Abstract
Yeast surface display has become an increasingly popular tool for protein engineering and library screening applications. Recent advances have greatly expanded the capability of yeast surface display, and are highlighted by cell-based selections, epitope mapping, cDNA library screening, and cell adhesion engineering. In this review, we discuss the state-of-the-art yeast display methodologies and the rapidly expanding set of applications afforded by this technology.
Keywords: Yeast surface display, protein engineering, library screening
Introduction
The expression of recombinant proteins incorporated into the cell wall of Saccharomyces cerevisiae, termed yeast surface display (YSD), has now been practiced for over a decade with noteworthy success. YSD offers selected advantages relative to other display technologies, most notably eukaryotic expression of the heterologous target protein, making it an outstanding tool for display and engineering of many proteins that are difficult to produce in other display formats. In the past several years, YSD has expanded beyond its original applications of immobilizing recombinant enzymes and affinity maturating single-chain Fv antibody fragments (scFvs). Below we discuss the attributes of YSD technology with a particular focus on novel YSD techniques and their application to new protein targets.
Yeast surface display systems
In the first section, we describe the various technological aspects that need to be considered when selecting a YSD platform, including the various anchors and yeast that can be used for surface display, effects of protein display orientation, and methods for library generation.
Anchors for display on Saccharomyces cerevisiae
Yeast surface display was first demonstrated as a method to immobilize enzymes and pathogen-derived proteins for vaccine development. The α-galactosidase gene from Cyamopsis tetragonoloba was fused to the C terminal half of α-agglutinin, a cell wall anchored mating protein in S. cerevisiae1 (Table 1). Increased stability was seen for the enzyme when linked to the cell wall, compared with direct secretion of the full α-galactosidase enzyme into the media. Early work also used the flocculin Flo1p as an anchor to attach α-galactosidase to the cell wall, with similar results2. Both α-agglutinin and flocculin, along with cell wall proteins such as Cwp1p, Cwp2p, Tip1p, and others, belong to the glycosylphosphatidylinositol (GPI) family of cell wall proteins that can be used directly for display3. These proteins are directed to the plasma membrane via GPI anchors and subsequently are linked directly to the cell wall through a β-1,6-glucan bridge for incorporation into the mannoprotein layer3. These large intact proteins as well as their C-terminal fragments have been demonstrated to mediate display of a range of heterologous proteins upon protein fusion. If instead, noncovalent display is desired, a Flo1p fragment has been used as the anchor protein. The protein of interest can be fused C-terminal to the flocculation functional domain of Flo1p, which is thought to interact with cell wall mannan chains to form a noncovalent adhesion3. Display of proteins on S. cerevisiae via these anchors has been reviewed recently by Ueda and colleagues3.
Table 1.
Selected proteins of interest engineered using yeast surface display
| Protein | Anchor | Anchor position* | Significance/functionality evolved |
|---|---|---|---|
| α-galactosidase1 | α-agglutinin (C-terminal half) | C-term | First protein displayed on yeast, increased stability over soluble enzyme |
| Human UDP-glucose receptor12 | Native mammalian GPCR | N/A | Expression of native mammalian protein used for YSD, directed evolution of GPCR to alter ligand specificity |
| R. oryzae lipase (ROL)15 | Flo1p, flocculation functional domain | N-term | Expression of ROL in P. pastoris achieved higher cell density and increased thermal stability over expression in S. cerevisiae |
| Anti-CD3ε scFvs17 | Aga2p | N-term | Showed increased affinity of scFvs to for ligand over C-terminal fusion |
| Anti-streptavidin Fab21 | Aga2p | N-term | Demonstrated the ability to assemble oligomeric proteins by interchain disulfide bond |
| scFv libraries24, 25, 28 | Aga2p | N-term | Non-immune, semi-immune, and immune libraries used to select novel scFvs binding target ligand |
| EGF/EGFR fragments30, 57, 58 | Aga2p | N-term | Antibody epitope mapping of EGFR and affinity maturation of EGF against EGFR |
| West Nile virus viral envelope protein ectodomain59 | Aga2p | N-term | Epitope mapping of a neutralizing antibody |
| IL-2 49–51 | Aga2p | N-term | Higher affinity for receptor, important implications for IL-2 therapeutics |
| 4-4-20 scFv4, 5, 72 | Aga2p | N-term | Affinity maturation, first use of YSD for library screening, identification of yeast proteins that increase heterologous protein expression |
| T cell receptors41–44 | Aga2p | N-term | Evolution of single chain TCRs for higher affinity, advances in understanding of relationship between expression levels and protein stability |
| αLβ2 I domain47, 81 | Aga2p | C-term | Understanding structure/function relationship, increased binding affinity for ICAM-1 |
| Human cDNA library63, 64 | Aga2p | N-term | Identification of human proteins that are tumor related/bind to phosphorylated peptides |
| Metal chelating peptides76 | α-agglutinin (C-terminal half) | C-term | Capture copper ions on yeast surface |
N-term and C-term corresponds to the constructs shown in Figure 1 A and 1 B, respectively
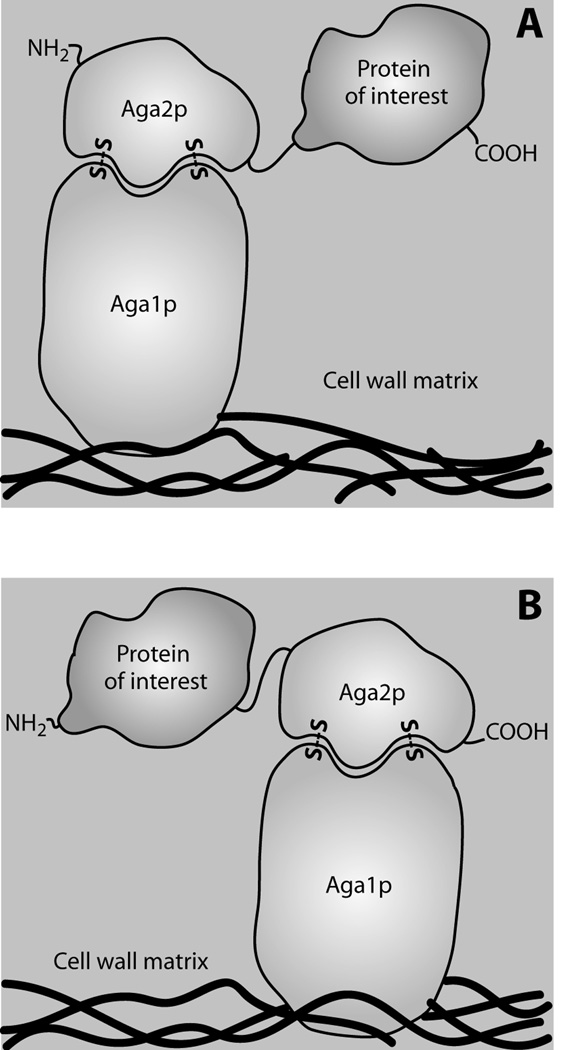
The a-agglutinin system developed by Wittrup et al 4,5 uses Aga2p as the display fusion partner. A disulfide linkage between Aga1p, a GPI/β-1,6-glucan-anchored protein, and Aga2p anchors the protein to the cell wall. Thus, coexpression of Aga1p with an Aga2p fusion leads to cell wall-anchored protein on the surface of yeast via disulfide bonding (Figure 1). The majority of applications of YSD discussed here utilize the Aga2p anchor system, and in the following discussion it is therefore assumed that the Aga2p system was used unless otherwise stated.
Figure 1.
Mode of Aga2p linkage may affect expression and function of yeast surface displayed target proteins. (A) Expression construct that fuses the protein of interest by its N-terminus to the C-terminus of Aga2p, via a flexible linker. Extracellular secretion is thus directed by the native Aga2p signal peptide. (B) Construct that fuses the protein of interest by its C-terminus to the N-terminus of Aga2p. A signal peptide must therefore be included at the N-terminus of this construct to direct secretion. In both construct formats, Aga2p is bound to the Aga1p subunit, which is in turn covalently linked to cell wall glucans83. A similar cell wall linkage is used by most of the alternate anchoring fusions discussed herein. In both constructs, the protein of interest is typically flanked on its N- and C-terminus by epitope tags. The protein of interest can be oligomeric, in which case one subunit is expressed as an Aga2p fusion while others require a signal peptide to direct them to the secretory pathway for assembly.
The Pir (proteins with internal repeats) family of cell wall proteins from S. cerevisiae has more recently been exploited as a fusion protein for display because of its alternate linkage capabilities. Unlike the GPI-cell wall anchored proteins, the Pir family of proteins (Pir1-4) are attached to the cell wall through a previously unknown linkage6. Recent work suggests that the modification of a glutamic acid residue, originally encoded as a glutamine residue, with a pentahexose chain results in a novel linkage with β-1,3-glucan in the cell wall through an ester bond7. The Pir family has been used to display several enzymes, such as a-1,2-glycosyltransferase6, xylanase A8, and others9,10.
Yeast cells can also be used to engineer membrane-targeted proteins such as mammalian G protein-coupled receptors (GPCRs) without using anchors for surface localization. Because yeast pheromone receptors are GPCRs, the native GPCR pathway can be engineered to respond to the mammalian GPCR ligand through the yeast signaling pathway11. This system was used to create a novel chemical sensing yeast strain expressing an engineered human UDP-glucose receptor, a GPCR, with distinct but overlapping specificities12 (Table 1).
Display in methylotrophic strains
In previous work13–15, the yeast surface display platform has been extended to strains that can utilize methanol as their sole carbon and energy sources. Compared to the widely used yeast S. cerevisiae, these methylotrophic strains, including Pichia pastoris and Hansenula polymorpha, have superior fermentation characteristics such as growth on an economical carbon source and very high cell density culture. Methylotrophic strains are therefore well suited to applications that require large-scale fermentations, such as the generation of whole-cell biocatalysts16. The concept of whole-cell biocatalysts has been demonstrated by surface display of Rhizopus oryzae lipase in P. pastoris15 using the Flo1p anchor (Table 1) and by the display of the C-terminus of carboxymethylcellulase in H. polymorpha13 using the Cwp1p anchor. In the surface display of Rhizopus oryzae lipase in P. pastoris, it was found that the enzyme shows higher thermal stability than the soluble form15. A similar increase in thermal stability was also observed in lipase displayed on S. cerevisiae but to a lower extent15. It was reasoned that high levels of glycosylation, especially an increased degree of O-glycosylation in P. pastoris compared to S. cerevisiae, yielded the increased stability of the enzyme15. Therefore, it appears that the higher glycosylation level of P. pastoris could be leveraged to improve enzyme-based fermentations through stabilization.
Protein display orientation
For some proteins, the ability to control whether the N- or C-terminus is linked to the anchor protein can improve display or functional properties (Figure 1). For example, the flexibility of the Aga2p protein allows one to fuse proteins to either end, and this ability has proven important for successful display of some proteins. Two anti-CD3ε scFvs exhibited 30–100-fold reduced affinity for the target protein when expressed as a fusion to the C-terminus of Aga2p17 (Table 1, Figure 1A). However, when expressed as a fusion to the N-terminus of Aga2p, the scFvs' binding affinities were restored (Figure 1B). Class II major histocompatibility complex (MHC), discussed below, can be expressed as a non-covalent heterodimer or as a single-chain species. When the C-terminal Aga2p β chain fusion was co-expressed with soluble MHC α chain, little reactivity was seen with conformationally specific antibodies recognizing either the α or β chains18. Therefore, to achieve display as a C-terminal fusion, directed evolution was required19,20. However, efficient display of class II MHC was observed when the β chain was expressed as a fusion to the N-terminus of Aga2p, similar to its native topography as a type I transmembrane protein18. Thus, the fusion topography appears in some cases to be critical to the ability to successfully express and display functional proteins.
Recent work has also shown that in addition to monomeric proteins, yeast can be used to display homo and heterodimeric proteins. In these cases, the eukaryotic yeast secretory machinery allows oligomeric mammalian proteins to assemble and export to the surface in a native-like conformation. A single vector encoding the heavy (HC) and light chains (LC) of an anti-steptavidin Fab was created with a HC-C-terminal Aga2p fusion and soluble LC under separate but identical GAL1 promoters21 (Table 1). HC and LC chains assemble and are stabilized through the native covalent interchain disulfide bond, and hence display as a full Fab fragment. Subsequently, affinity maturation was performed on the anti-SA Fab through error-prone PCR and successive rounds of fluorescence-activated cell sorting (FACS). Similarly, a catalytic Fab was displayed on the yeast surface and was shown to fully assemble and catalyze the formation of chloramphenicol from a chloramphenicol monoester derivative22. As the first example of noncovalent heterodimer expression, properly folded class II MHC was displayed using a system that coexpresses MHC α chain and MHC β chain-Aga2p fusion from a dicistronic plasmid employing the bidirectional GAL1-10 promoter18. As another example, homooligomeric streptavidin was expressed using two vectors, one encoding soluble streptavidin and the other encoding streptavidin fused to the C-terminus of Flo1p. While yeast expressing the anchored-streptavidin subunit alone showed low binding affinity to biotinylated fluorescein isothiocyanate (FITC), the construct expressing both soluble and Flo1p anchored streptavidin showed significant biotin-FITC binding23. These studies demonstrate the utility of yeast display for expression of a range of complex, oligomeric proteins.
Methods for library generation
A major application of YSD is expression and screening of combinatorial libraries. Beyond standard PCR-based methods for generating diversity, a number of other methods have been used to create diverse protein libraries that can be screened by surface display. As one example, scFvs against a number of different small molecule and protein targets were isolated from a 109 member nonimmune scFv library comprised of assembled heavy and light chain genes mimicking the natural germline diversity found in human B-lymphocytes24 (Table 1). If instead, it is desired to bias a library toward a particular antigen, pseudo-immune and immune libraries of scFv can be created. Cloned cDNA from healthy donors showing higher than normal antibody titer against the death receptor 5 (DR5) antigen were used to create a pseudo-immune library25. Screening both the smaller pseudo-immune library of 2 × 106 diversity and the aforementioned nonimmune library against DR5 showed that it was possible to isolate specific high-affinity clones from the pseudo-immune library, despite the significantly reduced size of the library. Library diversity can also be enhanced by taking advantage of the ability of haploid yeast to efficiently mate leading to the formation of a diploid organism26. Mutagenized subunits of dimeric proteins can therefore be cloned into yeast shuttle vectors with different selectable markers, transformed separately into stable a and α mating type haploid strains, and these strains can be mated to combine the two genetic sublibraries. Hufton and colleagues first demonstrated this approach by mating a and α yeast separately expressing heavy and light chains of Fab fragments to create a combined Fab library of over 109 clones. Screening this library led to isolation of Fab with high affinity (Kd = 6 nM) toward the target streptavidin antigen26. Similarly, by using peripheral blood lymphocyte RNA from a human patient immunized with pentavalent botulinum toxoid, separate immune libraries of 3 × 106 VH and 5 × 105 VK genes, both biased toward the target antigen, were created27. Subsequent mating of these libraries led to the creation of a combined VH/Vκ library consisting of 3 × 109 unique members enabling the isolation of sub-nanomolar Fabs against botulinum toxins27. In addition, antibody genes cloned from a patient with high HIV-1 neutralizing Ab titers were used to create a library of 107 scFv clones28, and many novel anti-HIV antibodies were isolated from this library. Importantly, this study provided a direct comparison between phage and yeast display library selection efficiencies, by cloning the same HIV-1 immune scFv library into both phage and yeast display formats28. Here, yeast display sampled the immune antibody repertoire considerably more fully than phage display, shown by the selection of twice as many novel antibodies, including all the scFv identified by phage display28.
The ability of yeast to efficiently recombine homologous DNA sequences can also be exploited to increase the diversity of a library. When two scFv genes that share 89.8% homology were mutated by PCR and co-transformed into yeast, a chimeric antibody library of 107 diversity was created through in vivo homologous recombination, showing evenly distributed crossover points throughout the two genes29.
A recent trend in library generation is to not only rely on random mutations, but also focus the library diversity to specific residues that are expected to dictate protein functions, thereby increasing the ability to probe more of the sequence space at functionally important positions. For example, it was noted in engineered mutants of epidermal growth factor (EGF), interleukin-2, and the EGF receptor ectodomain that isolated high affinity mutants were biased toward orthologous substitutions, or in other words, substitutions found in homologous proteins from other species30 (Table 1). Motivated by this observation, a library generation strategy to create artificial orthologous mutants, termed shotgun ortholog scanning mutagenesis (SOSM) was developed30. This method was used to isolate an epidermal growth factor (EGF) mutant with 30-fold increase in affinity for the receptor30.
Antibody fragments represent another target for focused libraries, since the antigen-antibody interaction is largely determined by residues in the complementarity determining region (CDR). Rajpal et al. generated a library of anti-TNF-α antibody D2E7 by replacing each residue of the six CDRs with one of nine amino acids that were expected to have representative chemical functionalities based on the side-chain functional groups31 These mutations were then combined to create a library with single mutations in one to three CDRs. Using this approach, mutants with 870-fold higher affinity were isolated from a relatively small library of 1.4 × 106 diversity31. It is anticipated that protein sequence analysis and modeling tools will continue to be leveraged for the generation of designer libraries, helping to reduce the sequence space to an experimentally amenable size.
Recent Applications of Yeast Surface Display
Various YSD technologies discussed in the previous section have greatly expanded the applications of YSD during the past few years. In this section, we summarize the recent advances in conventional applications such as affinity maturation, protein engineering for improved production and stability, as well as novel applications in cell-based selections, epitope mapping, cDNA library screening, cell adhesion molecule engineering, and selections against non-biological targets.
Affinity maturation
Protein affinity maturation has been one of the most successful applications of YSD. Initial studies led by Wittrup et al. used an anti-fluorescein scFv to show the effectiveness of YSD in protein affinity maturation4 (Table 1). Since each yeast cell is capable of displaying 104 – 105 scFv4, fluorescence from each cell can be readily detected and accurately quantified by flow cytometry. This feature of YSD allows not only precise and highly reproducible affinity measurement32, but also rapid enrichment of high-affinity populations within mutant libraries4,33,34. Mutant libraries are often screened under conditions where the binding interaction has reached equilibrium with a limiting concentration of soluble antigen to select mutants having higher affinity. However, kinetic selection methods have also been successfully applied to dramatically decrease the binding off-rate, and are desirable under conditions where the binding interaction is characterized by a dissociation rate constant lower than 10 nM34. In this way, an antibody with affinity among the highest reported so far (Kd = 48 fM)33 and dissociation half-time of seven days at 37 °C has been evolved35. A detailed methodological discussion on antibody affinity maturation and library selection using YSD can be found in Chao et al.36.
To date, antibodies against streptavidin21, carcinoembryonic antigen35, TNF-α31, botulinum neurotoxins37, lysozyme38, and CD3 diphtheria toxin39 have been engineered for higher affinity, for the purposes of effective tumor targeting, increased antagonistic activity, and sensitive toxin detection. A recent study showed that a single antibody can be engineered to have high affinity against two different but related subtypes of botulinum neurotoxin type A, that differ by seven amino acids in the structural epitope40. In this study, a highly targeted mutagenesis strategy was used to increase the affinity against one subtype by 1,250-fold while retaining the affinity against the other. This significant increase in cross-reactivity was reasoned to be due to specific mutations in the antigen-binding loops that enhance binding to one subtype while not affecting the binding of the second subtype. Therefore, this strategy could be generally applied to broaden antibody reactivity or conversely to better understand the determinants of antibody specificity.
Another major group of proteins that have been affinity engineered using YSD are T cell receptors (TCRs). Analogous to the scFv antibody construct, single-chain TCRs have been created, and directed evolution has resulted in more than 100-fold increases in affinity41, serving an important role in elucidating the origins of peptide specificity of TCRs and the biological interactions with antigen-presenting cells42–44 (Table 1). More recently, the Vβ region of TCR has been engineered for high affinity against toxic shock syndrome toxin-145 and staphylococcal enterotoxin B (SEB)46. A high affinity mutant of Vβ8 (Kd = 48 pM) with an approximately 3-million-fold increase in affinity against SEB inhibited SEB-mediated T-cell activation and completely neutralized SEB toxicity in animal models46.
Other examples of affinity maturation using YSD include integrin I domain47, epidermal growth factor (EGF)30, natural killer cell receptor48 and interleukin-2 (IL-2)49–51. In the affinity maturation of integrin I domain, the position of mutations that increase the affinity by 200,000-fold were located at key residues that transmit allosteric transitions, thereby providing structural insights on conformation-dependent binding to its ligand47 (Table 1). In the case of IL-2, high affinity binding of the IL-2 receptor complex was necessary since the therapeutic effect of IL-2 was greatly challenged by the rapid clearance of IL-2 after administration. By increasing the affinity of IL-2 by 15 to 30-fold, increased activity for T-cell proliferation was detected, which could improve the potency of IL-2 therapeutics49.
Cell-based selections
It was previously found that yeast cells displaying high-affinity single-chain T cell receptors (scTCRs) can form a cell-cell complex with an antigen-presenting cell52. In addition, effective screening of yeast-displayed antibody libraries against surface-immobilized ligands has been demonstrated using a magnetic bead capture method53. These results suggested that the YSD might be applied to screening yeast polypeptide libraries against cell surface ligands. Indeed, several recent reports show that such cell-based selection can be effectively performed54–56. The first approach was an extension of ‘panning’ selection methods to the YSD system56, wherein yeast cells displaying scFv were applied to a monolayer of mammalian cells and the binding population was enriched by washing away unbound yeast cells. The panning method was used with a naïve human scFv library24 to yield 34 unique scFv sequences that target the brain endothelial cell surface55. When the selected scFvs were secreted as a soluble protein, clear immunolabeling of, and in some cases endocytosis into, target cells was detected55. ScFvs with relatively high affinity (Kd = 82 nM) and/or avidity (Kd = 2.0 nM) were identified, indicating the advantage of multivalent YSD in screening for a wide range of affinities which could be important for lead molecule identification using experimentally attainable library sizes. Then, as discussed previously, these leads could be further optimized in terms of binding affinity.
As an alternative approach, a density centrifugation method was used to select yeast cell-mammalian cell conjugates54. Here, scTCR-displaying yeast cells that were specifically bound to mammalian lymphoid-derived cells were separated through centrifugation in a discontinuous density media. This method was effective in separating high affinity scTCRs from a Vα chain CDR3 library. In two rounds of selection against a novel peptide target, five high affinity mutants were isolated54. Whereas the panning methods rely on mammalian cells that grow on a support, the density centrifugation allows separation against cells in suspension. Collectively, these cell surface-based selections allow YSD library selection against complex cell surfaces, avoiding the need for target membrane protein expression and purification. In addition, when the library is selected against an unidentified target, the selected yeast cells may also be used as affinity reagents to directly immunoprecipitate and identify the target55.
Epitope mapping
Yeast surface display has been used for the mapping of antibody epitopes by displaying either the antibody fragment or the antigen. In pioneering work using epidermal growth factor receptor (EGFR) as a model, epitopes for a number of available antibodies were determined57,58 (Table 1). By displaying either stably folded EGFR fragments or randomly mutated full-length EGFR on the yeast surface, antibody binding to particular domains or strictly defined epitopes could easily be determined by flow cytometry57,58. Furthermore, heat denaturation of the EGFR fragments allowed for determination of linear versus conformational epitope binding. Binding sites for neutralizing antibodies against the West Nile virus viral envelope (E) protein were also determined using yeast surface display (Table 1). First, display of the entire ectodomain (domains I–III) or just domain III of the viral E protein was used to narrow down the binding region59. Error-prone PCR of the binding domain and library screening allowed for loss of function experiments to map contacting residues to domain III60 or domains I–II61. A similar method of screening for antibody binding utilized fragments of the West Nile virus NS1 glycoprotein to resolve binding epitopes60.
Conversely, scFvs displayed on yeast can also be used to determine binding epitopes. Siegel et al used three monoclonal antibodies with previously identified epitopes to characterize the epitopes of EGF binding scFvs that were isolated from a library62. Yeast cells displaying EGF-binding scFvs were incubated with EGF, and subsequently incubated with each of the three monoclonal antibodies. If the scFv epitopes overlap with those of the monoclonal antibodies, EGF binding to the scFvs was blocked62. It was thus possible to not only group the scFvs according to their epitopes but also sort epitope-specific scFvs against EGF62. As demonstrated by these examples, epitope mapping using YSD is highly effective and especially advantageous over conventional methods since it combines the capability of highly quantitative epitope interaction monitoring using flow cytometry and the eukaryotic protein processing machinery of yeast, which increases the possibility of correct epitope display.
Screening cDNA libraries
Yeast pose a significant advantage over most other display systems because of their ability to express mammalian proteins with reasonably high fidelity and with some level of post-translational modification. This property therefore allows for convenient screening of cDNA libraries derived from eukaryotic sources. To this end, the display of a human cDNA library on the surface of yeast has been used to screen libraries for cancer related antigens and proteins that are recognized by breast cancer serum63 (Table 1). After creating a cDNA library from breast carcinoma tumor tissue, the resulting library was displayed on yeast and incubated with serum antibodies isolated from patients to identify displayed proteins that were immunogenic and tumor-related63. While a number of known breast cancer-related antigens were isolated from the cDNA library, a previously unknown small breast epithelial mucin was isolated and found to have deletion mutation in the diseased tissue63. A human cDNA library was also screened to identify proteins that interact with phosphorylated, synthetic peptides derived from the EGF receptor or focal adhesion kinase (FAK). Two clones expressing proteins known to bind phosphorylated EGFR and three known to bind phosphorylated FAK were isolated from the library, and binding was shown to be phosphorylation dependent64. On a larger scale, it may also possible to use this method to identify specific protein-protein interactions across the proteome.
Protein engineering for improved production and stability
YSD has also been applied to the engineering of proteins for improved production and stability. Since surface-displayed and secreted proteins both follow the same folding and secretory pathway, it was hypothesized that the proteins having higher expression levels on the surface of yeast cells would have superior secretion efficiencies as well as folding properties. In an initial study, YSD was used to identify mutations in a poorly expressed scTCR to enable successful display of the scTCR65 (Table 1). It was also found that there was a strong correlation between surface display level and soluble secretion level of the mutant scTCRs66. In addition, scTCR mutants with higher surface expression possessed higher thermal tolerance in terms of decreased rates of thermal denaturation66. Subsequently, YSD has been applied for the engineering of other poorly expressed or unstable proteins such as class I and II MHC molecules19,20,42,67, epidermal growth factor receptor68, cancer-testis antigen NY-ESO-169, p55 tumor necrosis factor receptor70, and natural killer cell receptor48. These studies generally indicated the existence of a strong correlation between the levels of YSD and the secretion levels of the engineered proteins42,67,69,70. Interestingly, the mutations that enhanced surface expression in yeast also had the added property of increasing bacterial and mammalian secretion levels42,67,70. It was determined that an intrinsic protein property such as improved thermostability or enhanced folding was likely responsible for the increased production19,67,68,70. Importantly, in these examples, it was generally found that the surface display levels of the engineered protein were largely governed by alterations in the protein itself and not by the fusion scaffold. The wild-type protein construct was either display-incompetent65 or had a considerably lower expression level (~103 copies/cell)70 than the typical display levels of well-expressed fusion constructs (5–6 × 105 copies/cell) 4,71. Thus, when the studied protein at least in part regulates the surface expression levels, YSD can be successfully applied to engineer secretion levels and folding properties.
However, there are certain situations where the surface display level failed to correlate with the secretion efficiency. When cancer-testis antigen NY-ESO-1, which had nearly undetectable secretion level was mutated and displayed on yeast, mutant clones with high surface expression level were identified69. However, these NY-ESO-1 mutations did not increase the yeast secretion levels for the unfused NY-ESO-169. Instead, high secretion levels were detected only when Aga2p was retained as a fusion partner, indicating that the mutations and high display levels were Aga2p dependent. In addition, in a recent study to identify yeast genes that increase heterologous protein production72, it was found that coexpression of immunoglobulin heavy chain binding protein (BiP) and protein disulfide isomerase (PDI), both of which increase the secretion level of multiple unfused scFvs71,73, had no effect on the surface display level of the scFvs72. Here, the intracellular processing efficiency of the scFvs was altered by its fusion partner Aga2p such that it was no longer responsive to the same manipulations in cellular machinery72. In some cases however, when processing of the engineered protein was not affected by Aga2p fusion, an increase in surface expression was detected with PDI coexpression10.
In another study, it was found that protein display level failed to correlate with thermal stability. Mutants of three-helix bundle protein α3D that have very high but varying thermal stability showed identical display level74. Moreover, two progenitors of α3D that form highly stable but less highly ordered structures also showed similar surface expression level as the native form74. It is possible that the expression level of α3D, since it is extremely stable, may not be subject to processing and display limitations, or may be limited by the Aga2p fusion partner. Thus, while YSD can be used to engineer protein stability, there may be an upper limit determined by stability of the target protein that can make YSD approaches ineffective.
To decouple the effects of Aga2p on protein surface display, Wentz et al. used a selection pressure that reduced the surface display level of the Aga2p-target protein fusion but did not affect the display level of Aga2p alone72. Therefore, the fusion partner, rather than Aga2p, could act as the dominant determinant of display efficiency72. By elevating the induction temperature of the fusion construct above the optimal value of 20 °C, yeast genes that increase both the surface display and secretion level of scTCRs and scFvs were identified72.
Another possible solution to overcome stability or processing limitations described above is to utilize the yeast cell wall as a matrix to capture the secreted proteins, rather than relying on fusion to a cell surface anchor75. In this study, the yeast cell surface was modified with polyethylene glycol-fluorescein and secreted anti-fluorescein scFv 4m5.3 was captured as it was secreted. Using this method, yeast with three-fold higher secretion level of 4m5.3 above the background was enriched between 23–45 fold after one round of enrichment from mock libraries75. This type of capture method could be an effective alternative to the current YSD systems since it decouples any effect of anchor proteins on fusion display levels.
Non-biological targets
YSD has been used to bind or control assembly of a number of non-biological targets. To create yeast capable of adsorption and recovery of heavy metal ions, a strain of S. cerevisiae was engineered to display histidine hexapeptide on the surface in order to capture copper ions. In addition, the yeast strain also expressed a copper-responsive zinc-finger transcription factor related to cell aggregation76 (Table 1). Thus, in response to as low as 1 mM copper, the yeast were shown to both adsorb copper on the cell surface and self-aggregate to allow facile removal from the solution phase76. Moreover, screening of YSD peptide or antibody libraries against inorganic compounds such as semiconductor materials CdS, CdSe, ZnS, or ZnSe or gold, has been used to identify binding proteins77,78. Peptide tags have also been suggested as a method to couple proteins to metal oxides. A random peptide dodecamer YSD library was panned against synthetic sapphire (Al2O3) and clones were found that adhered specifically to three different phases of the metal oxide. One of the cloned peptides was used as a fusion to maltose binding protein (MBP) to show that the peptide-MBP fusion selectively bound sapphire 500–1000 fold over MBP alone79. Therefore, it seems that peptide or protein libraries could be screened against virtually any surface using YSD, and these applications suggest the potential for using displaying yeast as biosensors for non-biological targets.
Cell adhesion molecules
Several recent advances in the field of cell adhesion have resulted from application of YSD technologies. Yeast provide an excellent platform for cell adhesion studies because they are easy to manipulate and large enough to monitor cell adhesive behavior. By measuring angular and translational velocity, yeast expressing E-selectin were shown to roll, rather than slip, across sialyl-Lewis-x surfaces, providing the first evidence that selectin interactions mediate a true “rolling” motion80. Further work evaluating yeast rolling behavior showed that rolling velocity of the yeast expressing LFA-1 I domain, the binding domain of the molecule, on ICAM-1 surfaces was decorrelated from soluble binding affinity for the ligand81 (Table 1). Also using surface display of the LFA-1 I domain, an I domain mutant with 200,000-fold higher affinity for ICAM-1 over the wild-type molecule was engineered47. This mutant was shown to block lymphocyte adhesion and transmigration in in vitro systems. Additionally, an scFv isolated from a nonimmune surface display library24 against PSGL-1, the ligand that interacts with P-selectin to mediate rolling, was affinity matured and converted to a full antibody molecule82. Further work showed that this antibody could act as a potent anti-inflammatory agent.
Conclusions
As discussed above, YSD has developed into a powerful technology over the past decade and has enabled many applications in protein engineering and library screening (Table 1). Although the comparison of various molecular display technologies was beyond the scope of this review, one can see that YSD has gained a unique foothold in the protein engineering arena because of its eukaryotic protein processing machinery and the capability for quantitative flow cytometric screening. These attributes of YSD have proven their strength, especially in the fine-tuning of protein affinity and specificity, and have lead to novel applications such as human cDNA library screening. In addition, advances in library generation methods and library screening methods have further expanded the capability of YSD. Therefore, it is anticipated that YSD technology will continue to evolve and serve as an essential tool for protein engineering.
Acknowledgments
This work was in part supported by grants from the National Institutes of Health NS052649 to E.V.S., and National Science Foundation CAREER award 0239099 to E.T.B.. Y.K.C. is a recipient of a Genomic Sciences Training Program Fellowship funded through the National Institutes of Health, 5T32HG002760.
List of abbreviations
- cDNA
complementary DNA
- YSD
yeast surface display
- scFv
single-chain Fv antibody fragment
- GPI
glycosylphosphatidylinositol
- GPCR
G protein-coupled receptor
- MHC
major histocompatibility complex
- HC
heavy chain
- LC
light chain
- FACS
fluorescence-activated cell sorting
- Fab
antigen-binding fragment
- FITC
fluorescein isothiocyanate
- DR5
death receptor 5
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- SOSM
shotgun ortholog scanning mutagenesis
- CDR
complementarity determining region
- TCR
T cell receptor
- SEB
staphylococcal enterotoxin B
- IL-2
interleukin-2
- scTCR
single-chain T cell receptor
- FAK
focal adhesion kinase
- BiP
immunoglobulin heavy chain binding protein
- PDI
protein disulfide isomerase
- MBP
maltose binding protein
References
- 1.Schreuder MP, Brekelmans S, van den Ende H, Klis FM. Yeast. 1993;9:399–409. doi: 10.1002/yea.320090410. [DOI] [PubMed] [Google Scholar]
- 2.Schreuder MP, Mooren AT, Toschka HY, Verrips CT, Klis FM. Trends Biotechnol. 1996;14:115–120. doi: 10.1016/0167-7799(96)10017-2. [DOI] [PubMed] [Google Scholar]
- 3.Kondo A, Ueda M. Applied Microbiology and Biotechnology. 2004;64:28–40. doi: 10.1007/s00253-003-1492-3. [DOI] [PubMed] [Google Scholar]
- 4.Boder ET, Wittrup KD. Nat Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- 5.Boder ET, Wittrup KD. Methods Enzymol. 2000;328:430–444. doi: 10.1016/s0076-6879(00)28410-3. [DOI] [PubMed] [Google Scholar]
- 6.Abe H, Shimma Y, Jigami Y. Glycobiology. 2003;13:87–95. doi: 10.1093/glycob/cwg014. [DOI] [PubMed] [Google Scholar]
- 7.Ecker M, Deutzmann R, Lehle L, Mrsa V, Tanner W. J Biol Chem. 2006;281:11523–11529. doi: 10.1074/jbc.M600314200. [DOI] [PubMed] [Google Scholar]
- 8.Andres I, Gallardo O, Parascandola P, Javier Pastor FI, Zueco J. Biotechnology and bioengineering. 2005;89:690–697. doi: 10.1002/bit.20375. [DOI] [PubMed] [Google Scholar]
- 9.Abe H, Ohba M, Shimma Y, Jigami Y. FEMS yeast research. 2004;4:417–425. doi: 10.1016/S1567-1356(03)00193-4. [DOI] [PubMed] [Google Scholar]
- 10.Shimma Y, Saito F, Oosawa F, Jigami Y. Applied and Environmental Microbiology. 2006;72:7003–7012. doi: 10.1128/AEM.01378-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silverman L, Campbell R, Broach JR. Curr Opin Chem Biol. 1998;2:397–403. doi: 10.1016/s1367-5931(98)80015-x. [DOI] [PubMed] [Google Scholar]
- 12.Ault AD, Broach JR. Protein engineering, design & selection : PEDS. 2006;19:1–8. doi: 10.1093/protein/gzi069. [DOI] [PubMed] [Google Scholar]
- 13.Kim SY, Sohn JH, Pyun YR, Choi ES. Yeast (Chichester, England) 2002;19:1153–1163. doi: 10.1002/yea.911. [DOI] [PubMed] [Google Scholar]
- 14.Mergler M, Wolf K, Zimmermann M. Appl Microbiol Biotechnol. 2004;63:418–421. doi: 10.1007/s00253-003-1361-0. [DOI] [PubMed] [Google Scholar]
- 15.Tanino T, Fukuda H, Kondo A. Biotechnol Prog. 2006;22:989–993. doi: 10.1021/bp060133+. [DOI] [PubMed] [Google Scholar]
- 16.Kato M, Fuchimoto J, Tanino T, Kondo A, Fukuda H, Ueda M. Applied Microbiology and Biotechnology. 2007 doi: 10.1007/s00253-006-0835-2. [DOI] [PubMed] [Google Scholar]
- 17.Wang Z, Mathias A, Stavrou S, Neville DM., Jr Protein Eng Des Sel. 2005;18:337–343. doi: 10.1093/protein/gzi036. [DOI] [PubMed] [Google Scholar]
- 18.Boder ET, Bill JR, Nields AW, Marrack PC, Kappler JW. Biotechnol Bioeng. 2005;92:485–491. doi: 10.1002/bit.20616. [DOI] [PubMed] [Google Scholar]
- 19.Esteban O, Zhao H. J Mol Biol. 2004;340:81–95. doi: 10.1016/j.jmb.2004.04.054. [DOI] [PubMed] [Google Scholar]
- 20.Starwalt SE, Masteller EL, Bluestone JA, Kranz DM. Protein Eng. 2003;16:147–156. doi: 10.1093/proeng/gzg018. [DOI] [PubMed] [Google Scholar]
- 21.van den Beucken T, Pieters H, Steukers M, van der Vaart M, Ladner RC, Hoogenboom HR, Hufton SE. FEBS Lett. 2003;546:288–294. doi: 10.1016/s0014-5793(03)00602-1. [DOI] [PubMed] [Google Scholar]
- 22.Lin Y, Tsumuraya T, Wakabayashi T, Shiraga S, Fujii I, Kondo A, Ueda M. Applied Microbiology and Biotechnology. 2003;62:226–232. doi: 10.1007/s00253-003-1283-x. [DOI] [PubMed] [Google Scholar]
- 23.Furukawa H, Tanino T, Fukuda H, Kondo A. Biotechnology progress. 2006;22:994–997. doi: 10.1021/bp0601342. [DOI] [PubMed] [Google Scholar]
- 24.Feldhaus MJ, Siegel RW, Opresko LK, Coleman JR, Feldhaus JM, Yeung YA, Cochran JR, Heinzelman P, Colby D, Swers J, Graff C, Wiley HS, Wittrup KD. Nat Biotechnol. 2003;21:163–170. doi: 10.1038/nbt785. [DOI] [PubMed] [Google Scholar]
- 25.Lee HW, Lee SH, Park KJ, Kim JS, Kwon MH, Kim YS. Biochemical and biophysical research communications. 2006;346:896–903. doi: 10.1016/j.bbrc.2006.05.202. [DOI] [PubMed] [Google Scholar]
- 26.Blaise L, Wehnert A, Steukers MP, van den Beucken T, Hoogenboom HR, Hufton SE. Gene. 2004;342:211–218. doi: 10.1016/j.gene.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 27.Weaver-Feldhaus JM, Lou J, Coleman JR, Siegel RW, Marks JD, Feldhaus MJ. FEBS Lett. 2004;564:24–34. doi: 10.1016/S0014-5793(04)00309-6. [DOI] [PubMed] [Google Scholar]
- 28.Bowley DR, Labrijn AF, Zwick MB, Burton DR. Protein Eng Des Sel. 2007 doi: 10.1093/protein/gzl057. [DOI] [PubMed] [Google Scholar]
- 29.Swers JS, Kellogg BA, Wittrup KD. Nucleic Acids Res. 2004;32:e36. doi: 10.1093/nar/gnh030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cochran JR, Kim YS, Lippow SM, Rao B, Wittrup KD. Protein Eng Des Sel. 2006;19:245–253. doi: 10.1093/protein/gzl006. [DOI] [PubMed] [Google Scholar]
- 31.Rajpal A, Beyaz N, Haber L, Cappuccilli G, Yee H, Bhatt RR, Takeuchi T, Lerner RA, Crea R. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:8466–8471. doi: 10.1073/pnas.0503543102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.VanAntwerp JJ, Wittrup KD. Biotechnol Prog. 2000;16:31–37. doi: 10.1021/bp990133s. [DOI] [PubMed] [Google Scholar]
- 33.Boder ET, Midelfort KS, Wittrup KD. Proc Natl Acad Sci U S A. 2000;97:10701–10705. doi: 10.1073/pnas.170297297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boder ET, Wittrup KD. Biotechnol Prog. 1998;14:55–62. doi: 10.1021/bp970144q. [DOI] [PubMed] [Google Scholar]
- 35.Graff CP, Chester K, Begent R, Wittrup KD. Protein Eng Des Sel. 2004;17:293–304. doi: 10.1093/protein/gzh038. [DOI] [PubMed] [Google Scholar]
- 36.Chao G, Lau WL, Hackel BJ, Sazinsky SL, Lippow SM, Wittrup KD. Nature Protocols. 2006;1:755–768. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- 37.Razai A, Garcia-Rodriguez C, Lou J, Geren IN, Forsyth CM, Robles Y, Tsai R, Smith TJ, Smith LA, Siegel RW, Feldhaus M, Marks JD. J Mol Biol. 2005;351:158–169. doi: 10.1016/j.jmb.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 38.VanAntwerp JJ, Wittrup KD. J Mol Recognit. 1998;11:10–13. doi: 10.1002/(SICI)1099-1352(199812)11:1/6<10::AID-JMR381>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 39.Wang Z, Kim GB, Woo JH, Liu YY, Mathias A, Stavrou S, Neville DM., Jr Bioconjug Chem. 2007;18:947–955. doi: 10.1021/bc0603438. [DOI] [PubMed] [Google Scholar]
- 40.Garcia-Rodriguez C, Levy R, Arndt JW, Forsyth CM, Razai A, Lou J, Geren I, Stevens RC, Marks JD. Nat Biotechnol. 2007;25:107–116. doi: 10.1038/nbt1269. [DOI] [PubMed] [Google Scholar]
- 41.Holler PD, Holman PO, Shusta EV, O'Herrin S, Wittrup KD, Kranz DM. Proc Natl Acad Sci U S A. 2000;97:5387–5392. doi: 10.1073/pnas.080078297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weber KS, Donermeyer DL, Allen PM, Kranz DM. Proc Natl Acad Sci U S A. 2005;102:19033–19038. doi: 10.1073/pnas.0507554102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chlewicki LK, Holler PD, Monti BC, Clutter MR, Kranz DM. J Mol Biol. 2005;346:223–239. doi: 10.1016/j.jmb.2004.11.057. [DOI] [PubMed] [Google Scholar]
- 44.Holler PD, Kranz DM. Immunity. 2003;18:255–264. doi: 10.1016/s1074-7613(03)00019-0. [DOI] [PubMed] [Google Scholar]
- 45.Buonpane RA, Moza B, Sundberg EJ, Kranz DM. J Mol Biol. 2005;353:308–321. doi: 10.1016/j.jmb.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 46.Buonpane RA, Churchill HR, Moza B, Sundberg EJ, Peterson ML, Schlievert PM, Kranz DM. Nat Med. 2007;13:725–729. doi: 10.1038/nm1584. [DOI] [PubMed] [Google Scholar]
- 47.Jin M, Song G, Carman CV, Kim YS, Astrof NS, Shimaoka M, Wittrup DK, Springer TA. Proc Natl Acad Sci U S A. 2006;103:5758–5763. doi: 10.1073/pnas.0601164103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dam J, Guan R, Natarajan K, Dimasi N, Chlewicki LK, Kranz DM, Schuck P, Margulies DH, Mariuzza RA. Nat Immunol. 2003;4:1213–1222. doi: 10.1038/ni1006. [DOI] [PubMed] [Google Scholar]
- 49.Rao BM, Driver I, Lauffenburger DA, Wittrup KD. Mol Pharmacol. 2004;66:864–869. doi: 10.1124/mol.66.4.. [DOI] [PubMed] [Google Scholar]
- 50.Rao BM, Driver I, Lauffenburger DA, Wittrup KD. Biochemistry. 2005;44:10696–10701. doi: 10.1021/bi050436x. [DOI] [PubMed] [Google Scholar]
- 51.Rao BM, Girvin AT, Ciardelli T, Lauffenburger DA, Wittrup KD. Protein Eng. 2003;16:1081–1087. doi: 10.1093/protein/gzg111. [DOI] [PubMed] [Google Scholar]
- 52.Shusta EV, Holler PD, Kieke MC, Kranz DM, Wittrup KD. Nat Biotechnol. 2000;18:754–759. doi: 10.1038/77325. [DOI] [PubMed] [Google Scholar]
- 53.Yeung YA, Wittrup KD. Biotechnol Prog. 2002;18:212–220. doi: 10.1021/bp010186l. [DOI] [PubMed] [Google Scholar]
- 54.Richman SA, Healan SJ, Weber KS, Donermeyer DL, Dossett ML, Greenberg PD, Allen PM, Kranz DM. Protein Eng Des Sel. 2006;19:255–264. doi: 10.1093/protein/gzl008. [DOI] [PubMed] [Google Scholar]
- 55.Wang XX, Cho YK, Shusta EV. Nat Methods. 2007;4:143–145. doi: 10.1038/nmeth993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang XX, Shusta EV. J Immunol Methods. 2005;304:30–42. doi: 10.1016/j.jim.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 57.Chao G, Cochran JR, Wittrup KD. J Mol Biol. 2004;342:539–550. doi: 10.1016/j.jmb.2004.07.053. [DOI] [PubMed] [Google Scholar]
- 58.Cochran JR, Kim YS, Olsen MJ, Bhandari R, Wittrup KD. J Immunol Methods. 2004;287:147–158. doi: 10.1016/j.jim.2004.01.024. [DOI] [PubMed] [Google Scholar]
- 59.Oliphant T, Engle M, Nybakken GE, Doane C, Johnson S, Huang L, Gorlatov S, Mehlhop E, Marri A, Chung KM, Ebel GD, Kramer LD, Fremont DH, Diamond MS. Nature medicine. 2005;11:522–530. doi: 10.1038/nm1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chung KM, Nybakken GE, Thompson BS, Engle MJ, Marri A, Fremont DH, Diamond MS. Journal of virology. 2006;80:1340–1351. doi: 10.1128/JVI.80.3.1340-1351.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oliphant T, Nybakken GE, Engle M, Xu Q, Nelson CA, Sukupolvi-Petty S, Marri A, Lachmi BE, Olshevsky U, Fremont DH, Pierson TC, Diamond MS. J Virol. 2006;80:12149–12159. doi: 10.1128/JVI.01732-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Siegel RW, Coleman JR, Miller KD, Feldhaus MJ. J Immunol Methods. 2004;286:141–153. doi: 10.1016/j.jim.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 63.Wadle A, Mischo A, Imig J, Wullner B, Hensel D, Watzig K, Neumann F, Kubuschok B, Schmidt W, Old LJ, Pfreundschuh M, Renner C. International journal of cancer.Journal international du cancer. 2005;117:104–113. doi: 10.1002/ijc.21147. [DOI] [PubMed] [Google Scholar]
- 64.Bidlingmaier S, Liu B. Mol Cell Proteomics. 2006;5:533–540. doi: 10.1074/mcp.M500309-MCP200. [DOI] [PubMed] [Google Scholar]
- 65.Kieke MC, Shusta EV, Boder ET, Teyton L, Wittrup KD, Kranz DM. Proc Natl Acad Sci U S A. 1999;96:5651–5656. doi: 10.1073/pnas.96.10.5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shusta EV, Kieke MC, Parke E, Kranz DM, Wittrup KD. J Mol Biol. 1999;292:949–956. doi: 10.1006/jmbi.1999.3130. [DOI] [PubMed] [Google Scholar]
- 67.Jones LL, Brophy SE, Bankovich AJ, Colf LA, Hanick NA, Garcia KC, Kranz DM. J Biol Chem. 2006;281:25734–25744. doi: 10.1074/jbc.M604343200. [DOI] [PubMed] [Google Scholar]
- 68.Kim YS, Bhandari R, Cochran JR, Kuriyan J, Wittrup KD. Proteins. 2006;62:1026–1035. doi: 10.1002/prot.20618. [DOI] [PubMed] [Google Scholar]
- 69.Piatesi A, Howland SW, Rakestraw JA, Renner C, Robson N, Cebon J, Maraskovsky E, Ritter G, Old L, Wittrup KD. Protein Expr Purif. 2006;48:232–242. doi: 10.1016/j.pep.2006.01.026. [DOI] [PubMed] [Google Scholar]
- 70.Schweickhardt RL, Jiang X, Garone LM, Brondyk WH. J Biol Chem. 2003;278:28961–28967. doi: 10.1074/jbc.M212019200. [DOI] [PubMed] [Google Scholar]
- 71.Nakamura Y, Shibasaki S, Ueda M, Tanaka A, Fukuda H, Kondo A. Appl Microbiol Biotechnol. 2001;57:500–505. doi: 10.1007/s002530100802. [DOI] [PubMed] [Google Scholar]
- 72.Wentz AE, Shusta EV. Appl Environ Microbiol. 2006 doi: 10.1128/AEM.02427-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shusta EV, Raines RT, Pluckthun A, Wittrup KD. Nat Biotechnol. 1998;16:773–777. doi: 10.1038/nbt0898-773. [DOI] [PubMed] [Google Scholar]
- 74.Park S, Xu Y, Stowell XF, Gai F, Saven JG, Boder ET. Protein Eng Des Sel. 2006;19:211–217. doi: 10.1093/protein/gzl003. [DOI] [PubMed] [Google Scholar]
- 75.Rakestraw JA, Baskaran AR, Wittrup KD. Biotechnol Prog. 2006;22:1200–1208. doi: 10.1021/bp0600233. [DOI] [PubMed] [Google Scholar]
- 76.Kuroda K, Ueda M, Shibasaki S, Tanaka A. Appl Microbiol Biotechnol. 2002;59:259–264. doi: 10.1007/s00253-002-1014-8. [DOI] [PubMed] [Google Scholar]
- 77.Peelle BR, Krauland EM, Wittrup KD, Belcher AM. Acta Biomater. 2005;1:145–154. doi: 10.1016/j.actbio.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 78.Peelle BR, Krauland EM, Wittrup KD, Belcher AM. Langmuir. 2005;21:6929–6933. doi: 10.1021/la050261s. [DOI] [PubMed] [Google Scholar]
- 79.Krauland EM, Peelle BR, Wittrup KD, Belcher AM. Biotechnol Bioeng. 2007 doi: 10.1002/bit.21341. [DOI] [PubMed] [Google Scholar]
- 80.Bhatia SK, Swers JS, Camphausen RT, Wittrup KD, Hammer DA. Biotechnol Prog. 2003;19:1033–1037. doi: 10.1021/bp025756b. [DOI] [PubMed] [Google Scholar]
- 81.Pepper LR, Hammer DA, Boder ET. J Mol Biol. 2006;360:37–44. doi: 10.1016/j.jmb.2006.04.049. [DOI] [PubMed] [Google Scholar]
- 82.Swers JS, Widom A, Phan U, Springer TA, Wittrup KD. Biochem Biophys Res Commun. 2006;350:508–513. doi: 10.1016/j.bbrc.2006.08.197. [DOI] [PubMed] [Google Scholar]
- 83.Dranginis AM, Rauceo JM, Coronado JE, Lipke PN. Microbiol Mol Biol Rev. 2007;71:282–294. doi: 10.1128/MMBR.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]