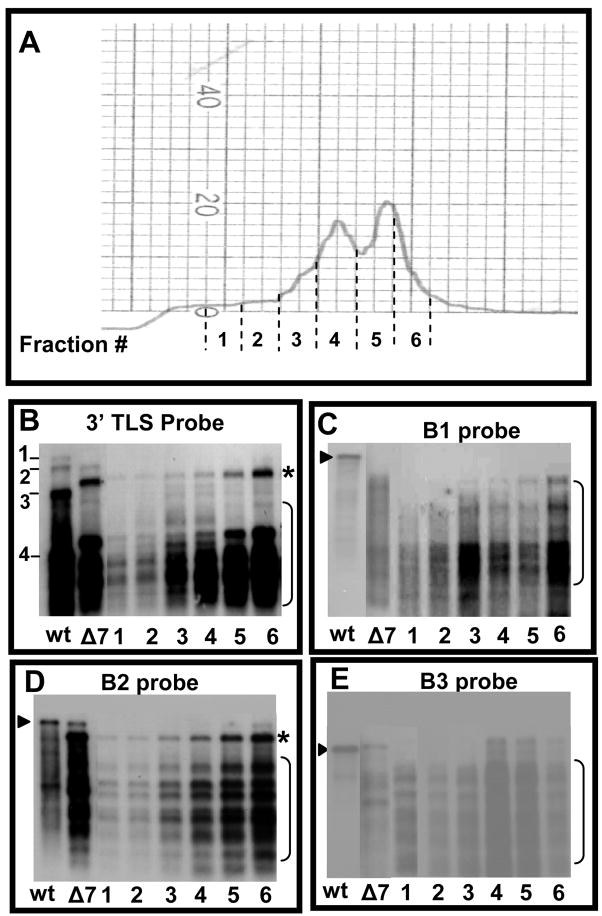
To analyze RNA profiles of the two distinct virion populations assembled in vivo with the CP subunits of B3/Δ7aa variant, partially purified virions were subjected to sucrose density gradient centrifugation. A total of six fractions encompassing the faster and slower sedimenting peaks were collected (Fig. 3A). RNA extracted from each fraction was subjected to Northern blot hybridization using riboprobes complementary to either the 3′ TLS region or specific for BMV genomic RNA. The results are summarized in Fig. 3(B–E). In each case, irrespective of the riboprobe probe, hybridization signal for fractions encompassing the slower sedimentation (#3 and #4) was always weaker than those of the faster sedimentation (#5 and #6) (Fig. 3, B–E). In accordance with Northern blot results from un-fractionated virion preparations, none of the fractions contained full-length B1 when the blots were hybridized with a riboprobe specific for B1. However this riboprobe detected faster migrating RNAs in all six fractions (Fig. 3C).
Figure 3.
Fractionation and RNA analysis of polymorphic virions of Δ7aa. (A) Partially purified virions of Δ7aa were subjected density gradient centrifugation as described under Fig. 1D legend. Individual fractions corresponding to areas indicated by dotted lines were collected, concentrated by Centricon-100 columns and RNA was extracted by SDS-phenol/chloroform method. (B–E) Northern blot hybridization. RNA extracted from the each of the six fractions corresponding to Δ7aa slow and fast sedimenting peaks were subjected to multiple Northern blot hybridizations. RNA isolated from unfractionated virions of wt and Δ7aa was used as controls. To maintain uniformity among the RNA sample loaded, each lane contained approximately 100 ng of virion RNA and the blot was hybridized with indicated 32P-labelled riboprobes. The arrow head indicates respective genomic full length RNA whereas the bracketed region indicates truncated RNA species.
Hybridization with riboprobes specific for B2 detected full-length RNA in faster sedimenting fraction # 6 (Fig. 3D; indicated by arrow head) while its presence in fraction #5 was evident only in blots exposed for longer period of time (data not shown). All six fractions contained the faster migrating truncated B2 species (indicated by a bracket in Fig. 3D). Riboprobe specific for B3 detected full-length B3 as well as faster migrating truncated species in three faster sedimenting fractions (#4, 5 and 6; Fig. 3E). No full length B3 was present in the slower sedimenting fractions although truncated RNAs were detected (# 1–3; Fig. 3E). Collectively, the data suggested that the faster sedimenting particles (measuring at 28 nm) packaged near full length as well as truncated species while slower sedimenting particles (measuring at 26 nm) packaged truncated RNA species. Furthermore the data also suggest that CP subunits of Δ7aa preferentially packaged truncated RNA species over the full length (see Discussion).
Induction of Δ7aa infection via agroinfiltration
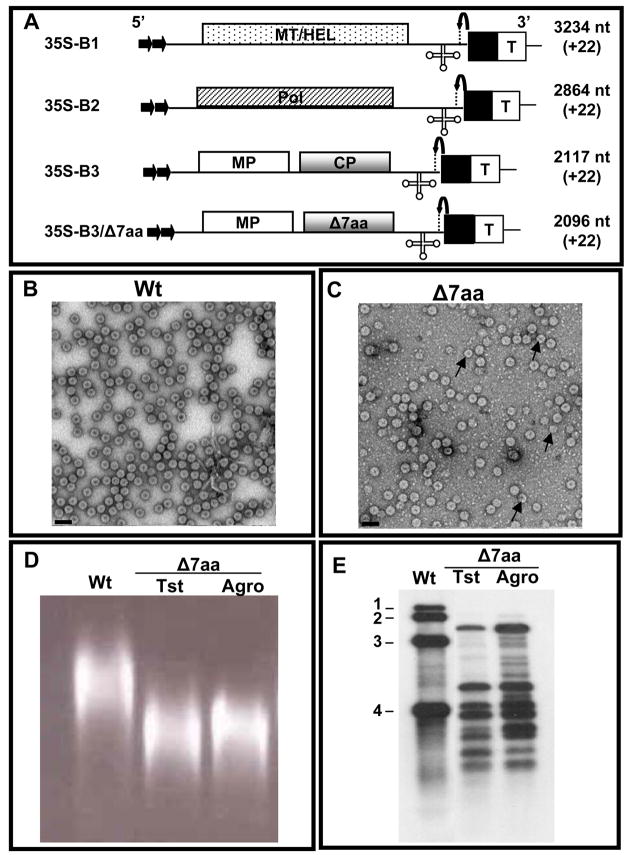
In order to get more insight on the molecular interaction between the CP subunits of variant Δ7aa and viral RNA during packaging, in vitro assembly assays would be ideal. However such experiments are impeded by insufficient virus yield resulting from transcript inoculations (Table 1). To circumvent this problem, we used an agroinfiltration approach to express Δ7aa CP subunits in Nicotiana benthamiana plants by engineering the deletion into the CP ORF of agroinfiltratable clone of B3 (35S-B3/Δ7aa; Fig. 4A). Virions purified from leaves agroinfiltrated with a mixture of inoculum containing 35S−B1+35S−B2+35S−B3/Δ7aa displayed properties similar to those recovered from transcript inoculations with respect to virion polymorphism (Fig. 4C), electrophoretic mobility pattern in agarose gels (Fig. 4D) and virion RNA profile (Fig. 4E). These observations suggested that Δ7aa infection induced by agroinfiltration faithfully mimicked that of transcripts. As compared to transcript inoculations, the yield of Δ7aa virions was increased approximately 4 fold (Table 1).
Figure 4.
Biological activity of Δ7aa induced by agroinfiltration. (A) Characteristics of T-DNA plasmids of wt BMV genomic RNAs and B3/Δ7aa used for Agrobacterium-mediated transient expression in plants. The 35S-B1, 35S-B2 and 35S-B3 constructs contain full-length cDNA copies of BMV genomic RNAs 1 (B1), 2 (B2) and 3 (B3) respectively (Annamalai and Rao, 2005). Filled arrows at the 5′ end represent the location of double 35S promoter (35S) whereas filled square and T respectively denote ribozyme sequence cassette derived from satellite tobacco ring spot virus and the 35S-polyadenylation terminator signals. Bent arrow at the 3′ end represents ribozyme cleavage site. The lengths of wt BMV RNAs and the number of non viral nucleotides left after self-cleavage by ribozyme (shown in bracket) are indicated. (B and C) Electron micrographic images of partially purified virions of wt (B) and Δ7aa (C) from agroinfiltrated N. benthamiana plants. In panel C, the smaller size particles are indicated by the arrow head. Bar = 50 nm. (D) Virion mobility profiles. Partially purified virions of Δ7aa from plant infected either with in vitro transcripts (Tst) or by agroinfiltration (agro) were subjected agrose gel electrophoresis as described under Fig. 1 legend. Agroinfiltration derived wt virion preparation was used as a control. (E) Northern blot analysis. RNA isolated from virions of Δ7aa purified from plants infected either with in vitro transcripts (Tst) or by agroinfiltration (agro) was subjected to Northern blot hybridization with a riboprobe complementary to the 3′ TLS as described under Fig. 2 legend. Virion RNA of wt BMV was used as a marker.
Stability of Δ7aa virions
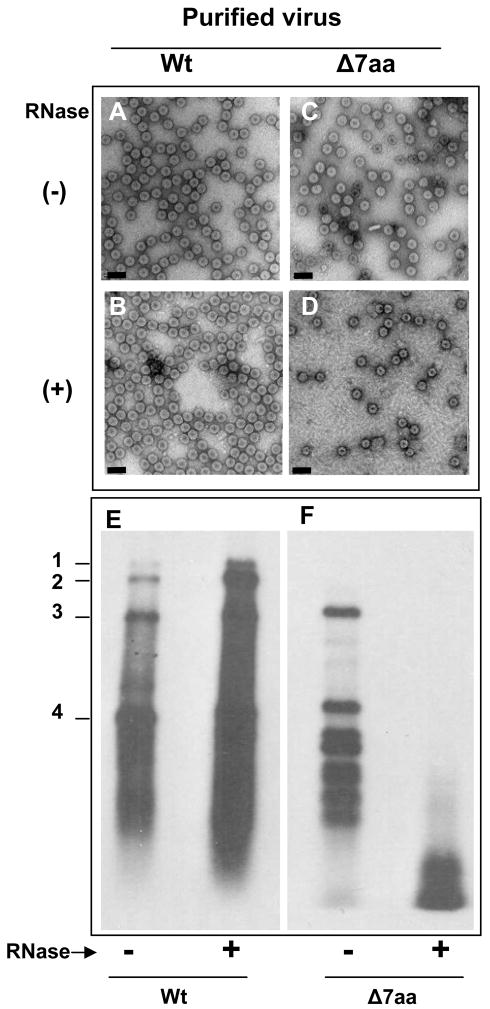
To examine the stability of Δ7aa, purified virions were subjected to RNase treatment. As a control, purified virions of wt BMV were also subjected to similar to treatment. The effect of this treatment on virion morphology and packaged RNA was respectively assessed by EM and Northern blot hybridization. Results are summarized in Fig. 5. RNase treatment had no discernable effect on either the morphology (Fig. 5A & B) or the packaged RNA of control wt BMV preparations (Fig. 5E). Interestingly, although RNase treatment did not affect the morphology of Δ7aa virions (Fig. 5C & D) but degraded the packaged RNA (Fig. 5F).
Figure 5.
RNase sensitivity of purified virions. Electron micrographic images of purified virions of wt and Δ7aa following RNase treatment (panels B and D). Untreated virions are shown in panels A and C. Bar = 50 nm. Approximately 50–100μg/ml of purified virions of wt and 7aa were treated with 1μ μg/μl of RNase A for 30 min at 30°C. Following RNase treatment, each preparation was divided into two aliquots. One aliquot was subjected to EM examination while the other was used to extract RNA for Northern blot hybridization (E and F). Preparation of virion samples for EM examination and conditions of Northern blot hybridization are as described under the legends of Fig 1 and 2.
Coat protein subunits of Δ7aa are assembly defective in vitro
From the data presented above (Figs. 1–4), it is obvious that Δ7aa CP subunits preferentially package truncated RNA species, although full-length BMV RNAs have been found in total RNA preparations (Fig. 2A). But the question that needs to be addressed is where did BMV RNA get fragmented? Is it prior to or after packaging? To find answers to these questions, first it is imperative to test whether CP subunits of Δ7aa are competent to package full-length BMV genomic RNAs using in vitro assembly assays. Bromovirus virions have been assembled in vitro under a variety conditions (Choi et al., 2002; Choi and Rao, 2003; Zhao et al., 1995). Although no empty virions have been found in vivo (Rao, 2001; Rao, 2006), empty virions or RNA-containing virions or both can be assembled in vitro under different physiological conditions. Thus, when CP and RNA are allowed to reassemble in buffer A (50 mM NaOAc, pH 4.8, 0.1M NaCl, 0.2mM PMSF) both empty and RNA containing virions are formed. By contrast, reassembly in buffer B (50 mM Tris-HCl, pH 7.2, 50 mM NaCl, 10 mM KCl, 5 mM MgCl2, and 1 mM DTT) results in the assembly of only RNA containing virions. To test the assembly competence of Δ7aa CP subunits in the presence and absence of RNA, in vitro assembly assays were performed using buffers A and B. Virions of Δ7aa purified from agroinfiltrated plants were dissociated into CP subunits and two in vitro reassembly experiments were performed. In the first experiment, dissociated CP subunits of wt and Δ7aa were independently allowed to reassemble using buffer A (i.e. in the absence of RNA). In the second experiment reassembly was performed in the presence of two sets of RNA preparations. In the first set, CP subunits of either wt or Δ7aa were allowed to reassemble with preparations of either wt BMV virion RNA or in vitro transcripts corresponding to each of the three BMV genomic RNA. In the second set, RNA preparation recovered from purified virions of Δ7aa was used as substrate. The results of these two independent experiments are summarized in Table 2. Irrespective of the physiological conditions and RNA substrates, efficient assembly of virions was observed only with CP subunits of wt BMV but not with Δ7aa (Table 2). Collectively, these results suggest that CP subunits of Δ7aa are defective in reassembly of either empty virions or RNA containing virions.
Table 2.
In vitro reassembly of BMV RNA and CP subunits of wild type and Δ7aa
| Substratea | CP subunitsb |
|||
|---|---|---|---|---|
| BMV |
CCMV |
|||
| Wild type | Δ7aa | Wild type | Δ7aa | |
| Assembly @ pH 4.8c (buffer A) | ||||
| CP only | + | − | NT | NT |
| Assembly @ pH7.2d (buffer B) | ||||
| CP only | − | − | − | − |
| BMV RNAe | + | − | + | + |
| B1f | + | − | NT | NT |
| B2f | + | − | NT | NT |
| B3f | + | − | NT | NT |
| BMV Δ7aae | + | − | NT | NT |
| CCMV RNAe | NT | NT | + | + |
| CCMV Δ7aae | NT | NT | + | + |
In vitro assembly was performed as described previously (Choi et al., 2002; Zhao et al., 1995)
Either CP or the indicated RNA was used as substrates.
CP subunits were isolated from purified virions using CaCl2 method.
Under these conditions either empty or RNA containing virions are assembled.
Under these conditions only RNA containing virions are assembled.
Virion RNA
in vitro transcripts
(+), Presence of virion assembly; (−), Absence of virion assembly; NT, not tested
Mutation Δ7aa abolished RNA-protein interactions
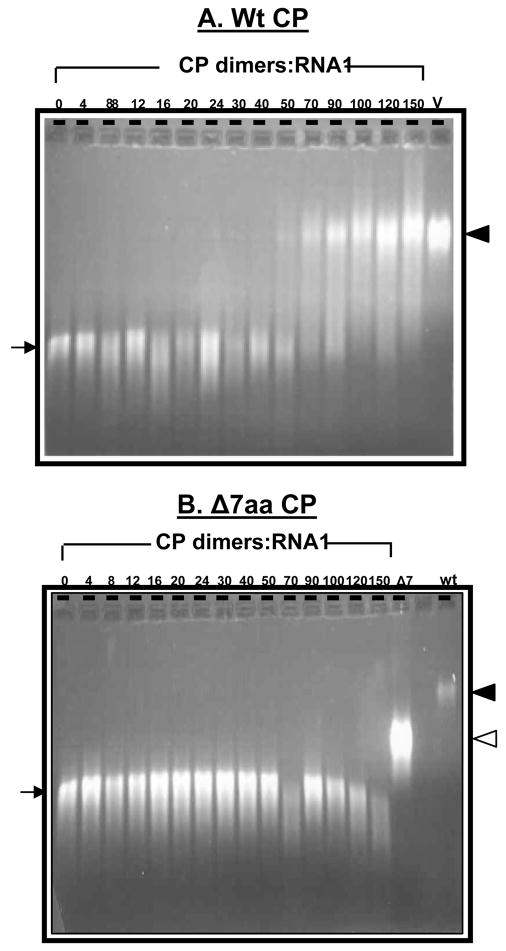
In vitro reassembly data presented above convincingly demonstrates that CP subunits of Δ7aa were defective in the formation of virions. However it is possible that the CP subunits of Δ7aa could have interacted with RNA to form a ribonucleoprotein complex, but failed to assemble into virions. To verify this possibility, polymerization kinetics between BMV RNA and either wt or Δ7aa CP subunits was examined by gel shift assays. Stoichiometric ratios of wt and mutant CP subunits were titrated against a constant concentration (30 nM) of B1 or B2 or B3 transcripts and incubated for 20 min (short time) and 24 hr (long time). A representative example of a typical interaction between wt CP:B1 and Δ7aa CP:B1 is shown in Fig. 6. As observed previously for CCMV (Annamalai et al., 2005; Johnson et al., 2004), increasing concentrations of wt CP dimers progressively retarded the migration of B1 (Fig. 6A). Agarose gel analysis of reaction products revealed that the migration of CP-B1 complex at stoichiometric ratio of 90 dimers and beyond per B1 molecule paralleled that of native virions (Fig. 6A). By contrast, titration of Δ7aa CP subunits against B1 showed no retardation irrespective of the concentration of the CP dimers (Fig. 6B). Similar results were obtained when transcripts of either B2 or B3 were titrated with Δ7aa CP subunits (Data not shown).
Figure 6.
Gel retardation analysis of RNAs by wt and Δ7aa CP subunits. Approximately 30 nM of desired BMV RNA transcript was titrated with the indicated amounts of CP dimers of either wt (A) Δ7aa (B) for 20 min at 20°C. After incubation the samples were loaded on to 1% agarose gel prepared and electrophoresed in TAE buffer. The relative electrophoretic mobility of purified virion samples from wt BMV (V) and Δ7aa (Δ7) infections are respectively indicated by filled and empty arrow heads shown to the right. The arrow to the left indicates mobility of pattern of free-viral RNA transcript.
Δ7aa mutation affected the structure of the CP subunits
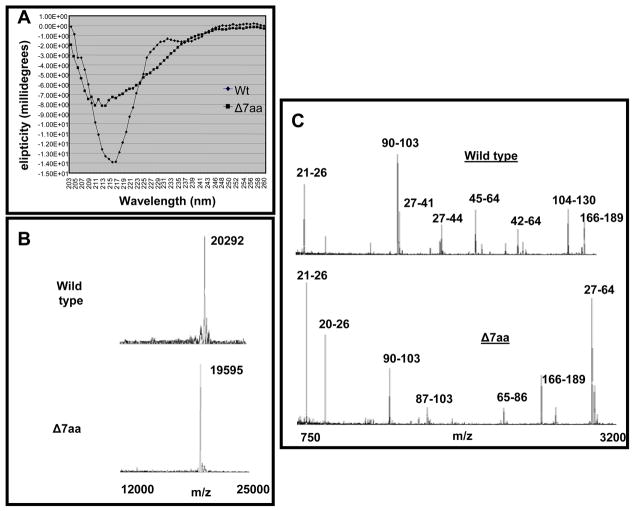
One of the reasons for the observed inability of Δ7aa CP subunits to promote RNA binding (Fig. 6B) is that the engineered deletion could have altered the structural conformation of the protein. To verify this possibility, dissociated CP subunits of wt and Δ7aa were subjected to circular dichroism spectroscopy in the far-UV spectral region (190–250 nm). The CD spectral data (Fig. 7A) indicate that the N-terminal deletion of seven residues caused a significant change in the secondary structure of the CP, with the most notable changes being the amount of alpha helices formed. Clearly in the structural context, the CP of Δ7aa appears to be different from that of wt.
Figure 7.
Biochemical characterization of wt and Δ7aa virions (A) Circular dichroism of wt and Δ7aa CP. (B) Mass spectrometry of wt and Δ7aa virions. Wt and Δ7aa virions yielded single peaks with a value of 20292 Da and 19595 Da respectively. The difference in the relative mass confirms the presence of 7 amino acid dele2tion in Δ7aa. (C) MALDI-TOF analysis of peptides released from wt and Δ7aa following trypsin treatment. The prominent peaks are labeled with corresponding polypeptide fragments of indicated amino acid residues (see text for details).
Wt and Δ7aa capsid protein dynamics assessed by MALDI-TOF
A very promising approach in distinguishing crystallographically identical virions is the application of limited proteolysis combined with peptide mapping and analysis by Matrix-Assisted laser Desorption/Ionization-Time Of Flight (MALDI-TOF) mass spectrometry (Bothner et al., 1999). In this assay, the site specific proteolytic degradation of a protein results in a set of digestion fragments which are subsequently mass analyzed by MALDI-TOF. The resulting digestion fragments provide structural information concerning the individual capsid proteins as well as their protein-protein interactions, because available cleavage sites are dependent on both the tertiary and quaternary protein structure. Cleavage sites residing on the exterior of the virus will be most accessible to the enzyme and therefore, be among the first digestion fragments observed. Because proteolysis is performed in solution and MALDI-TOF analysis can detect different protein conformational species, this method can contribute to an understanding of the dynamic domains within the virus structure (Bothner et al., 1999). Consequently we have applied this approach to determine whether virions of Δ7aa are structurally different from those of wt. Virion preparations of Δ7aa and wt BMV were treated with trypsin and the digested products were subjected to MALDI-TOF analysis.
MALDI-TOF of undigested wt BMV and Δ7aa CP virions revealed the expected mass-to-charge (m/z) values of 20,292 and 19,595, respectively (Fig. 7B). The 1:10 digestion of wt and Δ7aa CP produced slightly different cleavage patterns (1:100 same as 1:10, data not shown). As was observed for CCMV virions (Speir et al., 2006), the majority of the cleavages of wt and Δ7aa virions mapped to the N terminus of the CP. Four fragments (27-41, 27-44, 45-64, and 42-64) were present in wt yet absent in Δ7aa (Fig. 7C). These fragments were not released from Δ7aa due to the deletion of the two lysine residues at positions 42 and 45 in Δ7aa (Fig. 7C). The Δ7aa mutant also had a very prominent 27-64 fragment which also resulted from the loss of the two lysine residues at position 42 and 45 (Fig. 7C). Two additional fragments (65-86 and 87-103) found in Δ7aa were not observed in wt. Another fragment of 104-130 found in wt but not in Δ7aa could have resulted from a different folding pattern of the mutant CP exposing some additional arginine or lysine residues and not allowing access to others.
Fragment 104-130 starts in the DE loop on the particle exterior, and includes the E and F beta strands. It terminates on the FG loop, also on the exterior of the virus. For B and C subunits, this region is adjacent (4–5 angstroms distance) to a turn in the extended N-terminal (residues 36-38) just after the beta hexamer from a quasi 6-fold related subunit, and for all subunits is also near (5–7 angstroms) the last residue of the C-terminal from quasi 3-fold related subunits. Fragment 65-86 starts in the BC loop on the exterior, and includes the C-β strand and the CD helices that form the quasi 3-fold inter-subunit contacts. It terminates on the interior of the particle near the quasi 3-fold axis. Fragment 87-103 starts where the previous terminates, and includes the D-β strand. It terminates on the particle exterior in the DE loop. Residues 86-87 are on the particle interior next to the quasi 3-fold axis, and are adjacent (5.5–7 angstroms) to the C-terminal of the same subunit (175–176).
Based on the positions of these fragments, the deletion mutant may have propagated changes in structure to cause the different cleavage patterns. Each fragment is either next to a portion of the N-terminal, C-terminal or both, which have nearby regions that form the dimer clamp at the site of the deletion mutant. Therefore the extended termini and polypeptides near to them are most likely to be affected by the mutation. Residues 86-87 are on the particle interior but next to the quasi 3-fold axis, which has been show to open dramatically in the expanded CCMV particle. The deletion mutant has apparently made these residues more accessible to proteases compared to the wild-type particle probably through movement of the C-terminal and a more dynamic quasi 3-fold opening. Concurrently the same deletion mutant has somehow restricted access to the two highly exposed sites on the particle surface at residues 104 and 130. These residues have more pronounced contact with both extended termini structures that are dependent on the mutated region; thus, they may undergo sufficient conformational changes to prevent cleavage.
Δ7aa has no detectable effect on CCMV CP
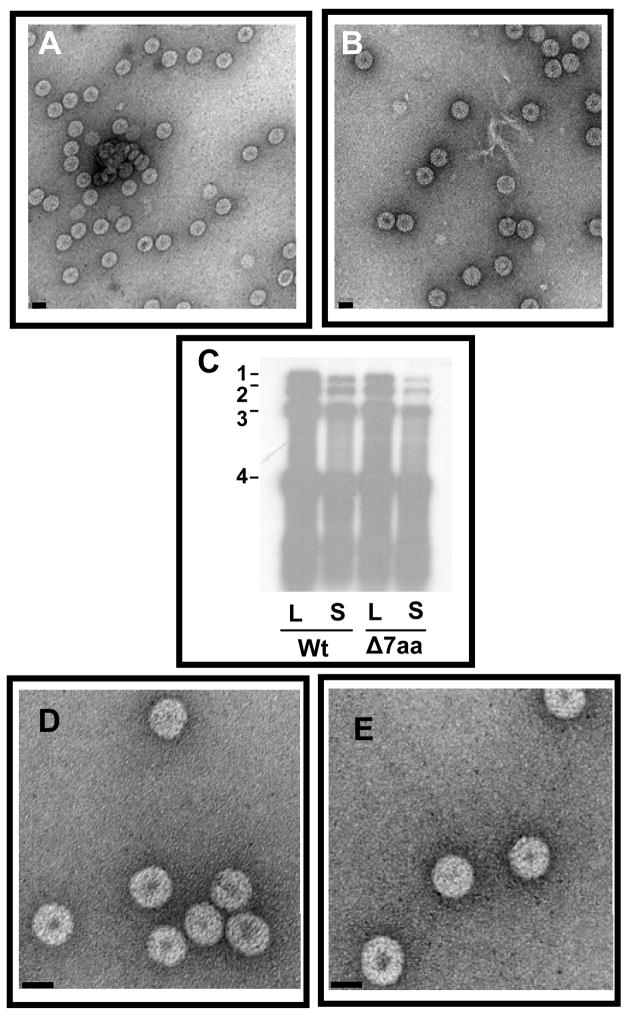
Since amino acids encompassing the first 50 N-terminal regions are highly conserved between BMV and CCMV, we were curious to verify the effect of Δ7aa mutation on CCMV CP with respect to RNA packaging in vivo and in vitro. Therefore, a variant of CCMV RNA 3 clone harboring the Δ7aa mutation was constructed and tested for its biological activity in cowpea plants by co-inoculating with wt counterparts. All cowpea plants inoculated with Δ7aa induced characteristic reddish local lesions followed by systemic chlorotic mottling on a time frame similar to control plants inoculated with wt CCMV (data not shown). Purified virions of CC3/Δ7aa were indistinguishable in morphology from wt CCMV (Fig. 8A and B) and Northern blot hybridization of CC3/Δ7aa virion RNA preparations exhibited wt profile (Fig. 8C). Unlike the CP subunits of BMV Δ7aa, those of CCMV were competent for reassembly in vitro (Fig. 8D and E). Taken together, these observations indicated that despite being structurally similar viruses, Δ7aa mutation can exert distinctly different effects.
Figure 8.
Characteristic features of CC3/Δ7aa. Electron micrograph images of purified virions of wt CCMV (A) and CC3/Δ7aa (B). Bar=50 nm. (C) Northern blot analysis of virion progeny. Virion RNA was isolated from local (L) and systemic (S) leaves of cowpea infected with indicated inoculum and analyzed by Northern hybridization with a riboprobe complementary to 3′TLS region of CCMV RNA. Conditions of hybridization are as described under Fig. 2 legend. The position of four CCMV RNAs is shown to the left. (D and E). In vitro reassembly. The electron micrographs shown in panel D and E represent the in vitro reassembled virion with CP subunits of wt and Δ7aa respectively. CP subunits of wt CCMV and variant Δ7aa were isolated and assembled in vitro with CCMV virion RNA. Bar = 20 nm.
DISCUSSION
We found that deletion of N-proximal 41–47 amino acids (Δ7aa) from the highly conserved putative RNA binding domain of BMV CP exhibited several unique features not observed previously with respect to virus assembly and RNA packaging in vivo and in vitro. These are (i) preferential packaging in vivo of truncated RNAs over full-length genomic RNAs; (ii) assembly of polymorphic virions in vivo, a feature that has not been observed when BMV CP is expressed in replication-dependent manner, and (iii) incompetence to form virions in vitro. In addition we also observed that CCMV CP harboring an identical 7 amino acid deletion did not exhibit any detectable effects on either virus assembly or RNA packaging.
Role of N-proximal region of bromovirus coat protein in virus assembly
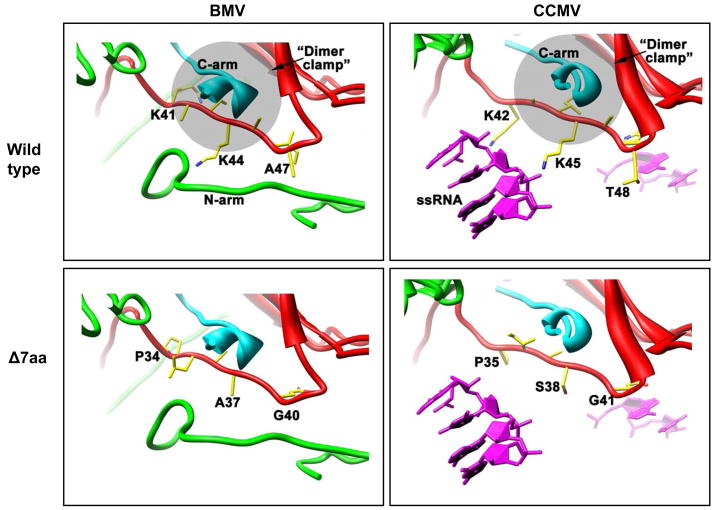
Previous studies characterized several CP variants of BMV and CCMV (Choi et al., 2000; Choi and Rao, 2000a; Rao and Grantham, 1995; Rao and Grantham, 1996; Zhao et al., 1995). Since BMV has been shown to move between plant cells only in the assembled form (Rao, 1997; Schmitz and Rao, 1996), CP variants that are assembly defective in vitro are also defective in vivo and remain non-infectious (Rao and Grantham, 1995; Rao and Grantham, 1996). Thus the most fascinating feature of the variant Δ7aa characterized in this study is its ability to assemble into virions in vivo (Fig. 1D) but not in vitro (Table 2). The region encompassing N-proximal amino acids 41–47 is similar in structure between CCMV and BMV, and forms a part of the interior section of the capsid protein shell (Fig. 9). In both viruses, this N-proximal region supports a critical contact (the “clamp”) with the C-termini of 2-fold related subunits that ties the capsomeres together (also hypothesized to be the solution dimer). Indeed, disorder of the subunit N-termini in the crystal structures never goes past about residue 40, emphasizing the structural importance of this region. In CCMV, two Lys 42/45, one Trp47 and one Thr48 (Fig. 9) appear to interact with RNA. The two Lys 42/45 point downward toward bulk RNA density and the phosphate backbones of the small ordered RNA segments beneath each subunit, but their side chains were partially disordered and therefore modeled as their most likely rotamer-the extended state. The Trp47and Thr48, both located on the turn between the first β-strand and the extended N-terminus, directly interact with the ordered RNA pieces. Thus, this region links RNA binding and critical quaternary structural elements in one small area.
Figure 9.
Structure of the BMV & CCMV T=3 particles at the Δ7aa mutation site. The view is tangential to the protein shells and along the A subunit (cyan) C-terminus toward a quasi-2fold related B subunit (red) “clamp” created by a wall of the β-barrel on the right, and the N-arm on the bottom and left. The same interaction also occurs between two C subunits related by icosahedral 2-fold symmetry (not shown). The bottom of each image is the interior of the shell. Neither the BMV or CCMV main chain structures were modified; the mutated side chains in the Δ7aa panels were placed in the same position as the wt residues they replace. Side chains are colored by atom type (carbon – yellow, nitrogen – blue, oxygen – red). The wild-type structures are closely similar at this position and share the reciprocal dimer clamping of 2-fold symmetry related C-arms. Deletion of the 7 residue segments (42KAIKAWT48 in CCMV; 41KAIKAIA47 in BMV) causes the shift of entirely different sequences (35PIASGQG41 in CCMV; 34PLAAGQG40 in BMV) that are normally polypeptide turns into the same region (Δ7aa panels). In particular, loss of the two lysines in each wild-type structure is not compensated for in the new sequence, and the additional potential flexibility may affect the clamp and dimer formation. The ordered RNA structure from CCMV is shown in magenta (each subunit has 2–4 bases of ssRNA underneath that stack with residue W47). Although no ordered RNA was seen in the BMV structure, the fully ordered N-termini of symmetry related BMV C subunits (green tubes running on bottom) occupies some of the same positions.
The two Lys 42/45 are conserved but Trp47 and Thr48 in CCMV are respectively substituted with Ile and Ala in BMV (Fig. 1A). Deletion of amino acid residues 41–47 would shift the sequence, but not necessarily disrupt the structure of this area (Fig. 9, Δ7aa). However, the resulting new sequence of this region (PLAAGQG) normally supports a turn in the extended N-terminus, and now is adjacent to the first turn of the N-terminus into the first β-strand of the subunit barrel. This area will now be more flexible and perhaps less structured, and will no longer have basic residues (Fig. 9, Δ7aa). This could affect the structure and dynamics of the N-termini and/or clamp necessary to package RNA and/or assemble particles competent for RNA packaging. It would certainly affect the turn that contacts the only ordered RNA segments found in the CCMV crystal structure. Our initial hypothesis assumed that BMV would have similar RNA contact points, given the structural identity between CCMV and BMV (Ding et al., 1995; Speir et al., 1995) and the lack of ordered RNA in the BMV crystal structure (Ding et al., 1995). Extrapolation of this predicted structural rearrangement to the observed defects in RNA packaging (Fig. 2) suggest that the region encompassing N-proximal amino acids 41–47 is critical for interacting with full length genomic RNAs. Virions recovered from plants inoculated with Δ7aa variant preferentially packaged truncated RNA species (Figs. 2 and 3). Altering this region did not completely block packaging of B2 or B3, but extensive fragmentation of these two RNA components suggest the engineered deletion weakens the interaction between CP and B2/B3. Consequently the weakened interaction could have rendered the capsid more susceptible to cellular nucleases resulting in extensive fragmentation.
Virion polymorphism
Virion polymorphism has been observed for icosahedral RNA viruses of plants (Lokesh et al., 2002; (Hsu et al., 2006)) and insects (Dong et al., 1998) when respective mutant coat proteins are expressed in vitro. Assembly of BMV CP into a 120 subunit capsids with T=1 symmetry was observed in vitro when the N-terminal 1–35 (Ding et al., 1995) or 1–63 amino acids (Cuillel et al., 1981) were lost due to protease activity. Likewise, formation in vivo of BMV virions with T=1 symmetry was observed only in the absence of homologous replication. For example, when the BMV CP was expressed via heterologous TMV based expression system in plants, assembly of both T=1 and T=3 virions was observed (Choi and Rao, 2000b). Similarly, autonomous expression of BMV CP mRNA in either a yeast based system (Krol et al., 1999) or in plants via agroinfiltration (Annamalai and Rao, 2005), also resulted in the formation of polymorphic forms. In these two instances, formation of polymorphic virions was hypothesized to be controlled by the RNA size. Interestingly, virion polymorphism was not observed with wt BMV infection even when four differently sized RNA species are synthesized during replication (Rao, 2006). For example, transient expression of BMV CP resulted in the formation of polymorphic virions of two distinct sizes packaging cellular RNA (Annamalai and Rao, 2005). However complementation with functional viral replicase not only abolished virion polymorphism but enhanced the specificity of packaging such that the mature virions exclusively contained viral progeny (Annamalai and Rao, 2005). Collectively this data suggests that an interaction between viral replicase and CP might play regulatory role to ensure that only one kind of virion population, T=3 particles, is assembled.
Prior to this work, numerous variants of BMV CP harboring either point mutations or deletions up to 18 amino acids have been expressed in vivo; matured virions never exhibited virion polymorphism (Choi et al., 2000; Choi and Rao, 2000a; Rao and Grantham, 1995; Rao and Grantham, 1996). Thus, variant Δ7aa represents the first example for BMV CP to display virion polymorphism in the presence of homologous replication. One likely explanation is that 41-47 amino acids from the N-terminus makes critical contact with the C-terminus to maintain the optimal “molecular switches” for precise protein-protein interaction required for the assembly of T=3 icosahedral virions. Thus, deletion of 41-47 amino acids disrupts the interaction between the N- and C- termini resulting in virion polymorphism. Therefore, the polymorphic phenotype exhibited by variant Δ7aa suggests that virion polymorphism can also be controlled by the CP, in addition to the RNA size. This hypothesis is further supported by the observed virion polymorphism in flock house virus (FHV), a non enveloped virus with a bipartite genome, when a region encompassing the N-terminal 31 amino acids was deleted from the FHV CP (Dong et al., 1998).
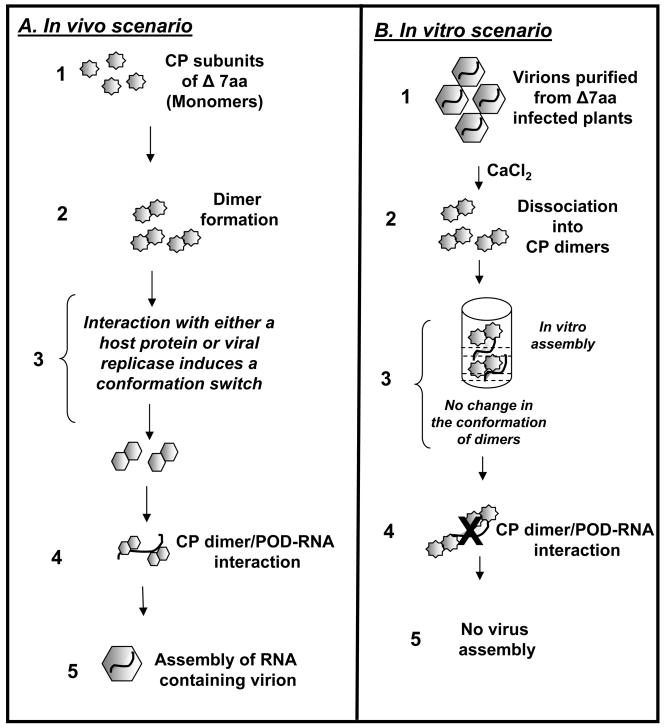
A chaperon mediated model for the assembly of variant Δ7aa in vivo
The assembly of bromovirus icosahedral virions involves three basic steps: dimerization of CP subunits, followed by the formation of pentamers of dimers, and finally cooperative addition of dimers to yield T=3 capsids (Zlotnick et al., 2000). In vitro assembly of RNA containing BMV virions suggests that no components other than CP subunits and RNA are required. In this respect variant Δ7aa is exceptional since it was competent to assemble in vivo but not in vitro (Table 2). The disruption of secondary structure by the engineered deletion as evidenced by the CD spectral analysis (Fig. 7A) explains why the CP subunits of Δ7aa failed to bind RNA in gel shift assays (Fig. 6). Then, the question that needs to be addressed is how did variant CP subunits interact with viral RNA and assemble into polymorphic virions in vivo? We offer a chaperon mediated model (Fig. 10) to explain this unusual in vivo assembly behavior of variant Δ7aa. As schematically shown Fig. 10, following the translation of the CP subunits of Δ7aa monomers (Step 1), we speculate that either a host factor or viral replicase functioning as chaperons interacts with CP dimers to restore the “molecular switch” and maintain optimal conformation permitting interaction with viral RNA and assembly into virions. This chaperon mediated conformational switch model explains why the CP subunits of variant Δ7aa are incompetent for assembly in vitro (Fig. 10B). Agroinfiltration experiments are in progress to identify whether in vivo packaging of Δ7aa is mediated through a host factor or viral replicase.
Figure 10.
Schematic illustration of chaperon mediated packaging model for BMV Δ7aa variant. (A) In vivo scenario. Following the translation of the CP subunits of Δ7aa dimerization occurs. We hypothesize that an interaction with either a host factor or viral replicase functioning as a chaperon induces a conformational switch to mutant CP dimers to promote RNA-CP dimer or pentamer of dimers (POD) interaction followed by completion of virion assembly. (B) In vitro scenario: In vitro dissociated capsids yield dimers in solution (Krol et al., 1999; Zhao et al., 1995). Absence of chaperon like molecules (host factor or viral replicase) failed to restore the optimal CP subunit conformation. This would block RNA-CP dimer or pentamer of dimers (POD) interaction and subsequent virion assembly.
Materials and Methods
Wild type and variant plasmids
Full-length cDNA clones corresponding to the three genomic RNAs of BMV, pT7B1, pT7B2 and pT7B3 from which infectious genomic RNAs 1 (B1), 2 (B2) and 3 (B3) can respectively be transcribed in vitro have been described previously (Dreher et al., 1989). CP variant Δ7aa (Fig. 1A) was constructed by deleting a sequence encompassing the 7 amino acids 41–47 of BMV CP ORF in the plasmid pT7B3 (Dreher et al., 1989) using polymerase chain reaction (PCR) with appropriate 5′ and 3′ oligonucleotide primers.
Agrotransformants and agroinfiltration
The construction and characterization of T-DNA based plasmids for efficient transient expression of full-length BMV genomic RNAs 1 (35S-B1), 2 (35S-B2) and 3 (35S-B3) have been described previously (Annamalai and Rao, 2005; Annamalai and Rao, 2006b). Each plasmid contains, in sequential order, a double 35S promoter, cDNA complementary to respective full-length BMV RNAs, a ribozyme sequence of satellite tobacco ring spot virus (Rz) and a 35S-terminator (Fig. 1B). The agroinfiltration procedure was performed as described (Annamalai and Rao, 2006a; Guo and Ding, 2002). Briefly, Agrobacterium strain EHA105 containing desired transformant was initially streaked on LB plate containing the antibiotics (kanamycin 50 μg/ml, rifampicin 10 μg/ml) and incubated at 28°C. A single colony was inoculated into a 2 ml LB medium with above antibiotics, and grown at 28°C for 48 hours with vigorous shaking. One ml of the culture was transferred to 50 ml LB medium containing the above antibiotics, 10 mM MES (pH 5.6) and 40 μM acetosyringone. After incubation at 28°C for 16 hours with vigorous shaking, OD600 of the culture was allowed to reach 1.0. The bacteria were spun down at 2000 ×g for 10 minutes, the pellet was resuspended in 50 ml 10 mM MgCl2, and then 125 μl 100 mM acetosyringone was added. To maintain a uniform concentration of Agrobacteriumi in co-infiltration experiments involving two or more Agrobacterium cultures, the OD600 of each culture was adjusted to 1.0. The final inoculum was prepared by mixing equal volumes of each culture. The bacteria were kept at room temperature for at least 3 hours without shaking. These cultures were then infiltrated into abaxial surface of the fully expanded N. benthamiana leaves using 1-cc syringe without a needle. pCASS4 (Annamalai and Rao, 2005) was used as an empty vector (EV) as well as to balance the inoculum concentration.
Progeny analysis and packaging assays
Total RNA from agroinfiltrated leaves were extracted using hot phenol method (Annamalai and Rao, 2007) and the RNA pellet was suspended in RNAse-free water. Virions were purified from agroinfiltrated leaves as described previously (Rao and Grantham, 1995). For Northern blot analysis (Annamalai and Rao, 2005) samples of virion RNA (0.5 μg) or plant total RNA (5 μg) were dried in a microfuge and suspended in 10 μl of sample buffer (10xMOPSbuffer/formamide/formaldehyde/H2O in a ratio of 1:1.8:5:2.2 respectively), heated at 65° for 10 min and electrophoresed in 1.2% agarose-formaldehyde gel (Sambrook and Russel, 2001). Following a 3 hr electrophoresis, fractionated RNA was transferred to a nylon membrane with a VacuGene XL blotting unit (Phramacia Biotech). The blot was then processed for pre-hybridization and hybridization using 32P-labeled riboprobes corresponding to either 3′ conserved tRNA-ilke structure (TLS) (Rao et al., 1989) or MP ORF or CP ORF or sequences specific for B1 or B2 as described previously (Choi et al., 2000). CP samples were analyzed by Western blots as described previously (Osman et al., 1997).
Coat protein preparation, in vitro assembly assays and electron microscopy
BMV virions were purified from wt infected leaves as described (Choi and Rao, 2000a; Choi and Rao, 2003). Preparation of CP subunits and in vitro assembly assays using either in buffer A (50 mM NaOAc, pH 4.8, 0.1M NaCl, 0.2mM PMSF) or buffer B (50 mM Tris-HCl, pH 7.2, 50 mM NaCl, 10 mM KCl, 5 mM MgCl2, and 1 mM DTT) was as described previously (Choi and Rao, 2000a; Zhao et al., 1995). Each assembly reaction (100 μl) contained 14 pmoles of genomic RNA and 3 μg of purified BMV CP subunits. Following purification of assembled virions by Centricon-100, each in vitro assembled virion preparation was spread on glow-discharged grids and negatively stained with 1% uranyl acetate prior to examining with a FEI Tecnai12 transmission electron microscope (Annamalai et al., 2005). Images were recorded digitally and arranged using Photoshop. For estimating the percentage of virions other than the native form, an area measuring 7.5 μm2 on each electron micrographic image was visually count the different-size virions (Choi and Rao, 2000b).
RNase sensitivity of purified virions
Purified virions preparations of wt BMV and Δ7aa in virus suspension buffer were treated with 1μg/μl of RNase A for 30 min at 30°C. Following RNase treatment each preparation was divided into two lots. One lot was subjected to EM examination and the other lot was used to extract viral RNA and subjected to Northern blot hybridization.
Gel retardation assays
The interaction between varying molar ratios of either wt or mutant CP dimers and desired genomic RNA component was analyzed as described previously (Johnson et al., 2004). Approximately 0–150 CP dimers were titrated against a constant RNA concentration in a typical 20 μl reaction containing 50mM MOPS (pH 7.2), 150 mM NaCl and 2 mM MgCl2. The reaction samples were then subjected to 1% agarose gel electrophoresis in TAE buffer (Sambrook and Russel, 2001), stained with ethidium bromide and photographed with BioRad Gel documentation system. In these assays, a sample of native wt BMV virions purified from symptomatic leaves was always co-electrophoresed as a control.
CD Spectral analysis
Virus was dissociated into CP subunits and the RNA was removed by performing dialysis against in vitro assembly buffer (Choi et al., 2002). The CP was diluted to .5μg/μl in BMV assembly buffer. 300μl of sample was used in the cuvette. BMV assembly was used as a blank. Samples were read 8 times. The 8 reading were then standardized (combined) and the baseline (buffer) was subtracted out. The following parameters were used: Bandwidth- 1.0nm; Slit width-auto; Time constant− 2.0 seconds; Near UV readings − 200–260nm.
MALDI-TOF
Virus was diluted to 1μg/μl in 10mM Tris-Hcl, 1mM EDTA buffer. Samples were treated at 1:10 and 1:100 ratios of trypsin:virus. Trypsin-GOLD Mass Spectrometry grade 1μg/μl in 50mM acetic acid was obtained from Promega. Trypsin digests were incubated for 15 minutes at 25°C. 5μl of digest samples (and 5μl of untreated virus) were put into speed vac ~10 minutes until lyophilized-these samples were taken to Chemistry for MALDI analysis.
Acknowledgments
The authors thank Cheng Kao, James Ig, Padmanaban Annamalai and Melissanne de Wispelaere for helpful discussions and Surekha Acharya for editing the manuscript. This study was supported by a grant GM 064465-01A2 from the National Institutes of Health (A.L.N.R) and Public Health Service grant GM54076 from the National Institutes of Health (J.E.J).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Annamalai P, Apte S, Wilkens S, Rao AL. Deletion of highly conserved arginine-rich RNA binding motif in cowpea chlorotic mottle virus capsid protein results in virion structural alterations and RNA packaging constraints. J Virol. 2005;79:3277–88. doi: 10.1128/JVI.79.6.3277-3288.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annamalai P, Rao AL. Replication-independent expression of genome components and capsid protein of brome mosaic virus in planta: a functional role for viral replicase in RNA packaging. Virology. 2005;338:96–111. doi: 10.1016/j.virol.2005.05.013. [DOI] [PubMed] [Google Scholar]
- Annamalai P, Rao AL. Delivery and Expression of functional viral RNA genomes in planta by agroinfiltration. In: Downey T, editor. Current Protocols in Microbiology. 1. 16B. John Wiley & Sons Inc; 2006a. pp. 2.1–2.15. [DOI] [PubMed] [Google Scholar]
- Annamalai P, Rao AL. Packaging of brome mosaic virus subgenomic RNA is functionally coupled to replication-dependent transcription and translation of coat protein. Journal of Virology. 2006b;80:10096–10108. doi: 10.1128/JVI.01186-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annamalai P, Rao AL. In Vivo Packaging of Brome Mosaic Virus RNA3, but Not RNAs 1 and 2, Is Dependent on a cis-Acting 3′ tRNA-Like Structure. J Virol. 2007;81:173–81. doi: 10.1128/JVI.01500-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancroft JB. The self-assembly of spherical plant viruses. Adv Virus Res. 1970;16:99–134. doi: 10.1016/s0065-3527(08)60022-6. [DOI] [PubMed] [Google Scholar]
- Bancroft JB, Bracker CE, Wagner GW. Structures derived from cowpea chlorotic mottle and brome mosaic virus protein. Virology. 1969;38:324–35. doi: 10.1016/0042-6822(69)90374-2. [DOI] [PubMed] [Google Scholar]
- Bothner B, Schneemann A, Marshall D, Reddy V, Johnson JE, Siuzdak G. Crystallographically identical virus capsids display different properties in solution. Nat Struct Biol. 1999;6:114–6. doi: 10.1038/5799. [DOI] [PubMed] [Google Scholar]
- Choi SK, Yoon JY, Ryu KH, Choi JK, Palukaitis P, Park WM. Systemic movement of a movement-deficient strain of Cucumber mosaic virus in zucchini squash is facilitated by a cucurbit-infecting potyvirus. J Gen Virol. 2002;83:3173–8. doi: 10.1099/0022-1317-83-12-3173. [DOI] [PubMed] [Google Scholar]
- Choi YG, Grantham GL, Rao AL. Molecular studies on bromovirus capsid protein. Virology. 2000;270:377–85. doi: 10.1006/viro.2000.0312. [DOI] [PubMed] [Google Scholar]
- Choi YG, Rao AL. Molecular studies on bromovirus capsid protein. VII. Selective packaging on BMV RNA4 by specific N-terminal arginine residuals. Virology. 2000a;275:207–17. doi: 10.1006/viro.2000.0513. [DOI] [PubMed] [Google Scholar]
- Choi YG, Rao AL. Packaging of tobacco mosaic virus subgenomic RNAs by Brome mosaic virus coat protein exhibits RNA controlled polymorphism. Virology. 2000b;275:249–57. doi: 10.1006/viro.2000.0532. [DOI] [PubMed] [Google Scholar]
- Choi YG, Rao AL. Packaging of brome mosaic virus RNA3 is mediated through a bipartite signal. J Virol. 2003;77:9750–7. doi: 10.1128/JVI.77.18.9750-9757.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuillel M, Jacrot B, Zulauf M. A T=1 capsid formed by the protein of brome mosaic virus in the presnce of trypsin. Virology. 1981;110:63–72. doi: 10.1016/0042-6822(81)90008-8. [DOI] [PubMed] [Google Scholar]
- Ding B, Li Q, Nguyen L, Palukaitis P, Lucas WJ. Cucumber mosaic virus 3a protein potentiates cell-to-cell trafficking of CMV RNA in tobacco plants. Virology. 1995;207:345–53. doi: 10.1006/viro.1995.1093. [DOI] [PubMed] [Google Scholar]
- Dong XF, Natarajan P, Tihova M, Johnson JE, Schneemann A. Particle polymorphism caused by deletion of a peptide molecular switch in a quasiequivalent icosahedral virus. J Virol. 1998;72:6024–33. doi: 10.1128/jvi.72.7.6024-6033.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher TW, Rao AL, Hall TC. Replication in vivo of mutant brome mosaic virus RNAs defective in aminoacylation. J Mol Biol. 1989;206:425–38. doi: 10.1016/0022-2836(89)90491-9. [DOI] [PubMed] [Google Scholar]
- Guo HS, Ding SW. A viral protein inhibits the long range signaling activity of the gene silencing signal. Embo J. 2002;21:398–407. doi: 10.1093/emboj/21.3.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C, Singh P, Ochoa W, Manayani DJ, Manchester M, Schneemann A, Reddy VS. Characterization of polymorphism displayed by the coat protein mutants of tomato bushy stunt virus. Virology. 2006;349:222–9. doi: 10.1016/j.virol.2006.02.038. [DOI] [PubMed] [Google Scholar]
- Johnson JM, Willits DA, Young MJ, Zlotnick A. Interaction with capsid protein alters RNA structure and the pathway for in vitro assembly of cowpea chlorotic mottle virus. J Mol Biol. 2004;335:455–64. doi: 10.1016/j.jmb.2003.10.059. [DOI] [PubMed] [Google Scholar]
- Kao CC, Quadt R, Hershberger RP, Ahlquist P. Brome mosaic virus RNA replication proteins 1a and 2a from a complex in vitro. J Virol. 1992;66:6322–9. doi: 10.1128/jvi.66.11.6322-6329.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol MA, Olson NH, Tate J, Johnson JE, Baker TS, Ahlquist P. RNA-controlled polymorphism in the in vivo assembly of 180-subunit and 120-subunit virions from a single capsid protein. Proc Natl Acad Sci U S A. 1999;96:13650–5. doi: 10.1073/pnas.96.24.13650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokesh GL, Gowri TD, Satheshkumar PS, Murthy MR, Savithri HS. A molecular switch in the capsid protein controls the particle polymorphism in an icosahedral virus. Virology. 2002;292:211–23. doi: 10.1006/viro.2001.1242. [DOI] [PubMed] [Google Scholar]
- Miller WA, Dreher TW, Hall TC. Synthesis of brome mosaic virus subgenomic RNA in vitro by internal initiation on (−)-sense genomic RNA. Nature. 1985;313:68–70. doi: 10.1038/313068a0. [DOI] [PubMed] [Google Scholar]
- Osman F, Grantham GL, Rao AL. Molecular studies on bromovirus capsid protein. IV. Coat protein exchanges between brome mosaic and cowpea chlorotic mottle viruses exhibit neutral effects in heterologous hosts. Virology. 1997;238:452–9. doi: 10.1006/viro.1997.8849. [DOI] [PubMed] [Google Scholar]
- Rao AL. Molecular studies on bromovirus capsid protein. III. Analysis of cell-to-cell movement competence of coat protein defective variants of cowpea chlorotic mottle virus. Virology. 1997;232:385–95. doi: 10.1006/viro.1997.8579. [DOI] [PubMed] [Google Scholar]
- Rao AL. Bromoviruses. In: Murry OCMaTD., editor. Encyclopedia of Plant Pathology. John Wiley & Sons; Mississauga, Ontario, Canada: 2001. pp. 155–158. [Google Scholar]
- Rao AL. Genome Packaging by Spherical Plant RNA Viruses. Annu Rev Phytopathol. 2006;44:61–87. doi: 10.1146/annurev.phyto.44.070505.143334. [DOI] [PubMed] [Google Scholar]
- Rao AL, Dreher TW, Marsh LE, Hall TC. Telomeric function of the tRNA-like structure of brome mosaic virus RNA. Proc Natl Acad Sci U S A. 1989;86:5335–9. doi: 10.1073/pnas.86.14.5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao AL, Grantham GL. Biological significance of the seven amino-terminal basic residues of brome mosaic virus coat protein. Virology. 1995;211:42–52. doi: 10.1006/viro.1995.1377. [DOI] [PubMed] [Google Scholar]
- Rao AL, Grantham GL. Molecular studies on bromovirus capsid protein. II. Functional analysis of the amino-terminal arginine-rich motif and its role in encapsidation, movement, and pathology. Virology. 1996;226:294–305. doi: 10.1006/viro.1996.0657. [DOI] [PubMed] [Google Scholar]
- Sacher R, Ahlquist P. Effects of deletions in the N-terminal basic arm of brome mosaic virus coat protein on RNA packaging and systemic infection. J Virol. 1989;63:4545–52. doi: 10.1128/jvi.63.11.4545-4552.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russel DL. Molecular Cloning: A Laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- Schmitz I, Rao AL. Molecular studies on bromovirus capsid protein. I. Characterization of cell-to-cell movement-defective RNA3 variants of brome mosaic virus. Virology. 1996;226:281–93. doi: 10.1006/viro.1996.0656. [DOI] [PubMed] [Google Scholar]
- Speir JA, Bothner B, Qu C, Willits DA, Young MJ, Johnson JE. Enhanced local symmetry interactions globally stabilize a mutant virus capsid that maintains infectivity and capsid dynamics. J Virol. 2006;80:3582–91. doi: 10.1128/JVI.80.7.3582-3591.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speir JA, Munshi S, Wang G, Baker TS, Johnson JE. Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryo-electron microscopy. Structure. 1995;3:63–78. doi: 10.1016/s0969-2126(01)00135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Fox JM, Olson NH, Baker TS, Young MJ. In vitro assembly of cowpea chlorotic mottle virus from coat protein expressed in Escherichia coli and in vitro-transcribed viral cDNA. Virology. 1995;207:486–94. doi: 10.1006/viro.1995.1108. [DOI] [PubMed] [Google Scholar]
- Zlotnick A, Aldrich R, Johnson JM, Ceres P, Young MJ. Mechanism of capsid assembly for an icosahedral plant virus. Virology. 2000;277:450–6. doi: 10.1006/viro.2000.0619. [DOI] [PubMed] [Google Scholar]