Abstract
It has been well established that acute lung injury (ALI), and the more severe presentation of acute respiratory distress syndrome (ARDS), constitute complex traits characterized by a multigenic and multifactorial etiology. Identification and validation of genetic variants contributing to disease susceptibility and severity has been hampered by the profound heterogeneity of the clinical phenotype and the role of environmental factors, which includes treatment, on outcome. The critical nature of ALI and ARDS, compounded by the impact of phenotypic heterogeneity, has rendered the amassing of sufficiently powered studies especially challenging. Nevertheless, progress has been made in the identification of genetic variants in select candidate genes, which has enhanced our understanding of the specific pathways involved in disease manifestation. Identification of novel candidate genes for which genetic association studies have confirmed a role in disease has been greatly aided by the powerful tool of high-throughput expression profiling. This article will review these studies to date, summarizing candidate genes associated with ALI and ARDS, acknowledging those that have been replicated in independent populations, with a special focus on the specific pathways for which candidate genes identified so far can be clustered.
Keywords: lung injury, acute respiratory distress syndrome, genetic epidemiology, genetic association, polymorphism
major advances in the epidemiological study of acute lung injury (ALI) and its more severe form, acute respiratory distress syndrome (ARDS), reiterate the considerable heterogeneity that has long been observed in the response to injurious stimuli and their impact on the lung among the critically ill. Associations between genetic polymorphisms and lung injury-related phenotypes in some populations but not others reflects, to a certain extent, limitations related to statistical power and inconsistencies in phenotype definition, but also the profound role that environmental factors play as key determinants in the development (and manifestation) of ALI.
Despite the fact that, in the last few years, there has been a number of studies focusing on the genetics of ALI (11, 29, 30, 37, 73, 81), there remain significant challenges in elucidating the major genes involved in lung injury pathophysiology. First, ALI represents a continuum rather than a discreet group of phenotypes, and therefore lacks unique, readily measurable biomarkers for definitive diagnosis. Second, ALI arises from diverse precipitating factors in a critically ill population and subsequently exhibits a variable clinical course. Because of the phenotypic variance and likely complex gene-environment interactions, significant difficulties persist in elucidating the most likely candidate genes that govern susceptibility and severity of ALI out of the ∼20,000 potential genes in the human genome. Well-designed studies examining the association between genetic polymorphisms and susceptibility and severity of ALI should be conducted with careful consideration of selection of candidate genes, phenotype definition, power estimation, and sample size of the study population, selection of appropriate controls, and adjustment, as needed, for confounding factors including population stratification (33, 37, 81).
In this review, we summarize the role of genetic polymorphisms in the pathogenesis of lung injury based on a review of the literature as of 2008, with a special focus on target pathways. We further discuss the complexities of the ALI phenotype and role of microarray-based analyses of genome-wide gene expression in identifying suitable targets for candidate gene studies in ALI. Coassociations between candidate genes with ALI and other inflammatory phenotypes (sepsis and asthma) are presented as a thought-provoking approach toward testing the common variant/multiple disease hypothesis. Throughout this paper, we highlight strengths and weaknesses of studies conducted to date, for the purpose of underscoring the importance of consideration for ideal study design and attributes associated with the basic principles of genetic epidemiology.
ALI and ARDS: Complex Etiology and Heterogeneity of the Phenotype
Both ALI and ARDS are devastating clinical syndromes affecting over 200,000 patients in the United States each year (80). Despite the survival benefit of advances in therapy such as low tidal volumes and lung protective ventilation, mortality remains high, ranging between 25% and 50%, with an average of 38.5% (13, 79, 80). The most common underlying causes of ALI include direct injury to the lung (e.g., “pulmonary” ALI), such as pneumonia, aspiration of gastric contents, pulmonary contusion, fat or air emboli, near drowning, inhalation injury, or reperfusion injury, as well as indirect injury (e.g., “extrapulmonary”), such as sepsis, burn, severe trauma with multiple transfusions, cardiopulmonary bypass, drug overdose or toxic ingestion, or acute pancreatitis (94). The many inciting injuries that can lead to ALI, compounded by the multigenic and multifactorial features that are characteristic of a complex trait such as ALI, are the basis for the heterogeneity of disease manifestations and outcomes.
To optimize the dissection of the genetic basis for ALI, a major focus has been on candidate genes that are prominently involved in the primary pathophysiology of the disease, which includes widespread endothelial and epithelial disruption leading to increased permeability, protein-rich pulmonary edema, neutrophilic inflammation, and surfactant dysfunction (67, 95). A unifying theme regardless of the etiological basis for disease is an imbalance between pro- and anti-inflammatory cytokines, oxidants and antioxidants, procoagulants and anticoagulants, neutrophil recruitment/activation and clearance, and proteases and protease inhibitors (94). The pathogenesis of ALI is similar to that of sepsis, as both disease states involve uncontrolled host defense responses that lead to inflammation, endothelial damage, enhanced coagulation, diminished fibrinolysis, and fibroproliferation (58). Sepsis is associated with the highest risk of progression to ALI, ∼40% (95). Many stimuli can trigger sepsis, but gram-negative bacteria (containing endotoxin or LPS) are responsible for most clinical sepsis cases (71). Genes and polymorphisms involved in multiple processes of the bacteria-induced cellular response have been associated with susceptibility and severity of the sepsis phenotype, and, therefore, by association, represent potential candidates of relevance for susceptibility to lung injury.
Table 1 summarizes the most current review of literature through 2008 and includes all genes that were identified as a potential ALI locus for susceptibility and/or outcome in conventional association studies. Unfortunately, and due to the fact that non-significant associations are less likely to be published in the scientific arena, it is not possible to determine to what extent these studies have been truly confirmed, and, for every significant association, how many others have failed to observe the same relationship (16, 74). An area of particular discourse (and disagreement) is what constitutes the optimal control group for a reference population of ALI or other severe disease phenotypes such as sepsis (81, 84). The published studies to date on ALI, as summarized in Table 1, have utilized mostly population-based healthy individuals (9 out of 26 studies) or sick controls at risk for development of ALI but whom do not develop ALI (13 out of 26 studies). The fundamental principle behind selection of a control group is to insure that the control group does not have the disease of interest, or any associated intermediate phenotypes (84), and that the control group has had equal opportunity of exposure to the relevant environmental factors that are associated with disease pathogenesis, but are clearly unaffected (70). True associations can also be missed when cases and controls are not matched by important confounders, such as stratification due to admixture, whereby the ethnic make-up of the case group is significantly different than the control group; this confounder, and its relevance in designing genetic epidemiology studies of lung injury, is discussed in detail elsewhere (11). Indeed, as demonstrated in our studies and studies by others, associations with specific polymorphisms may be unique to certain ethnic groups (i.e., MYLK polymorphisms and associations among African Americans as summarized in Refs. 33 and 24), and for this reason, it is not advisable to “mix” ethnic groups in a single case-control or cohort group.
Table 1.
Summary of polymorphisms in candidate genes significantly associated with ALI and/or ARDS
| Function/Pathway | Gene (Symbol) | Variant | Ref. (Year) | Population (Cases:Controls/Control Type) |
Significant Association |
|
|---|---|---|---|---|---|---|
| Susceptibility | Severity/Outcome | |||||
| Respiratory gaseous exchange | Surfactant, pulmonary-associated protein B (SFTPB) | Intron 4 repeats | 64 (1996) | EA (15:21)/population-based | ARDS: 1580C | |
| T1580C | 54 (2000) | EA (52:46:25 “others”)/both | ARDS: 1580C | |||
| Intron 4 repeats | 39 (2004) | EA (72:117)/at-risk | ARDS in females | |||
| T1580C | 77 (2004) | EA + AA (12:390)/at-risk | ARDS: 1580C | |||
| Cytokine | IL-6 (IL6) | G-174C | 61 (2002) | EA (96:88, 174, 1906)/both | C allele and CC genotype associated with mortality | |
| Haplotypes | 87 (2005) | European (228 ALI) | ALI: haplotypes −174C/1753C/2954G, G/G/G, or G/C/C | |||
| 3 SNPs | 73 (2005) | EA (98:84)/population-based | ALI: 3-SNP haplotype | |||
| 6 SNPs | 31 (2008) | European (67:96)/population-based | ALI: 6-SNP haplotype | |||
| IL-8 (IL8) | A-251T | 44 (2007) | EA (23:74)/at-risk | ARDS: −251A | ||
| IL-10 (IL10) | G-1082A | 38 (2006) | EA (211:429)/at-risk | ARDS risk: −1082GG | −1082GG genotype associated with lower mortality in ARDS | |
| 83 (2008) | European (100, prospective study) | Trauma-assoc. acute respiratory failure: protective −1082 GG | ||||
| Macrophage migration inhibitory factor (MIF) | 8 SNPs | 32 (2007) | EA (90 ALI:113 sepsis:85 controls); AA (61 ALI:69 sepsis:88 controls)/population-based | Sepsis-associated ALI: 3′ haplotype | ||
| PreB cell colony-enhancing factor 1 (PBEF1) | T-1001G, C-1543T | 97 (2005) | EA (87 ALI:100 sepsis: 84 controls)/population-based | ALI risk: −1001G & −1001G:1543C haplotype; ALI protection: T-1543 | ||
| 10 (2007) | EA (375:787)/at-risk | ALI in septic & noninfectious ARDS: −1001G & −1001G:C-1543 haplotype | ||||
| Tumor necrosis factor (TNF/TNFA), lymphotoxin α (LTA/ TNFB) | G-308A (in TNFA) 1/2 Ncol (in TNFB) | 40 (2005) | EA (237:476)/at-risk | ARDS: −308A, −308A:TNFB1 haplotype | −308A allele conferred increased mortality in young ARDS patients | |
| VEGF A (VEGF/ VEGFA) | C936T | 66 (2005) | EA (117:137:103 respiratory failure)/both | ARDS: CT & TT genotypes | ARDS severity: CT & TT genotypes | |
| C-460T, C405G, C936T | 99 (2007) | EA (394:859)/at-risk | ARDS mortality: 936TT and haplotype TCT | |||
| Blood pressure regulation/ proteolysis | Angiotensin I converting enzyme 1 (ACE) | DIP | 60 (2002) | EA (96:88, 174 and 1906)/both | ARDS: deletion homozygotes | ARDS: deletion homozygotes |
| DIP | 48 (2006) | Asian (101:138)/both | ARDS mortality: ACE deletion | |||
| DIP | 2 (2007) | EA (84:200)/population-based | ARDS mortality: ACE deletion | |||
| Endothelial barrier regulation | Myosin light chain kinase (MYLK) | 28 SNPs in EA; 25 SNPs in AA | 33 (2006) | EA (92 ALI:114 sepsis:85 controls); AA (46 ALI:46 sepsis:61 controls)/population-based | ALI in EA: one SNP and one haplotype ALI in AA: 2 haplotypes | |
| 17 tagging SNPs | 24 (2008) | EA (48:83); AA (43:99)/at-risk | Trauma-assoc. ALI in AA: 3 variants and multiple haplotypes | |||
| Oxidant/antioxidant | Nuclear factor (erythroid-derived 2)-like 2 (NFE2L2 or NRF2) | C-617A | 63 (2007) | EA:AA =1:1 (30 cases & 60 matched controls; 2 controls for each case subject)/at-risk | Trauma-assoc. ALI: −617A | |
| Extracellular superoxide dismutase 3 (SOD3 or EC-SOD) | 28 SNPs | 8 (2008) | EA (251:179; 157:179)/population-based | Infection-associated ALI mortality: protective GCCT haplotype | ||
| NAD(P)H:quinine oxidoreductase 1 (NQO1) | 3 SNPs | 78 (2008) | EA and AA (93:185)/at-risk | Trauma-assoc. ALI: protective AC genotype at position −1221 | ||
| Innate immune response | Mannose-binding lectin (protein C) 2 (MBL2) | Codon −221, 52, 54, and 57 | 41 (2007) | EA (212:442)/at-risk | ARDS: homozygotes for variant codon 54B allele | ARDS severity: homozygotes for variant codon 54B allele |
| Transcription regulation | Nuclear factor of κ light polypeptide gene enhancer in B cells inhibitor, α (NFKBIA) | A-881G, C-826T, C-297T | 100 (2007) | EA (382:828)/at-risk | ARDS: GTC haplotype | |
| Nuclear factor of κ light polypeptide gene enhancer in B cells 1 (NFKB1) | −94ins/del ATTG | 3 (2007) | European (103 ARDS) | ARDS severity: D allele (homozygote deletion or heterozygote) | ||
| Iron ion homeostasis | Ferritin light polypeptide (FTL), heme oxygenase (decycling) 2 (HMOX2) | 3 SNPs in FTL; 5 SNPs in HMOX2 | 52 (2008) | EA (104:193)/population-based | Risk extrapulmonary ARDS: FTL −3381 GG; decreased risk pulmonary ARDS: HMOX2 haplotype | |
| Blood coagulation | Urokinase (PLAU) | 6 SNPs | 7 (2008) | EA (252:175)/population-based | ALI mortality: 6-SNP haplotype | |
| Coagulation factor V (F5) | Arg506Gln | 4 (2008) | European (106) | ARDS mortality: protective heterozygous genotype | ||
ALI, acute lung injury; ARDS, acute respiratory distress syndrome; DIP, deletion/insertion polymorphism in intron 16 of ACE gene defined by the absence (deletion, D) or presence (insertion, I) of a 287-bp repeat; EA, European American; AA, African American.
An obvious shortcoming of using healthy controls for studying associations with severe disease is that the significant associations may represent nothing more than “critical illness” genes, not necessarily the specific clinical trait of interest (i.e., ALI). However, how one defines “sick controls” is also problematic because they are potentially heterogeneous. As pointed out by Sapru and colleagues (81), hospital-based at-risk controls run the risk of eliminating the control group that actually contributes to the case group, at least in the context of ALI/ARDS, where the disease is rapidly evolving and misclassification of phenotype is a serious risk. As discussed by Tracey and Warren (90), even controlling for the outcome “severe sepsis” is problematic given that the disease is caused by different microbes affecting different organs but is classified as a single diagnosis. A solution they put forth is to select patients and controls on the basis of clinical and molecular (including genomic) data. However, the practicality of this solution given the a priori difficulties of providing informed consent to severely ill patients (and often surrogate consentees) and the huge costs of screening controls for these parameters before actually enrolling them is questionable. The case-cohort study design, in which a group of at-risk patients is followed for the development of disease (which mitigates selecting a control group), has been advocated (24, 78), but again, this approach is relatively costly as it requires the screening of many more patients to achieve a sufficiently powered case group (15).
In terms of power, this is perhaps one of the greatest concerns of the majority of publications on genetic studies of ALI to date. Sufficient power is established on the basis of a variety of factors, including the effect size of the causative genotype on the trait of interest, frequency of the risk allele, the degree of linkage disequilibrium (LD) between the genetic marker(s) typed and the actual causal allele, and the extent to which the allele frequencies match between the risk allele and genetic marker(s) tested (82). While there is no uniform figure that can be applied across studies, it is generally accepted that well-powered association studies are based on hundreds of samples (and depending on the number of variants to be studied, thousands). As illustrated by Table 1, less than 40% (12 out of 31) of the studies conducted have been performed in study groups where the cases and controls each exceeded 100 or the cohort exceeded 200, and very few of these papers (10, 24, 31, 41) addressed adequacy of power in the Methods section of the manuscript, assuring the reader that power was sufficient for detecting true associations.
Genomic Studies in Lung Injury: A Tool for the Discovery of Novel Candidate Genes
The sporadic nature of ALI has precluded one of the most conventional approaches for elucidating genes in a disease for which a strong genetic basis is suspected but not understood: genome-wide linkage mapping. Linkage mapping is a powerful approach whereby families with both affected and unaffected members are examined for marker loci that segregate with or are transmitted along with genes that are linked to a disease of interest. Whole genome approaches have the advantage of making no assumptions about the candidacy of particular loci. They require, however, families with multiple individuals having the disease trait, and given the acute nature of lung injury, this approach has not been feasible.
Because of these limitations, candidate gene-based approaches have been the preferred mode of study in the genetic epidemiology of lung injury, and for the practical reasons described above; these types of studies have been conducted among unrelated lung injury cases and controls selected on the basis of not having lung injury. In this approach, the major challenge is choosing the genes most likely to be involved in the trait, because lung injury is such a complex pathophysiological process involving a variety of molecular pathways. Microarray approaches allow the characterization and quantification of the global levels of transcript abundance, or expression profile, of all known genes in the human genome. Similar to the genome-wide linkage approach used in genetic studies, microarray-based analyses of genome-wide gene expression have the potential to provide an unbiased view of multiple molecular pathways simultaneously, thus allowing an integrated view of disease pathogenesis.
Several examples exist to date whereby a previously unknown candidate gene for lung injury was identified using high-throughput expression profiling, and subsequently demonstrated, through genetic association studies, to be relevant to disease (e.g., PBEF1) (97). One example of this approach is that by Grigoryev and colleagues (43), in which they utilized gene expression data derived from multiple animal models of mechanical ventilation and shear stress and identified five key biological process (ontologies), which included inflammation and immune responses, chemotaxis, cell proliferation, and blood coagulation. Using this same approach, it is possible to derive and prioritize a total of 85 candidate genes significantly differentially expressed in mechanical ventilation or LPS-induced ALI models across different species (mouse, canine, and human). Of these, 54 genes are affected by mechanical ventilation, 51 genes are affected by LPS, and 20 candidate genes are commonly differentially expressed in both mechanical ventilation and LPS-induced ALI models (unpublished data). The bioprocesses affected by a particular stimulus were subsequently identified using MAPPFinder software (27), which can link the gene expression data to the Gene Ontology (GO; http://www.geneontology.org) hierarchy. As a proof of concept, this approach has successfully identified gene ontologies previously linked to ALI, as well as novel ones that offer new insights into disease pathogenesis. Twenty major ontologies are listed according to the number of supporting evidences in the literature in Table 2 below.
Table 2.
Major biological processes identified by MAPPFinder based on 20 common ALI candidate genes and number of relevant literature references
| GO Name |
PubMatrix Terms |
||
|---|---|---|---|
| Acute lung injury | ARDS | Total | |
| Transport | 1,760 | 600 | 2,360 |
| Development | 1,476 | 619 | 2,095 |
| Inflammatory response | 829 | 289 | 1,118 |
| Nitric oxide biosynthesis | 266 | 92 | 358 |
| Phospholipid metabolism | 243 | 78 | 321 |
| Blood coagulation | 232 | 78 | 310 |
| Apoptosis | 246 | 57 | 303 |
| Cell motility | 244 | 54 | 298 |
| Transcription | 222 | 50 | 272 |
| Cell adhesion | 195 | 58 | 253 |
| Signal transduction | 187 | 36 | 223 |
| Chemotaxis | 156 | 32 | 188 |
| Cell communication | 150 | 29 | 179 |
| Collagen catabolism | 124 | 36 | 160 |
| Immune response | 118 | 32 | 150 |
| Heart development | 82 | 28 | 110 |
| Phagocytosis | 91 | 16 | 107 |
| Arginine metabolism | 80 | 13 | 93 |
| Muscle contraction | 56 | 18 | 74 |
| Cell cycle | 60 | 11 | 71 |
Shown are the number of references found by PubMatrix (http://pubmatrix.grc.nia.nih.gov) citing biological processes and relevant terms.
In addition to the identification of candidate genes by using the data to build detailed pathways of genes involved in lung injury, expression profiling using high-throughput microarray technology has the potential to distinguish disease subtypes, to follow the expression of genes across a time- and/or dosage-dependent gradient when exposed to a stimulus to reveal the complex control processes taking place in an individual cell, and it is also useful in validating genetic studies (34). A thorough review of genomic studies in lung injury was recently presented by Wurfel (96), and there are increasing numbers of reports demonstrating the utility of this tool.
Despite the enormous promise of this revolutionary technology, there are potential pitfalls and inherent difficulties in interpreting data from microarray-based, genome-wide expression studies. These may include difficulties in defining normal expression because of 1) heterogeneity of disease in the patients from which the samples were selected; 2) heterogeneity of the target tissue; 3) difficulties in establishing statistically valid comparability; and 4) cross platform variability (50, 93). Human subject research in ALI using high-throughput expression profiling is constrained given that collection of lung biopsies from critically ill patients is not feasible on a large scale, certainly to the extent that collection of material such as bronchoalveolar lavage-derived cell collection is from smokers and patients with other types of lung disease such as scleroderma or acute lung transplant rejection (96).
Established Genetic Associations in Lung Injury: A Pathway-Oriented Approach
With completion of the International HapMap Project (1), there has been a remarkable increase in the number of genetic association studies performed for complex traits, especially common complex lung diseases such as asthma (74, 92). Sadly, this has not been the case for some of the devastating lung diseases, including ALI and ARDS. Consider that, based on an updated review of the literature on genetic associations with lung injury, only 31 genetic association studies have been published, and of those, associations between polymorphisms in only 21 genes and ALI/ARDS phenotypes have been confirmed (11, 30, 37, 51, 73, 81), as summarized in Table 1.
The cellular and molecular mechanisms of ALI involve endothelial injury, epithelial injury, cytokine-mediated inflammation and injury, neutrophil-mediated injury, oxidant-mediated injury, ventilator-induced injury, coagulation pathway and fibrosing alveolitis (94). Recently, the role of altered iron mobilization and decompartmentalization in ALI was also reported (51). With this backdrop, it is reasonable that many of the candidate genes studied to date fall within these specific pathways. Some of the most compelling associations have been between specific gene polymorphisms and susceptibility and/or outcomes of ALI in genes encoding surfactant proteins (SPs) (39, 54, 64, 77); angiotensin converting enzyme (ACE) (2, 48, 60); proinflammatory cytokines [TNFA/TNFB (40), IL6 (31, 61, 73, 87), IL8 (44), PBEF1 (10, 97), MIF (32), and VEGF (66, 99)] and anti-inflammatory cytokine IL10 (38, 83). The most replicated genes include IL6, SFTPB, and ACE. Associations observed between variants in the myosin light chain kinase gene (MYLK), which plays a key role in endothelial cell barrier dysfunction, and ALI have been recently replicated in African American patients with ALI after major trauma (24, 33). Several associations have been reported between polymorphisms in genes encoding innate immune receptors (CD14) (91) and signaling molecule mannose-binding lectin (MBL) (41). Associations between transcription regulator NF-κB (NFKB1) and inhibitor NFKBIA and ARDS have also been reported recently (3, 100). After the first gene in regulating oxidant-mediated response in ALI-transcription factor NRF2 was reported (63), two additional genes in oxidant/antioxidant pathway were reported (SOD3/EC-SOD and NQO1) (8, 78). Recently the role of genes in blood coagulation pathway (PLAU and F5) (4, 7) has been reported, and genes involved in altered iron handling (i.e., excessive iron-catalyzed oxidative stress) may also play a role in ALI (52). A detailed summary of these candidate genes associated with lung injury is presented here.
Polymorphisms in cellular messenger genes.
Cytokines are peptides produced by diverse types of cells (including immune, endothelial, and nervous cells), which act as cell-to-cell messengers to regulate diverse processes, including immunity, inflammation, and cellular response to adverse conditions (85). Genetic variations in the proinflammatory cytokines TNFA, TNFB, MBL, and IL6 and the anti-inflammatory cytokine IL10 are the most extensively studied polymorphisms in relation to sepsis (75). A number of these polymorphisms have also been implicated in ALI (Table 1). Pre-B cell colony-enhancing factor (PBEF1), also known as visfatin, was identified as a novel biomarker in ALI by extensive analysis of microarray-based, gene expression profiling data, as described above (97), and has since been shown to exert three distinct activities related to cellular energetics and innate immunity. PBEF1 can be strongly induced by inflammatory stimuli in cells involved in innate immunity, specifically neutrophils, monocytes, and macrophages, as well as in endothelial and epithelial cells (57). Novel PBEF1 variants were identified through resequencing of PBEF1 exons (including exon-intron boundaries) and 2 kb of the 5′ UTR (97). Subsequent genotyping was conducted in a European American case-control population focusing on two promoter SNPs (T-1001G and C-1543T) that were physically close to mapped transcription factor binding sites. The T-1001G polymorphism was associated with an increased risk of sepsis-associated ALI, whereas the variant C-1543T polymorphism was associated with a protective effect. These findings were eventually replicated in an independent European American ARDS population (10), and since the initial report of an association between PBEF1 polymorphisms and disease, genetic associations have also been reported for other diseases that share an inflammation basis, such as type 2 diabetes (9).
As suggested above, associations with other cytokine genes are summarized in Table 1, indicating replication of association at both the gene level, and, in several cases, at the level of the variant. Upon close examination, however, seemingly contradictory results have been reported for several of these variants, rendering it difficult to determine whether true replication has been provided or not, and underscoring a phenomenon that is not unique to genetics studies of ALI. For example, Gong et al. (38) reported that the high IL-10-producing −1082GG genotype was associated with the development of ARDS, but only in the presence of a significant interaction between the −1082GG genotype and age. Elsewhere, Schroeder et al. (83) reported in a study from Europe that carriers of the same −1082GG genotype were less likely to develop acute respiratory failure after major trauma. One explanation for this phenomenon was attributed to “ascertainment bias” (23), owed to the manner by which subjects were selected into the study, interactions with clinical variables (in this case, age) that potentially modify genetic associations, or because of the diverse etiological mechanisms (i.e., sepsis, pneumonia, trauma, massive transfusion) that predisposed patients to the condition under study. Certainly, as in the case of these two studies, replication of a disease-marker association where the risk allele is reversed in the follow-up study, also referred to as a “flip-flop” association (53), has been increasingly observed as more and more association studies are performed in general, and have raised the question of whether these associations are confirmations or contradictions. Lin and colleagues (53) argue that, certainly in the case of opposite alleles for the same disease in the same ethnic group, particularly for a genuine causal variant, a flip-flop association is unlikely. However, for studies performed in different populations (which is usually the case in replication studies, and certainly the case for the Gong et al. vs. the Schroeder et al. studies on IL10 G-1082A), flip-flop associations may highlight heterogeneity due to differences in the genetic background of different populations, which is discussed in greater detail in the next section. The Gong et al. study was a nested case-control study focused on cases of those of European descent living in Boston with at least one defined risk factor for ARDS (sepsis, pneumonia, trauma, massive transfusion, or aspiration) and intensive care unit-based controls with the same risk factors but without ARDS; the primary outcome was organ failure and 60-day mortality. The Schroeder et al. study was a relatively small cohort study of those of European ancestry living in Berlin, Germany, with multiple trauma, where the primary outcome measure was acute respiratory failure. Moreover, the variant of interest is not functional and is therefore likely to be in linkage disequilibrium with the true (causal) variant. Thus, the differences in associations observed in this particular example cannot be reconciled without further efforts toward replication of the IL10 G-1082A variant in other independent populations selected for the same phenotypes.
Polymorphisms in endothelial-mediated lung injury-related genes.
Both in vivo and in vitro studies have demonstrated that lung endothelial cell barrier dysfunction plays a key role in the pathophysiology of ALI. The nonmuscle endothelial cell myosin light chain kinase (EC MLCK), encoded by the myosin light chain kinase (MYLK) gene, is centrally involved in cytoskeleton rearrangement participating in apoptosis, inflammation, cell trafficking, cell division, and angiogenesis (28). Lung edema formation in ALI, which is caused by injury to the lung microvascular endothelium, also involves the MLCK-dependent cytoskeletal rearrangement. We first explored MYLK as a novel candidate gene in sepsis and sepsis-associated ALI because of its pivotal role in the disease process, and through sequencing, polymorphisms were identified for which significant associations were observed for both severe sepsis and sepsis-associated ALI (33). A novel observation in our initial report was the difference in risk-conferring haplotypes according to ethnicity in two case-control populations (African Americans vs. European Americans) ascertained under identical protocols. Upon close examination, we observed striking differences in genetic structure, or LD, of the two populations according to the 28 SNPs that were genotyped, and considered these differences, as well as potential interactions with other (untyped) genetic markers or environmental risk factors, as an explanation for the greater risk of ALI in the African American group.
The association between MYLK variants and the development of ALI was recently replicated in a study of trauma-induced ALI patients, although the risk allele for each of the three functional variants (Thr335Thr, Pro147Ser, His21Pro) was reversed from our original report. In comparisons between associations in the combined ethnic groups vs. the individual ethnic groups, Christie and colleagues (24) noted that associations appeared to be driven by the African Americans, a finding that supported our original report. Although modestly powered to evaluate each ethnic group independently (i.e., less than 100 individuals in each case or control group), it is important to highlight the group's own contention that differences in the pathophysiology of ALI between major trauma and sepsis as an etiological factor could account for the flip-flop effect described in the section above. As described elsewhere, the flip-flop effect is not expected in the event that the association is for a genuine causal variant and in samples ascertained from a common population in the same manner (53); however, if we consider that 1) even though the three MYLK variants are functional variants, there is no evidence as of yet that they are “causal”; 2) although the populations showing the strongest association in both studies are African American, there is known heterogeneity among African Americans in the U.S. dependent on where they reside (76); and 3) the outcome of interest in the two studies was distinctly different (i.e., sepsis-associated vs. trauma-associated ALI), it is perhaps not surprising that inconsistent patterns of association are observed across the studies.
ALI is characterized by overlapping phases: exudative (early/ acute) and fibroproliferative (subacute/late phase) (58, 94). Animal studies of ALI suggested that circulating endothelial precursor cells are critical in the repair of alveolar endothelium in the late fibroproliferative phase of ALI (14). Higher lung levels of VEGF, a mitogen that specifically acts on endothelial cells and has various effects, are associated with recovery from ALI (89). VEGF is also a chemoattractant for endothelial precursors. Medford et al. (66) first reported that the variant allele of the C/T SNP at +936 of the 3′ UTR of the gene associated with lower VEGF plasma levels and conferred increased risk for the development of the disease, whereas Zhai (99) et al. did not find any increased risk of ALI associated with the C936T polymorphism but reported only an association with the risk of mortality among patients with ARDS. These results provide grounds for future research aimed at investigating whether or not host genetic factors also influence the recovery of ALI.
Polymorphisms in innate immune receptor and signaling molecule genes.
Sepsis describes a complex clinical syndrome as a result of a systemic inflammatory response to live bacteria and/or bacterial products, and sepsis is the most common clinical risk factor for indirect lung injury (95). During the past decade, functional and association studies involving genetic polymorphisms in essential host defense genes, including innate immune receptors and signaling molecules, cytokines and coagulation factors, have provided important insights into the mechanisms involved in the pathogenesis of sepsis and its induced organ dysfunction (6). A number of genetic associations have been reported between sepsis and polymorphisms in innate immune receptor genes, including TLR4 (5, 18, 55), CD14 (36, 86), and MBL (35, 42, 86). In several of these studies, the specific polymorphism associated with the trait has also been associated with ALI/ARDS, i.e., the variant B allele of codon 54 of MBL gene (41). TLRs function as the pattern recognition receptors through which the innate immune system recognizes microbial pathogens, and TLR4 is one of the most extensively studied. Genetic association between two coding SNPs, Asp299Gly and Thr399Ile, and increased susceptibility to bacterial infection and severe sepsis/septic shock have been reported (5, 18, 55). Similarly, CD14 is a ubiquitous pattern recognition receptor, specific for LPS and a variety of other ligands, and is expressed primarily on immune and endothelial cells. CD14 also acts as a coreceptor for TLR4 (86). A soluble form (sCD14) is shed by monocytes to facilitate LPS signaling for all other cells (49). The relatively common C-260T variant (initially identified as C-159T) functions by decreasing the affinity of Sp protein binding and subsequently enhancing transcriptional activity, and is associated with increased sCD14, which is markedly increased in bronchoalveolar lavage of patients with ARDS (62). In a Canadian study, the T allele was associated with increased prevalence of gram-negative infections and sepsis (86), and in a French multicenter study, an association with sepsis mortality was reported (36). MBL is a circulating serum protein that recognizes and binds carbohydrate structures on the surface of infectious agents and activates the complement system through MBL-associated serine proteases (88). Evidence for association with sepsis was found in several studies (35, 42, 86), and recently Gong and colleagues (41) reported that patients homozygous for the variant codon 54B allele (54BB) had increased odds of ARDS when compared with heterozygotes and homozygotes for the wild-type allele, especially among patients with septic shock. Given the prominent role that sepsis plays in lung injury etiology, it is not surprising that genes associated with sepsis and severity of sepsis are likely candidates for ALI.
Polymorphisms in oxidant-mediated injury genes.
Studies have shown that the transcription factor NRF2 appears to be an essential regulatory element in response to oxidant injury (19–21). Positional cloning studies in inbred mice identified Nrf2 as a candidate susceptibility gene for hyperoxia-induced lung inflammation and injury, a model of ALI (19, 20). Marzec and coworkers (63) performed resequencing to scan the entire coding region, and the proximal 1 kb of the promoter and genotyping was performed on two promoter SNPs (−617 and −651); they subsequently found that patients who were homozygous for the −617 minor “A” allele had a significantly higher risk for developing ALI after major trauma relative to patients with the wild type (−617 CC). Although this is a relatively new group of candidate genes in the field of lung injury genetic epidemiology, there has been a tremendous number of genetic associations between genes associated with defense against oxidant stress and other complex lung diseases, including glutathione synthesis-related genes [e.g., GCLC and cystic fibrosis (65)] and superoxide dismutase (SOD) genes [e.g., SOD3 and COPD (98)]. Recent work by Arcaroli et al. (8) confirmed the association between haplotypes in extracellular superoxide dismutase 3 gene (SOD3 or EC-SOD) and mortality in patients with infection-associated ALI, and another report suggested that the A-1221C variant of the NAD(P)H:quinine oxidoreductase 1 (NQO1), a phase II antioxidant gene, conferred higher risk for trauma-associated ALI (78).
Polymorphisms in blood coagulation pathway genes.
Polymorphisms in genes encoding proteins of the coagulation pathways [i.e., protein C and plasminogen activator inhibitor 1 (PAI-1)] are associated with altered levels of their respective mediators and phenotypes of disordered coagulation (81). Recently, Arcaroli et al. (7) reported the association between haplotypes in urokinase gene (PLAU), which encodes a serine protease that cleaves plasminogen to generate plasmin, a potent mediator of fibrinolysis, and outcomes (both 60-day mortality and ventilator-free days) from ALI. Growing evidence also supported that a factor V Leiden (F5) genotype influenced survival in patients with ARDS (4). Thus, this group of candidates represents a novel area in the field of lung injury genetics.
Coassociations between ALI Candidate Gene Variants and Lung Inflammatory Phenotypes: The Common Variant/Multiple Disease Hypothesis
Sepsis is one of the main risk factors leading to the development of ALI, and, as demonstrated above, polymorphisms in genes previously implicated in sepsis function as strong candidate genes for lung injury. As summarized, several sepsis-associated genes, such as ACE, IL6, TNFA/TNFB, and MBL, have already been associated with ALI susceptibility. Additional sepsis-associated genes, such as coagulation pathway-related polymorphisms, were also involved in ALI (4, 7, 81). In contrast, genes of relevance for risk of ALI (e.g., MYLK and MIF) may also constitute potential candidates for susceptibility to other distinct, but possibly related, clinical syndromes, such as allergic asthma (34). Macrophage migration inhibitory factor (MIF) is a proinflammatory cytokine central to the response to endotoxemia, and a putative biomarker in ALI. As a pleiotropic lymphocyte and macrophage cytokine, MIF is likely to play an important role in innate immunity. Haplotypes in the 3′ end of the MIF gene were associated with severe sepsis and sepsis-associated ALI (32). Evidence for significant association between the functional MIF gene promoter SNP (−173G/C) and −794 [CATT]5-8 repeat polymorphisms and atopy was similarly reported in a Japanese population (46), and a significant association between asthma and the 5-CATT MIF allele was subsequently confirmed by Mizue et al. (68). Collectively, these findings support the “common variant/multiple disease” hypothesis, which is an extension of the common disease/common variant hypothesis, and suggests that certain disease genes may not be disease specific and may contribute to related clinical phenotypes (34). Indeed, these sorts of examples of genetic association of the same gene (or allele) to multiple, related disorders abound (http://geneticassociationdb.nih.gov). Given the prominent role that certain pathways, such as host defense, play across various complex lung diseases, it is likely that many more coassociations will be observed in the near future.
General Insights into the Pathophysiological Pathways Related to Lung Injury
For complex inflammatory diseases such as ALI, it is anticipated that many genes will be involved and multiple variants will contribute to the alteration of gene function and expression. Because the effects of these genes/polymorphisms are typically relatively modest, it is generally assumed that variants in multiple genes cooperate in an additive or synergistic manner to impact disease risk. This phenomenon is referred to as “epistasis.” One approach toward characterizing potential gene-gene interactions and systematically evaluating the role of ALI genes/polymorphisms in disease susceptibility is pathway analysis, whereby hierarchical clustering of genes is performed considering their functional closeness in a pathway or risks imparted independently from other genes in the pathway. Using the program Ingenuity Pathways Analysis (Ingenuity Systems: www.analysis.ingenuity.com), Loza and coworkers (56) identified 1,027 key inflammation-related genes targeting various phases of inflammation responses and associated 17 subpathways that can further facilitate pathway-focused genetic association analyses for inflammatory diseases. The Ingenuity Pathways knowledge base is a web-based entry tool developed by Ingenuity Systems to characterize genes according to the predefined canonical pathway(s) into which they fit, and also to investigate the extent to which genes identified in a particular study (e.g., microarray-based studies) are in shared networks and may cooperate in a synergistic or an additive manner to impact risk of disease.
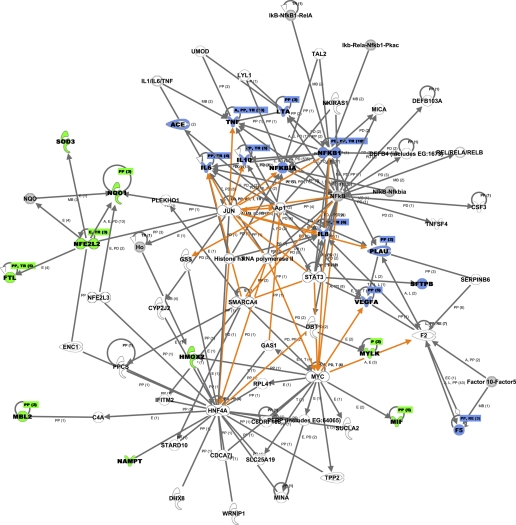
As a proof of concept, we evaluated the 21 genes that have been well established as ALI/ARDS genes and summarized in Table 1 using the Ingenuity Pathway Analysis. Consequently, two major networks were revealed (Table 3). Twelve genes (ACE, IL6, IL8, IL10, TNF, LTA, NFKB1, NFKBIA, SFTPB, VEGFA, PLAU, F5) that have previously been associated with lung injury were clustered into a major network related to 1) “immune and lymphatic system development and function”; 2) tissue morphology; and 3) cellular movement. Moreover, an additional 24 related genes/molecules belong to the same network. Using the multiplex literature search tool, PubMatrix (http://pubmatrix.grc.nia.nih.gov), we found evidence supporting the involvement of 9 out of the 24 related genes (Ap1, DBT, DEFB4, F2, Histone h3, MICA, RNA polymerase II, STAT3, and TNFSF4) in ALI/ARDS. Furthermore, another nine genes previously associated with ALI [FTL, HMOX2, MBL2, MIF, MYLK, PBEF1 (NAMPT), NFE2L2, SOD3, NQO1] were in the second major network related to “cellular compromise, molecular transport, hematological system development and function.” Among the 26 related genes in the same network, evidence was found in the literature for eight genes (C4A, ENC1, Ho, JUN, MINA, MYC, NQO, PPCS) supporting their involvement in ALI/ARDS pathogenesis. The top 5 “diseases and disorders” and “molecular and cellular functions” suggested by the two networks derived from 21 ALI genes were also summarized in Table 3. Notably, the top molecular and cellular functions represented in current association studies fit within the major biological processes underlying ALI development suggested by the common ALI candidate genes derived from diverse across species microarray experiments (Table 2). While this list is not intended to be a comprehensive summary of all genes potentially related to ALI pathology, it serves as an example of the power of this approach in selecting optimal candidates for genetic association studies. Figure 1 illustrates the overlay of the two major networks that are summarized in Table 3. Other tools such as MetaCore, a pathway analysis and data mining tool provided by GeneGo (http://www.genego.com), can serve a similar purpose.
Table 3.
Summary of the 2 networks and their relevant functions derived from the 21 published ALI genes
| Published ALI Genes/Molecules in Network | Other Related Genes/Molecules in Network | Associated Diseases and Disorders (Top 5) | Associated Molecular and Cellular Functions (Top 5) |
|---|---|---|---|
| Network 1 (12 genes): ACE, IL6, IL8, IL10, TNF, LTA, NFKB1, NFKBIA, SFTPB, VEGFA, PLAU, F5 | Ap1, CSF3, DBT, DEFB103A, DEFB4 (includes EG:1673), F2, Factor 10-Factor5, Histone h3, IκB-NfκB1-RelA, Iκb-Rela-Nfκb1-Pkac, IL1/IL6/TNF, LYL1, MICA, NFκB, NfκB-Nfκbia, NKIRAS1, REL/RELA/RELB, RNA polymerase II, SERPINB6, STAT3, TAL2, TNFSF4, UMOD | 1. Organismal injury and abnormalities; | 1. Cellular movement; |
| Network 2 (9 genes): FTL, HMOX2, MBL2, MIF, MYLK, PBEF1 (NAMPT), NFE2L2, SOD3, NQO1 | C4A, C6ORF108, CDCA7L, CYP2J2, DHX8, ENC1, GAS1, GSS, HNF4A, Ho, IFITM2, JUN, MINA, MYC, NFE2L3, NQO, PERP (includes EG:64065), PLEKHO1, PPCS, RPL41, SLC25A19, SMARCA4, STARD10, SUCLA2, TPP2, WRNIP1 | 2. Immunological disease; | 2. Cell death; |
| 3. Hematological disease; | 3. Lipid metabolism; | ||
| 4. Respiratory disease; | 4. Small molecule biochemistry; | ||
| 5. Inflammatory disease | 5. Cell-to-cell signaling and interaction |
Fig. 1.
Networks revealed through the Ingenuity Pathways Analysis based on 21 ALI genes established in acute lung injury genetic association studies. The major network of 36 genes (the 12 published ALI genes are highlighted in blue) was merged with the second network of 35 genes (the 9 genes previously associated with ALI are highlighted in green). Molecules highlighted in gray represent molecules that belong to a group (multiple specific molecules with a similar function) or a protein complex made up of multiple protein subunits in the network. Genes/molecules are connected according to their relationships, and the newly added relationships that connect the two networks are also highlighted in orange. The top 5 diseases/disorders related to genes/molecules in the two networks include 1) organismal injury and abnormalities; 2) immunological disease; 3) hematological disease; 4) respiratory disease; and 5) inflammatory disease. Top molecular and cellular functions suggested by the networks are cellular movement, cell death, lipid metabolism, small molecule biochemistry, and cell-to-cell signaling and interaction.
Methodology To Date for Genetic Association Studies in ALI
In the past three decades, substantial progress has been made in understanding the mechanisms that underlie ALI/ARDS. Although there has indeed been an increase in the number of published genetic association studies focusing on lung injury over the past few years, with increasing reliance on novel and potentially powerful tools such as microarray-based approaches and computationally intensive techniques for interpretation of findings, the field has far to go. Improvements in the methodology used in genetic association studies, including population selection, phenotype definition, determination of appropriate sample size, genotype data quality control, statistical methodologies to adjust confounding factors such as multiple testing and population stratification and study replication will greatly facilitate discovery of true associations important in the care of ALI/ARDS patients. From recently published studies, we do see increased reproducibility with 8 out of the 21 genes associated with all-cause ALI susceptibility and/or outcome were replicated. Another major advance is using haplotype tagging SNP (htSNP) to capture gene-wide common variations. The premise behind this approach is that in regions of high LD, and because many SNPs are frequently inherited together, only certain SNPs are selected to fully represent a candidate gene. Several htSNP methods exist, and the selection of which method to use is somewhat determined by the structure of the population under study (12, 17, 25). However, as highlighted in Table 1, of the studies published so far, very few have focused on non-white patient populations. Because ALI is typical of other complex lung disorders whereby ethnic disparities exist in terms of morbidity and mortality (11, 72), a greater focus on diverse populations is warranted. The role of ethnicity is only one component of the observed heterogeneity of disease manifestations and outcomes. ALI is characterized by overlapping phases: exudative (early/acute) and fibroproliferative (subacute/late phase) (58, 94). Some patients recover during the acute phase, but most progress to the subacute phase, which renders ALI/ARDS one of the major causes of pulmonary and nonpulmonary morbidity in patients after discharge (26, 58). It is reasonable that the variation in recovery may be related in part to the underlying genetic characteristics of the individual patient. The identification of genetic factors associated with susceptibility, severity, and outcome/recovery constitutes a necessary step toward personalized treatment. Finally, and importantly, of the published association studies in ALI summarized in Table 1, many studies have been underpowered to detect true false-negative results that could be one of the potential confounding factors. To be fair, this is understandable given the profound challenges in amassing large datasets of what is in actuality an uncommon disease. The use of derivation and validation subsets of patients is one solution to these limitations and a failure to reproduce findings to date (15). Flores et al. (29) evaluated the quality of 29 published positive associations for 16 genes in ALI based on current recommendations suggested for genetic association studies (16). Using a 10-point quality scoring system derived from 14 criteria, they found the studies had an intermediate quality score of 4.62, suggesting that more and better designed studies with larger sample sizes are needed, including the replication of previous findings, and should be more inclusive of population groups other than European descent (29).
Genome-wide association studies offer a potential solution to the limitation in candidate gene approaches; that is, that each causal variant in each candidate gene will only make a modest contribution to overall heritability (45). By screening dense panels of markers representing the most common genetic variation across the genome, it is assumed that many more risk variants will be identified than what could be accomplished using conventional, candidate gene-based studies. High-density genome-wide association (GWA) studies to date have demonstrated the success and robustness of this approach in identifying genetic variants or regions associated with nearly 40 complex diseases or traits (59), including complex lung diseases such as asthma (69). Although this approach has yet to be applied to ALI or sepsis, the limitations of HapMap-based GWA studies discovered to date from these preliminary successes should be carefully considered (22, 47, 59). GWA studies often require very large numbers of samples (i.e., thousands of cases and controls needed to obtain sufficient statistical power to identify the alleles of reasonable effect size); conversely, the GWA design has not proven successful for identifying rare and structural variants. Because of the manner by which the chips are designed (i.e., random “tagging” SNPs are selected as a priority over design of markers of causal variants), the correlations identified with disease do not typically implicate causation, and follow-up studies to identify the specific mutation are not always straightforward. Extensive follow-up and replication of findings in the discovery population are essential to rule out false positives. Given that ALI is not as prevalent as other complex traits such as asthma, and given the complex nature and heterogeneity of the phenotype, there are certainly obstacles with GWA studies in ALI. Nevertheless, it can be anticipated that more carefully designed primary and replication studies focused on candidate genes identified in known cellular pathways implicated in ALI pathogenesis, or genes supported by plausible biological hypothesis, will allow for alternatives to the GWA design and lead to identification of gene-gene and gene-environment interactions (i.e., infection, mechanical ventilation). It is reasonable that collaborative efforts towards establishing large DNA banks from ALI/ARDS patients will ultimately satisfy the requirements for successful GWA studies and integration of data across studies from “omics” (i.e., genomics and proteomics) to facilitate identification of novel targets and cellular pathways specific to the disease.
Conclusion
In summary, additional association studies based on novel candidate genes and a deeper appreciation for the relevant pathways involved in lung injury have advanced the field of genetic epidemiology of ALI and ARDS. The integration of novel tools for elucidating candidate genes, such as microarray-based approaches and computational approaches including pathway analysis, has broadened our understanding of the different groups of genes involved in disease manifestation and outcome. Originally designed for extracting new biological insight from high-throughput genomic studies of human disease, pathway analysis can also be a powerful approach to facilitate the detection of gene-gene interactions and pathway-focused genetic analysis.
Recent advances in genotyping approaches have already been utilized, and the most robust approach to date, genome-wide association studies, is yet to be tested in either ALI or sepsis. Ultimately, a clearer understanding of the genetic determinants conferring risk to ALI, ARDS, and other associated phenotypes, will pave the way toward a more personalized approach of therapy and resolution of disease.
GRANTS
This review represents the efforts of investigators participating in the National Heart, Lung, and Blood Institute-funded SCCOR on “Molecular Approaches to Ventilator-Associated Lung Injury” (P50-HL-073994) and the projects entitled “Genetic Modifiers in Acute Lung Injury” and “Genomics and Genotyping Core.” K. C. Barnes was supported in part by the Mary Beryl Patch Turnbull Scholar Program.
Acknowledgments
Special thanks to Dr. Dmitry Grigoryev for his work on the expression profiling data. Pat Oldewurtel provided technical assistance in the submission of this manuscript.
REFERENCES
- 1.The International HapMap Project. Nature 426: 789–796, 2003. [DOI] [PubMed] [Google Scholar]
- 2.Adamzik M, Frey U, Sixt S, Knemeyer L, Beiderlinden M, Peters J, Siffert W. ACE I/D but not AGT (−6)A/G polymorphism is a risk factor for mortality in ARDS. Eur Respir J 29: 482–488, 2007. [DOI] [PubMed] [Google Scholar]
- 3.Adamzik M, Frey UH, Rieman K, Sixt S, Beiderlinden M, Siffert W, Peters J. Insertion/deletion polymorphism in the promoter of NFKB1 influences severity but not mortality of acute respiratory distress syndrome. Intensive Care Med 33: 1199–1203, 2007. [DOI] [PubMed] [Google Scholar]
- 4.Adamzik M, Frey UH, Riemann K, Sixt S, Lehmann N, Siffert W, Peters J. Factor V Leiden mutation is associated with improved 30-day survival in patients with acute respiratory distress syndrome. Crit Care Med 36: 1776–1779, 2008. [DOI] [PubMed] [Google Scholar]
- 5.Agnese DM, Calvano JE, Hahm SJ, Coyle SM, Corbett SA, Calvano SE, Lowry SF. Human toll-like receptor 4 mutations but not CD14 polymorphisms are associated with an increased risk of gram-negative infections. J Infect Dis 186: 1522–1525, 2002. [DOI] [PubMed] [Google Scholar]
- 6.Arcaroli J, Fessler MB, Abraham E. Genetic polymorphisms and sepsis. Shock 24: 300–312, 2005. [DOI] [PubMed] [Google Scholar]
- 7.Arcaroli J, Sankoff J, Liu N, Allison DB, Maloney J, Abraham E. Association between urokinase haplotypes and outcome from infection-associated acute lung injury. Intensive Care Med 34: 300–307, 2008. [DOI] [PubMed] [Google Scholar]
- 8.Arcaroli JJ, Hokanson JE, Abraham E, Geraci M, Murphy JR, Bowler RP, Dinarello CA, Silveira L, Sankoff J, Heyland D, Wischmeyer PE, Crapo JD. EC-SOD haplotypes are associated with acute lung injury and mortality. Am J Respir Crit Care Med 179: 105–112, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bailey SD, Loredo-Osti JC, Lepage P, Faith J, Fontaine J, Desbiens KM, Hudson TJ, Bouchard C, Gaudet D, Perusse L, Vohl MC, Engert JC. Common polymorphisms in the promoter of the visfatin gene (PBEF1) influence plasma insulin levels in a French-Canadian population. Diabetes 55: 2896–2902, 2006. [DOI] [PubMed] [Google Scholar]
- 10.Bajwa EK, Yu CL, Gong MN, Thompson BT, Christiani DC. Pre-B-cell colony-enhancing factor gene polymorphisms and risk of acute respiratory distress syndrome. Crit Care Med 35: 1290–1295, 2007. [DOI] [PubMed] [Google Scholar]
- 11.Barnes KC Genetic determinants and ethnic disparities in sepsis-associated acute lung injury. Proc Am Thorac Soc 2: 195–201, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265, 2005. [DOI] [PubMed] [Google Scholar]
- 13.Bersten AD, Edibam C, Hunt T, Moran J. Incidence and mortality of acute lung injury and the acute respiratory distress syndrome in three Australian states. Am J Respir Crit Care Med 165: 443–448, 2002. [DOI] [PubMed] [Google Scholar]
- 14.Burnham EL, Taylor WR, Quyyumi AA, Rojas M, Brigham KL, Moss M. Increased circulating endothelial progenitor cells are associated with survival in acute lung injury. Am J Respir Crit Care Med 172: 854–860, 2005. [DOI] [PubMed] [Google Scholar]
- 15.Cardon LR, Bell JI. Association study designs for complex diseases. Nat Rev Genet 2: 91–99, 2001. [DOI] [PubMed] [Google Scholar]
- 16.Chanock SJ, Manolio T, Boehnke M, Boerwinkle E, Hunter DJ, Thomas G, Hirschhorn JN, Abecasis G, Altshuler D, Bailey-Wilson JE, Brooks LD, Cardon LR, Daly M, Donnelly P, Fraumeni JF Jr, Freimer NB, Gerhard DS, Gunter C, Guttmacher AE, Guyer MS, Harris EL, Hoh J, Hoover R, Kong CA, Merikangas KR, Morton CC, Palmer LJ, Phimister EG, Rice JP, Roberts J, Rotimi C, Tucker MA, Vogan KJ, Wacholder S, Wijsman EM, Winn DM, Collins FS. Replicating genotype-phenotype associations. Nature 447: 655–660, 2007. [DOI] [PubMed] [Google Scholar]
- 17.Chi PB, Duggal P, Kao WH, Mathias RA, Grant AV, Stockton ML, Garcia JG, Ingersoll RG, Scott AF, Beaty TH, Barnes KC, Fallin MD. Comparison of SNP tagging methods using empirical data: association study of 713 SNPs on chromosome 12q14.3–12q2421 for asthma and total serum IgE in an African Caribbean population. Genet Epidemiol 30: 609–619, 2006. [DOI] [PubMed] [Google Scholar]
- 18.Child NJ, Yang IA, Pulletz MC, de Courcy-Golder K, Andrews AL, Pappachan VJ, Holloway JW. Polymorphisms in Toll-like receptor 4 and the systemic inflammatory response syndrome. Biochem Soc Trans 31: 652–653, 2003. [DOI] [PubMed] [Google Scholar]
- 19.Cho HY, Jedlicka AE, Reddy SP, Kensler TW, Yamamoto M, Zhang LY, Kleeberger SR. Role of NRF2 in protection against hyperoxic lung injury in mice. Am J Respir Cell Mol Biol 26: 175–182, 2002. [DOI] [PubMed] [Google Scholar]
- 20.Cho HY, Jedlicka AE, Reddy SP, Zhang LY, Kensler TW, Kleeberger SR. Linkage analysis of susceptibility to hyperoxia. Nrf2 is a candidate gene. Am J Respir Cell Mol Biol 26: 42–51, 2002. [DOI] [PubMed] [Google Scholar]
- 21.Cho HY, Reddy SP, Debiase A, Yamamoto M, Kleeberger SR. Gene expression profiling of NRF2-mediated protection against oxidative injury. Free Radic Biol Med 38: 325–343, 2005. [DOI] [PubMed] [Google Scholar]
- 22.Christensen K, Murray JC. What genome-wide association studies can do for medicine. N Engl J Med 356: 1094–1097, 2007. [DOI] [PubMed] [Google Scholar]
- 23.Christie JD Interleukin-10, age and acute lung injury genetics: the action is in the interaction. Eur Respir J 27: 669–670, 2006. [DOI] [PubMed] [Google Scholar]
- 24.Christie JD, Ma SF, Aplenc R, Li M, Lanken PN, Shah CV, Fuchs B, Albelda SM, Flores C, Garcia JG. Variation in the myosin light chain kinase gene is associated with development of acute lung injury after major trauma. Crit Care Med 36: 2794–2800, 2008. [DOI] [PubMed] [Google Scholar]
- 25.Daly MJ, Rioux JD, Schaffner SF, Hudson TJ, Lander ES. High-resolution haplotype structure in the human genome. Nat Genet 29: 229–232, 2001. [DOI] [PubMed] [Google Scholar]
- 26.Davidson TA, Caldwell ES, Curtis JR, Hudson LD, Steinberg KP. Reduced quality of life in survivors of acute respiratory distress syndrome compared with critically ill control patients. JAMA 281: 354–360, 1999. [DOI] [PubMed] [Google Scholar]
- 27.Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. MAPPFinder: using gene ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol 4: R7, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dudek SM, Garcia JG. Cytoskeletal regulation of pulmonary vascular permeability. J Appl Physiol 91: 1487–1500, 2001. [DOI] [PubMed] [Google Scholar]
- 29.Flores C, Del Mar Pino-Yanes M, Villar J. A quality assessment of genetic association studies supporting susceptibility and outcome in acute lung injury. Crit Care 12: R130, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flores C, Ma SF, Maresso K, Ahmed O, Garcia JG. Genomics of acute lung injury. Semin Respir Crit Care Med 27: 389–395, 2006. [DOI] [PubMed] [Google Scholar]
- 31.Flores C, Ma SF, Maresso K, Wade MS, Villar J, Garcia JG. IL6 gene-wide haplotype is associated with susceptibility to acute lung injury. Transl Res 152: 11–17, 2008. [DOI] [PubMed] [Google Scholar]
- 32.Gao L, Flores C, Fan-Ma S, Miller EJ, Moitra J, Moreno L, Wadgaonkar R, Simon B, Brower R, Sevransky J, Tuder RM, Maloney JP, Moss M, Shanholtz C, Yates CR, Meduri GU, Ye SQ, Barnes KC, Garcia JG. Macrophage migration inhibitory factor in acute lung injury: expression, biomarker, and associations. Transl Res 150: 18–29, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao L, Grant A, Halder I, Brower R, Sevransky J, Maloney JP, Moss M, Shanholtz C, Yates CR, Meduri GU, Shriver MD, Ingersoll R, Scott AF, Beaty TH, Moitra J, Ma SF, Ye SQ, Barnes KC, Garcia JG. Novel polymorphisms in the myosin light chain kinase gene confer risk for acute lung injury. Am J Respir Cell Mol Biol 34: 487–495, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao L, Grant AV, Rafaels N, Stockton-Porter M, Watkins T, Gao P, Chi P, Munoz M, Watson H, Dunston G, Togias A, Hansel N, Sevransky J, Maloney JP, Moss M, Shanholtz C, Brower R, Garcia JG, Grigoryev DN, Cheadle C, Beaty TH, Mathias RA, Barnes KC. Polymorphisms in the myosin light chain kinase gene that confer risk of severe sepsis are associated with a lower risk of asthma. J Allergy Clin Immunol 119: 1111–1118, 2007. [DOI] [PubMed] [Google Scholar]
- 35.Garred P, Strom J, Quist L, Taaning E, Madsen HO. Association of mannose-binding lectin polymorphisms with sepsis and fatal outcome, in patients with systemic inflammatory response syndrome. J Infect Dis 188: 1394–1403, 2003. [DOI] [PubMed] [Google Scholar]
- 36.Gibot S, Cariou A, Drouet L, Rossignol M, Ripoll L. Association between a genomic polymorphism within the CD14 locus and septic shock susceptibility and mortality rate. Crit Care Med 30: 969–973, 2002. [DOI] [PubMed] [Google Scholar]
- 37.Gong MN Genetic epidemiology of acute respiratory distress syndrome: implications for future prevention and treatment. Clin Chest Med 27: 705–724, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gong MN, Thompson BT, Williams PL, Zhou W, Wang MZ, Pothier L, Christiani DC. Interleukin-10 polymorphism in position −1082 and acute respiratory distress syndrome. Eur Respir J 27: 674–681, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gong MN, Wei Z, Xu LL, Miller DP, Thompson BT, Christiani DC. Polymorphism in the surfactant protein-B gene, gender, and the risk of direct pulmonary injury and ARDS. Chest 125: 203–211, 2004. [DOI] [PubMed] [Google Scholar]
- 40.Gong MN, Zhou W, Williams PL, Thompson BT, Pothier L, Boyce P, Christiani DC. −308GA and TNFB polymorphisms in acute respiratory distress syndrome. Eur Respir J 26: 382–389, 2005. [DOI] [PubMed] [Google Scholar]
- 41.Gong MN, Zhou W, Williams PL, Thompson BT, Pothier L, Christiani DC. Polymorphisms in the mannose binding lectin-2 gene and acute respiratory distress syndrome. Crit Care Med 35: 48–56, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gordon AC, Waheed U, Hansen TK, Hitman GA, Garrard CS, Turner MW, Klein NJ, Brett SJ, Hinds CJ. Mannose-binding lectin polymorphisms in severe sepsis: relationship to levels, incidence, and outcome. Shock 25: 88–93, 2006. [DOI] [PubMed] [Google Scholar]
- 43.Grigoryev DN, Finigan JH, Hassoun P, Garcia JG. Science review: searching for gene candidates in acute lung injury. Crit Care 8: 440–447, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hildebrand F, Stuhrmann M, van Griensven M, Meier S, Hasenkamp S, Krettek C, Pape HC. Association of IL-8–251A/T polymorphism with incidence of acute respiratory distress syndrome (ARDS) and IL-8 synthesis after multiple trauma. Cytokine 37: 192–199, 2007. [DOI] [PubMed] [Google Scholar]
- 45.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet 6: 95–108, 2005. [DOI] [PubMed] [Google Scholar]
- 46.Hizawa N, Yamaguchi E, Takahashi D, Nishihira J, Nishimura M. Functional polymorphisms in the promoter region of macrophage migration inhibitory factor and atopy. Am J Respir Crit Care Med 169: 1014–1018, 2004. [DOI] [PubMed] [Google Scholar]
- 47.Hunter DJ, Kraft P. Drinking from the fire hose–statistical issues in genomewide association studies. N Engl J Med 357: 436–439, 2007. [DOI] [PubMed] [Google Scholar]
- 48.Jerng JS, Yu CJ, Wang HC, Chen KY, Cheng SL, Yang PC. Polymorphism of the angiotensin-converting enzyme gene affects the outcome of acute respiratory distress syndrome. Crit Care Med 34: 1001–1006, 2006. [DOI] [PubMed] [Google Scholar]
- 49.Jersmann HP Time to abandon dogma: CD14 is expressed by non-myeloid lineage cells. Immunol Cell Biol 83: 462–467, 2005. [DOI] [PubMed] [Google Scholar]
- 50.King HC, Sinha AA. Gene expression profile analysis by DNA microarrays: promise and pitfalls. JAMA 286: 2280–2288, 2001. [DOI] [PubMed] [Google Scholar]
- 51.Lagan AL, Melley DD, Evans TW, Quinlan GJ. Pathogenesis of the systemic inflammatory syndrome and acute lung injury: role of iron mobilization and decompartmentalization. Am J Physiol Lung Cell Mol Physiol 294: L161–L174, 2008. [DOI] [PubMed] [Google Scholar]
- 52.Lagan AL, Quinlan GJ, Mumby S, Melley DD, Goldstraw P, Bellingan GJ, Hill MR, Briggs D, Pantelidis P, du Bois RM, Welsh KI, Evans TW. Variation in iron homeostasis genes between patients with acute respiratory distress syndrome and healthy controls. Chest 133: 1302–1311, 2008. [DOI] [PubMed] [Google Scholar]
- 53.Lin PI, Vance JM, Pericak-Vance MA, Martin ER. No gene is an island: the flip-flop phenomenon. Am J Hum Genet 80: 531–538, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin Z, Pearson C, Chinchilli V, Pietschmann SM, Luo J, Pison U, Floros J. Polymorphisms of human SP-A, SP-B, and SP-D genes: association of SP-B Thr131Ile with ARDS. Clin Genet 58: 181–191, 2000. [DOI] [PubMed] [Google Scholar]
- 55.Lorenz E, Mira JP, Frees KL, Schwartz DA. Relevance of mutations in the TLR4 receptor in patients with gram-negative septic shock. Arch Intern Med 162: 1028–1032, 2002. [DOI] [PubMed] [Google Scholar]
- 56.Loza MJ, McCall CE, Li L, Isaacs WB, Xu J, Chang BL. Assembly of inflammation-related genes for pathway-focused genetic analysis. PLoS ONE 2: e1035, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luk T, Malam Z, Marshall JC. Pre-B cell colony-enhancing factor (PBEF)/visfatin: a novel mediator of innate immunity. J Leukoc Biol 83: 804–816, 2008. [DOI] [PubMed] [Google Scholar]
- 58.MacLaren R, Stringer KA. Emerging role of anticoagulants and fibrinolytics in the treatment of acute respiratory distress syndrome. Pharmacotherapy 27: 860–873, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest 118: 1590–1605, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marshall RP, Webb S, Bellingan GJ, Montgomery HE, Chaudhari B, McAnulty RJ, Humphries SE, Hill MR, Laurent GJ. Angiotensin converting enzyme insertion/deletion polymorphism is associated with susceptibility and outcome in acute respiratory distress syndrome. Am J Respir Crit Care Med 166: 646–650, 2002. [DOI] [PubMed] [Google Scholar]
- 61.Marshall RP, Webb S, Hill MR, Humphries SE, Laurent GJ. Genetic polymorphisms associated with susceptibility and outcome in ARDS. Chest 121: 68S–69S, 2002. [DOI] [PubMed] [Google Scholar]
- 62.Martin TR, Rubenfeld GD, Ruzinski JT, Goodman RB, Steinberg KP, Leturcq DJ, Moriarty AM, Raghu G, Baughman RP, Hudson LD. Relationship between soluble CD14, lipopolysaccharide binding protein, and the alveolar inflammatory response in patients with acute respiratory distress syndrome. Am J Respir Crit Care Med 155: 937–944, 1997. [DOI] [PubMed] [Google Scholar]
- 63.Marzec JM, Christie JD, Reddy SP, Jedlicka AE, Vuong H, Lanken PN, Aplenc R, Yamamoto T, Yamamoto M, Cho HY, Kleeberger SR. Functional polymorphisms in the transcription factor NRF2 in humans increase the risk of acute lung injury. FASEB J 21: 2237–2246, 2007. [DOI] [PubMed] [Google Scholar]
- 64.Max MP, Pison U, Floros J. Frequency of SP-B and SP-A1 gene polymorphisms in acute respiratory distress syndrome (ARDS). Appl Cardiopulm Pathophysiol 6: 111–118, 1996. [Google Scholar]
- 65.McKone EF, Shao J, Frangolias DD, Keener CL, Shephard CA, Farin FM, Tonelli MR, Pare PD, Sandford AJ, Aitken ML, Kavanagh TJ. Variants in the glutamate-cysteine-ligase gene are associated with cystic fibrosis lung disease. Am J Respir Crit Care Med 174: 415–419, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Medford AR, Keen LJ, Bidwell JL, Millar AB. Vascular endothelial growth factor gene polymorphism and acute respiratory distress syndrome. Thorax 60: 244–248, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mendez JL, Hubmayr RD. New insights into the pathology of acute respiratory failure. Curr Opin Crit Care 11: 29–36, 2005. [DOI] [PubMed] [Google Scholar]
- 68.Mizue Y, Ghani S, Leng L, McDonald C, Kong P, Baugh J, Lane SJ, Craft J, Nishihira J, Donnelly SC, Zhu Z, Bucala R. Role for macrophage migration inhibitory factor in asthma. Proc Natl Acad Sci USA 102: 14410–14415, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, Depner M, von Berg A, Bufe A, Rietschel E, Heinzmann A, Simma B, Frischer T, Willis-Owen SA, Wong KC, Illig T, Vogelberg C, Weiland SK, von Mutius E, Abecasis GR, Farrall M, Gut IG, Lathrop GM, Cookson WO. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature 448: 470–473, 2007. [DOI] [PubMed] [Google Scholar]
- 70.Morton NE, Collins A. Tests and estimates of allelic association in complex inheritance. Proc Natl Acad Sci USA 95: 11389–11393, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moss M Clinical Year in Review II: interstitial lung disease, sepsis, pulmonary infections, and sleep medicine. Proc Am Thorac Soc 4: 482–488, 2007. [DOI] [PubMed] [Google Scholar]
- 72.Moss M, Mannino DM. Race and gender differences in acute respiratory distress syndrome deaths in the United States: an analysis of multiple-cause mortality data (1979–1996). Crit Care Med 30: 1679–1685, 2002. [DOI] [PubMed] [Google Scholar]
- 73.Nonas SA, Finigan JH, Gao L, Garcia JG. Functional genomic insights into acute lung injury: role of ventilators and mechanical stress. Proc Am Thorac Soc 2: 188–194, 2005. [DOI] [PubMed] [Google Scholar]
- 74.Ober C, Hoffjan S. Asthma genetics 2006: the long and winding road to gene discovery. Genes Immun 7: 95–100, 2006. [DOI] [PubMed] [Google Scholar]
- 75.Papathanassoglou ED, Giannakopoulou MD, Bozas E. Genomic variations and susceptibility to sepsis. AACN Adv Crit Care 17: 394–422, 2006. [DOI] [PubMed] [Google Scholar]
- 76.Parra EJ, Marcini A, Akey J, Martinson J, Batzer MA, Cooper R, Forrester T, Allison DB, Deka R, Ferrell RE, Shriver MD. Estimating African American admixture proportions by use of population-specific alleles. Am J Hum Genet 63: 1839–1851, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Quasney MW, Waterer GW, Dahmer MK, Kron GK, Zhang Q, Kessler LA, Wunderink RG. Association between surfactant protein B + 1580 polymorphism and the risk of respiratory failure in adults with community-acquired pneumonia. Crit Care Med 32: 1115–1119, 2004. [DOI] [PubMed] [Google Scholar]
- 78.Reddy AJ, Christie JD, Aplenc R, Fuchs B, Lanken PN, Kleeberger SR. Association of human NAD(P)H:quinone oxidoreductase 1 (Nqo1) polymorphism with development of acute lung injury. J Cell Mol Med. In press. [DOI] [PMC free article] [PubMed]
- 79.Rubenfeld GD Epidemiology of acute lung injury. Crit Care Med 31: S276–S284, 2003. [DOI] [PubMed] [Google Scholar]
- 80.Rubenfeld GD, Caldwell E, Peabody E, Weaver J, Martin DP, Neff M, Stern EJ, Hudson LD. Incidence and outcomes of acute lung injury. N Engl J Med 353: 1685–1693, 2005. [DOI] [PubMed] [Google Scholar]
- 81.Sapru A, Wiemels JL, Witte JS, Ware LB, Matthay MA. Acute lung injury and the coagulation pathway: potential role of gene polymorphisms in the protein C and fibrinolytic pathways. Intensive Care Med 32: 1293–1303, 2006. [DOI] [PubMed] [Google Scholar]
- 82.Schaid DJ Power and sample size for testing associations of haplotypes with complex traits. Ann Hum Genet 70: 116–130, 2006. [DOI] [PubMed] [Google Scholar]
- 83.Schroeder O, Schulte KM, Schroeder J, Ekkernkamp A, Laun RA. The −1082 interleukin-10 polymorphism is associated with acute respiratory failure after major trauma: a prospective cohort study. Surgery 143: 233–242, 2008. [DOI] [PubMed] [Google Scholar]
- 84.Silverman EK, Palmer LJ. Case-control association studies for the genetics of complex respiratory diseases. Am J Respir Cell Mol Biol 22: 645–648, 2000. [DOI] [PubMed] [Google Scholar]
- 85.Steinke JW, Borish L. 3. Cytokines and chemokines. J Allergy Clin Immunol 117: S441–S445, 2006. [DOI] [PubMed] [Google Scholar]
- 86.Sutherland AM, Walley KR, Russell JA. Polymorphisms in CD14, mannose-binding lectin, and Toll-like receptor-2 are associated with increased prevalence of infection in critically ill adults. Crit Care Med 33: 638–644, 2005. [DOI] [PubMed] [Google Scholar]
- 87.Sutherland AM, Walley KR, Manocha S, Russell JA. The association of interleukin 6 haplotype clades with mortality in critically ill adults. Arch Intern Med 165: 75–82, 2005. [DOI] [PubMed] [Google Scholar]
- 88.Takahashi K, Ezekowitz RA. The role of the mannose-binding lectin in innate immunity. Clin Infect Dis 41, Suppl 7: S440–S444, 2005. [DOI] [PubMed] [Google Scholar]
- 89.Thickett DR, Armstrong L, Millar AB. A role for vascular endothelial growth factor in acute and resolving lung injury. Am J Respir Crit Care Med 166: 1332–1337, 2002. [DOI] [PubMed] [Google Scholar]
- 90.Tracey KJ, Warren HS. Human genetics: an inflammatory issue. Nature 429: 35–37, 2004. [DOI] [PubMed] [Google Scholar]
- 91.Tsai YJ, Gao L, Campbell M, Grant AV, Rafaels N, Mathias R, Chi P, Sevransky J, Garcia JGN, Shanholtz C, Maloney JP, Moss M, Martin G, Herr D, Aggarwal N, D'Alessio F, King L, Cheadle C, Beaty TH, Grigoryev DN, Brower R, Barnes KC. CD14 and TLR4: synergistic gene-gene interactions and expression in acute lung injury and mortality. Am J Respir Crit Care Med 176: In press.
- 92.Vercelli D Discovering susceptibility genes for asthma and allergy. Nat Rev Immunol 8: 169–182, 2008. [DOI] [PubMed] [Google Scholar]
- 93.Wang Y, Barbacioru C, Hyland F, Xiao W, Hunkapiller KL, Blake J, Chan F, Gonzalez C, Zhang L, Samaha RR. Large scale real-time PCR validation on gene expression measurements from two commercial long-oligonucleotide microarrays. BMC Genomics 7: 59, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ware LB Pathophysiology of acute lung injury and the acute respiratory distress syndrome. Semin Respir Crit Care Med 27: 337–349, 2006. [DOI] [PubMed] [Google Scholar]
- 95.Ware LB, Matthay MA. The acute respiratory distress syndrome. N Engl J Med 342: 1334–1349, 2000. [DOI] [PubMed] [Google Scholar]
- 96.Wurfel MM Microarray-based analysis of ventilator-induced lung injury. Proc Am Thorac Soc 4: 77–84, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ye SQ, Simon BA, Maloney JP, Zambelli-Weiner A, Gao L, Grant A, Easley RB, McVerry BJ, Tuder RM, Standiford T, Brower RG, Barnes KC, Garcia JG. Pre-B-cell colony-enhancing factor as a potential novel biomarker in acute lung injury. Am J Respir Crit Care Med 171: 361–370, 2005. [DOI] [PubMed] [Google Scholar]
- 98.Young RP, Hopkins R, Black PN, Eddy C, Wu L, Gamble GD, Mills GD, Garrett JE, Eaton TE, Rees MI. Functional variants of antioxidant genes in smokers with COPD and in those with normal lung function. Thorax 61: 394–399, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhai R, Gong MN, Zhou W, Thompson TB, Kraft P, Su L, Christiani DC. Genotypes and haplotypes of the VEGF gene are associated with higher mortality and lower VEGF plasma levels in patients with ARDS. Thorax 62: 718–722, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhai R, Zhou W, Gong MN, Thompson BT, Su L, Yu C, Kraft P, Christiani DC. Inhibitor kappaB-alpha haplotype GTC is associated with susceptibility to acute respiratory distress syndrome in Caucasians. Crit Care Med 35: 893–898, 2007. [DOI] [PubMed] [Google Scholar]