Abstract
Previously, we reported that expression of lipocalin-prostaglandin D synthase (L-PGDS) is inducible in macrophages and protects from Pseudomonas pneumonia. Here, we investigated the mechanism by which L-PGDS gene expression is induced in macrophages. A promoter analysis of the murine L-PGDS promoter located a binding site of PU.1, a transcription factor essential for macrophage development and inflammatory gene expression. A chromatin immunoprecipitation assay showed that PU.1 bound to the cognate site in the endogenous L-PGDS promoter in response to LPS. Overexpression of PU.1, but not of PU.1S148A, a mutant inert to casein kinase II (CKII) or NF-κB-inducing kinase (NIK), induced L-PGDS in RAW 264.7 cells. Conversely, siRNA silencing of PU.1 expression blunted productions of L-PGDS and prostaglandin D2 (PGD2). LPS treatment induced formation of the complex of PU.1 and cJun on the PU.1 site, but inactivation of cJun by treatment with JNK or p38 kinase inhibitor abolished the complex, and suppressed PU.1 transcriptional activity for L-PGDS gene expression. Together, these results show that PU.1, activated by CKII or NIK, cooperates with MAPK-activated cJun to maximally induce L-PGDS expression in macrophages following LPS treatment, and suggest that PU.1 participates in innate immunity through the production of L-PGDS and PGD2.
Keywords: prostaglandin, gene regulation, transcription factors, inflammation
lipocalin-type prostaglandin d synthase (L-PGDS) and hematopoietic-type prostaglandin D synthase (H-PGDS) are constitutively expressed in neuronal cells (59) and in hematopoietic cells including macrophages (27, 58), respectively. They convert prostaglandin H2 (PGH2), the end product of cyclooxygenase-1 and -2 (COX-1 and -2), to prostaglandin D2 (PGD2) (7). We demonstrated that L-PGDS expression is also inducible in macrophages and accounts for the majority of PGD2 produced by macrophages (25). Furthermore, we showed that L-PGDS expression protects from Pseudomonas pneumonia (25). These results suggest that induction of L-PGDS in macrophages plays an important role in innate immunity.
PGD2 is produced mainly by macrophages and mast cells (3, 8, 19, 30, 36) and functions either pro- or anti-inflammatory depending on the nature of inflammatory milieu. For instance, PGD2 exacerbates asthma (15, 37, 55), suggesting the proinflammatory role of PGD2. On the other hand, PGD2 suppresses lung inflammation in animal models of bleomycin and monosodium urate monohydrate crystal challenges (1, 22, 42) and facilitates resolution of acute inflammation (46). Thus, along with resolvins, protectins, lipoxins, and aspirin-triggered lipoxins, PGD2 is considered as a proresolving lipid molecule (54).
Macrophages are a key effector cell in innate immunity. They abundantly express Toll-like receptor 4 (TLR4), a receptor for LPS, and TLR4 plays an essential role in innate immunity (6, 32, 56), as evidenced by the observation that mice harboring defective TLR4 are more susceptible to bacterial infection (11, 39, 44). Binding of LPS to TLR4 activates IκB kinase (IKK) and mitogen-activated protein kinases (MAPKs), such as c-Jun NH2-terminal kinase (JNK) and p38 kinase, resulting in activation of NF-κB and cJun, respectively (20). In addition to this, TLR4 activates casein kinase II (CKII) via a less characterized pathway to induce PU.1 activity (34, 45). Activation of these transcription factors results in the expression of inflammatory genes including COX-2 (7).
PU.1, a member of the Ets transcription factor family, plays a critical role during the macrophage development (10, 12, 16, 38, 53). The essential function of PU.1 in the macrophage development has been highlighted in recent studies showing that PU.1 expression, in concert with other partner proteins, converts pro-T cells into macrophages (51) and fibroblasts into macrophage-like cells (14). In macrophages, PU.1 regulates expression of various inflammatory genes including TLR4 and COX-2 (9, 26, 33, 48–50), and the transcriptional activity of PU.1 is regulated by various kinases. CKII (34, 45) and NF-κB-inducing kinase (NIK) (2) phosphorylate PU.1 at Ser148, resulting in the increase of PU.1 transcriptional activity. p38 kinase and JNK have also been known to increase PU.1 activity, although it is controversial whether or not these kinases directly phosphorylate PU.1 (21, 60).
Our recent finding that L-PGDS expression in macrophages is inducible (25) led us to the hypothesis that a macrophage-specific mechanism regulates the induction of L-PGDS expression. To elucidate the mechanism, we analyzed the sequences of the murine L-PGDS promoter and located a putative PU.1 binding site. Here, we provide evidence showing that PU.1 is a critical factor in regulating L-PGDS expression and thus PGD2 production in macrophages. In addition, we show that PU.1 functionally cooperates with cJun at the PU.1 binding site of the endogenous L-PGDS promoter, in which two distinctive pathways, one running for cJun and the other for PU.1, are involved. Based on the results, we propose that PU.1 provides the macrophage-specific mechanism for producing L-PGDS and PGD2, which play an important role in inflammation and host immunity.
MATERIALS AND METHODS
Reagents.
TLR4-specific Escherichia coli LPS (1 μg/ml; Alexis Biochemical, San Diego, CA) was added to the cell culture media. Antibodies for murine L-PGDS and H-PGDS, PU.1, ETS-1/2, IgG, IκBα, p65, cJun, tubulin, and actin were obtained from Santa Cruz Biotechnology. Antibodies for COX-1 and COX-2 and the COX-1 specific inhibitor SC-506 were purchased from Cayman Chemical (Ann Arbor, MI). In final concentration, 10 μM SB-220025 (p38 inhibitor; Calbiochem, Darmstadt, Germany) and JNK inhibitor II (Calbiochem) were used in this study.
Animals.
Male and female wild-type mice (C57BL/6) weighing 20–28 g were used for this experiment, which was performed per the protocol ID: M/05/044 and approved by the Vanderbilt University Institutional Animal Care and Use Committee.
Bone marrow-derived macrophages and cell culture.
Bone marrow-derived macrophages (BMDM) were obtained as described elsewhere (41). In short, cellular material from femurs of mice ranging from 8 to 16 wk of age was cultured in 10% L929 cell-conditioned medium. A murine macrophage cell line, RAW 264.7, and HEK-293 cells (American Type Culture Collection, Rockville, MD) were cultured in DMEM containing 10% FBS.
DNA constructs.
pcDNA-M2 PU.1, -M2 PU.1S148A, NF-κB-firefly luciferase, and tk-Renilla luciferase reporter constructs were described previously (26). To generate a PU.1-firefly luciferase reporter construct, we used MR656 reporter construct that has the proximal 656-bp mannose receptor promoter inserted into pGL2 basic vector (Promega) (23). MR656, digested with XhoI and PvuII, was inserted with a linker sequence, which was generated by annealing two complementary oligonucleotides: 5′-T CGAGCTGCAGTTCCTGTTTTTCTAAccgcccCcatgtcACAGCTAGCTTCCTGTTTTTCTAAccgcccCcatgtgACACGCGTTCCTGTTTTTCTAAcccgcccCcatgtgACA-3′ and 5′-TGTCACATGGGGGCGGTTAGAAAAACAGGAACGCGTGTCACATGGGGGCGGTTAGAAAAACAGGAAGCTAGCTGTCACATGGGGGCGGTTAGAAAAACAGGAACTGCAGC-3′. The linker was composed of three repeats of the minimal mannose receptor promoter sequence that contains PU.1 (ttcctg, underlined), Sp.1 (ccgccc, shown in small caps), and USF (catgtg, shown in lowercase). To generate 3× PU.1-firefly luciferase construct that contains three repeats of the PU.1 binding site in the murine L-PGDS promoter, we inserted into the pGL2 basic vector a minimal promoter of cytomegalovirus (CMV), which lacks the enhancer that is part of the complete CMV promoter (Clontech), and a linker made of two complementary oligonucleotides: 5′-TCGAGTACCCACTTCCGGTAGCCCTACCCACTTCCGGTAGCCCTACCCACTTCCGGTAGCCGTAC-3′ and 5′-GGCTACCGGAAGTGGGTAGGGCTACCGGAAGTGGGTAGGGCTACCGGAAGTGGGTAC-3′. The linker contains three repeats of the putative PU.1 binding site, which is underlined, with flanking four nucleotides of the murine L-PGDS promoter. Resultant constructs were verified by sequencing and were prepared by Endotoxin-free MaxiPrep Kit (Qiagen). For transfection into macrophages, GenePORTER 2 (Gene Therapy Systems) was used.
PU.1 siRNA cell line.
RAW 264.7 cell line, in which PU.1 expression is epigenetically suppressed, was described in detail elsewhere (24).
Protein isolation and Western blot.
RIPA (radioimmunoprecipitation assay) cell lysis buffer [50 mM Tris·HCl (pH 8.0), 150 mM NaCl, 2 mM EDTA, 1% sodium orthovanadate, 1% Triton X-100, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate] was used to prepare total cell lysate. The amount of proteins in cell lysate was quantified by the Bradford assay (Bio-Rad). Membranous (13) and nuclear proteins (23) were prepared as described previously. For immunoprecipitation, 1–2 μg of antibodies was used. Immune complexes captured with 30 μl of Protein A-Sepharose (Invitrogen) were washed with RIPA buffer. Proteins of interest fractionated by SDS-PAGE were transferred to PDVF membrane (Bio-Rad), which was incubated with appropriate antibodies. A specific immune complex was revealed by enhanced chemiluminescence (ECL Plus, Amersham).
Fluorescent-activated cell sorting.
Macrophages (∼106 per sample) were scored by Trypan blue exclusion and preincubated with 5 μg of normal IgG (rat) for 30 min before staining with either phycoerythrin-conjugated rat anti-mouse TLR4/MD2 or isotypic IgG (Santa Cruz Biotechnology) at 4°C for 45 min in a medium (DMEM, 10% newborn calf serum). FACS was performed by a FACScan flow cytometer (Becton Dickinson, Mountain View, CA) and CellQuest software.
Chromatin immunoprecipitation assay.
Reagents were obtained from Upstate Biotechnology (Lake Placid, NY), and the assay was performed as described previously (28, 43, 47, 52). PCR conditions were as follows: 94°C 240 s; 30∼32 cycles at 94°C 40 s, 54°C 40 s and 72°C 60 s; final elongation at 72°C 10 min. PCR for the input was performed with 100 ng of genomic DNA. Primers used were 5′-atggagctgagtg ttctggg-3′ and 5′-tctggggc acaggatcattt-3′. This primer set covers the murine L-PGDS promoter segment from −945 to −776 nt, which contains the putative PU.1 binding site.
Prostanoids measurement.
PGD2 was measured by a LC-ESI-MS-MS as previously described (29). Liquid chromatographic (LC) separation was performed isocratically on a Phenomenex Luna 3-μm C18 5.0 × 0.2-cm column. The mass spectrometer (MS) was operated in positive-ion ESI mode. Detection of the analytes was accomplished by selected reaction monitoring, employing the following reactions: 370 → 317 (PGE2 and PGD2), 374 → 321 (PGD2-d4). The quantum was set to the following parameters: capillary V = 35 V; spray voltage = 4.3 kV; capillary temperature = 300°C; tube lens V = 137 V; sheath gas = 49 psi; auxiliary gas = 25 (no units); CID pressure = 1.0 mTorr. These values were observed to maximize the response of the SRM transitions employed. Collision energy was set to 13 eV for both reactions. Quantitation was accomplished by stable isotope dilution.
Luciferase assay.
Cells were cotransfected with NF-κB-luciferase and tk-Renilla luciferase constructs. At 48 h after transfection, the cells were treated with LPS (1 μg/ml) for 4 h before cell harvest. 10 μM of either JNK or p38 inhibitor was added to the culture media for 1 h before LPS treatment. Luciferase assay was performed with a dual luciferase assay kit and the manual of the manufacturer (Promega).
Statistical analysis.
For comparison among groups, paired or unpaired t-tests and one-way ANOVA tests were used (with the assistance of InStat, Graphpad Software, San Diego, CA). P values < 0.05 were considered significant. All experiments were performed at least three times independently.
RESULTS
LPS treatment of macrophages induces binding of PU.1 to the endogenous L-PGDS promoter, resulting in L-PGDS expression.
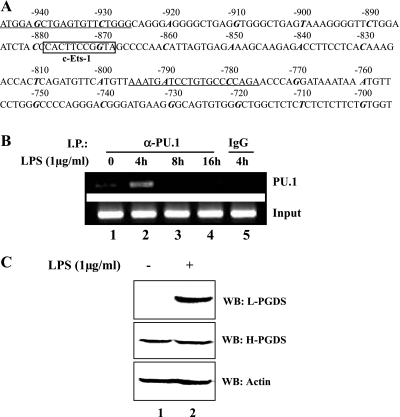
To investigate a macrophage-specific mechanism for L-PGDS induction following LPS treatment, we analyzed the murine L-PGDS promoter sequence by the TFSEARCH program (version 1.3; Tokyo Univ., Tokyo, Japan) and located a putative PU.1 binding site from −868 to −878 nt upstream of the transcription initiation site of the gene (Fig. 1A). To examine whether PU.1 recognizes the putative PU.1 site in vivo, we performed a chromatin immunoprecipitation (ChIP) assay. BMDM treated with a purified, TLR4-specific LPS (1 μg/ml) for various periods were fixed with formaldehyde, and the nuclear fractions of treated cells were collected, sonicated, and immunoprecipitated with α-PU.1 antibody. DNA fragments coprecipitated with the immune complex of PU.1, and the PU.1 antibody were eluted and amplified by PCR with a specific set of primers flanking the putative PU.1 binding site of the murine L-PGDS promoter (Fig. 1A). As shown in Fig. 1, LPS treatment induced binding of PU.1 to the putative site of the endogenous L-PGDS promoter (B), resulting in L-PGDS expression (C).
Fig. 1.
PU.1 binds to the endogenous lipocalin-prostaglandin D synthase (L-PGDS) promoter in response to LPS treatment. A: scheme represents the sequences of the murine L-PGDS promoter. Nucleotides were numbered backward from the transcription initiation site of the promoter. A putative PU.1 binding site (c-Ets-1) is shown in the box, and there is no other significant transcription factor binding site in this segment of the promoter. The 10th nucleotide in every 10-count is shown in italic bold. A set of primers flanking the putative PU.1 site, which was used for PCR for a chromatin immunoprecipitation (ChIP) assay, is underlined. B: bone marrow-derived macrophages (BMDM) were treated with LPS (1 μg/ml) for indicated time points for a ChIP assay. DNA bound to PU.1 was precipitated with an α-PU.1 antibody. For exclusion of nonspecific immunoprecipitation, an isotypic IgG was added to the lysate of the cells treated with LPS for 4 h (lane 5). Precipitated DNA and genomic DNA (bottom) were amplified by PCR with the same set of primers described in A. C: BMDM of C57BL/6 mice treated with either PBS (lane 1) or LPS (1 μg/ml) for 16 h (lane 2) were analyzed by Western blotting for L-PGDS, hematopoietic-type prostaglandin D synthase (H-PGDS), and actin after stripping the L-PGDS blot.
PU.1 regulates L-PGDS expression in macrophages.
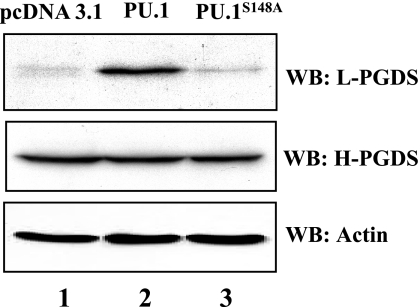
Next, to examine whether PU.1 affects L-PGDS expression in macrophages, we transfected RAW 264.7 cells with a plasmid encoding PU.1 or PU.1S148A, a mutant that impairs in transcriptional activity induced by CKII phosphorylation (34). At 48 h after transfection, total cell lysate was analyzed by Western blot. As shown in Fig. 2, overexpression of PU.1, but not of PU.1S148A, led to L-PGDS expression in macrophages without affecting the constitutive expression of H-PGDS. These results suggest that PU.1, activated by CKII pathway, induces L-PGDS expression.
Fig. 2.
Overexpression of PU.1, but not of PU.1S148A, supports L-PGDS expression in macrophages. RAW 264.7 cells were transfected with a host vector plasmid (lane 1) or a plasmid encoding PU.1 (lane 2) or PU.1S148A (lane 3). Total cell lysate of transfected cells was analyzed by Western blotting for L-PGDS, H-PGDS, and actin as internal controls.
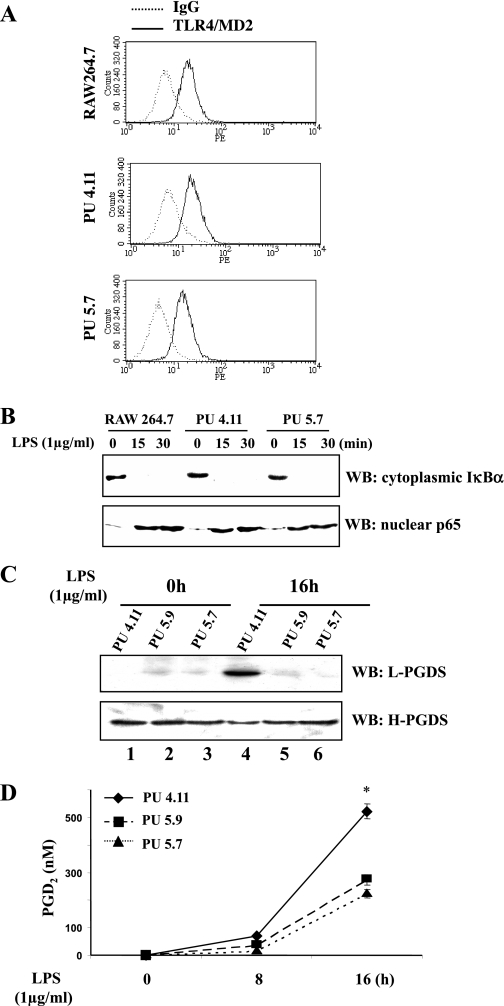
To examine whether PU.1 is required for L-PGDS expression, we used two independently isolated cell lines, PU5.7 and PU5.9, in which PU.1 expression was epigenetically suppressed by PU.1-specific siRNAs (24). First, although we previously showed that the PU.1 siRNA cell lines are similar to the parental cell line, RAW 264.7 cells, in the levels of the surface expression of TLR4/MD2 during LPS treatment and in the functionality of TLR4 signaling (24), we confirmed these results by measuring the cell surface TLR4/MD2 in an untreated condition by FACS analysis (Fig. 3A) and by determining the presence of nuclear RelA (p65) and the absence of cytoplasmic IκBα, an inhibitor of NF-κB, following LPS treatment by Western blot analyses (Fig. 3B). These results confirm our previous report that PU.1 neither affects the level of immunoreactive TLR4/MD2 on the cell surface nor TLR4-mediated signaling in response to LPS treatment.
Fig. 3.
PU.1 is required for induction of functional L-PGDS expression in macrophages. A: fluorescent-activated cell sorting (FACS) analysis was performed to examine cell surface TLR4/MD2. The dotted line represents the numbers of cells stained with a PE-conjugated isotypic IgG, and the solid line represents the cells stained with a PE-conjugated TLR4/MD2 antibody. B: the functionality of TLR4/MD2-mediated signaling was evaluated by determining the degradation of cytoplasmic IκBα and the nuclear translocation of p65 by Western blot analysis. C: along with the control cell line PU4.11 (lanes 1 and 4), the PU.1 siRNA cell lines PU5.9 (lanes 2 and 5) and PU5.7 (lanes 3 and 6) were treated with LPS for 16 h (lanes 4–6). Total cell lysate of treated cells was analyzed by Western blotting for L-PGDS and H-PGDS after stripping the L-PGDS blot. D: PGD2 produced by PU5.7, PU5.9, and PU4.11 was measured from the cell culture supernatant at the indicated time points. The results were from triplicate sets of experiments, and a similar experiment was performed three times independently. Each line represents the mean concentration ± SE (*P < 0.05 compared with PU5.7 or PU5.9).
Next, we treated PU5.7 and PU5.9 cells with LPS. At 16 h after LPS treatment, total cell lysate was analyzed by Western blot. As shown in Fig. 3C, silencing PU.1 resulted in a marked diminution in L-PGDS protein production. We also measured PGD2 in the culture media of those cell lines, and a much lower amount of PGD2 was present in the silenced PU.1 lines compared with the control (Fig. 3D). Although it is possible that silencing PU.1 affects the level of COX-2 expression and thus the amount of substrate for L-PGDS, our results showed that silencing PU.1 affects only early COX-2 expression (24) and does not limit the amount of substrate for L-PGDS (data not shown). Together, these results suggest that PU.1 is essential for L-PGDS expression.
PU.1 is functionally associated with cJun at the endogenous L-PGDS promoter in response to LPS.
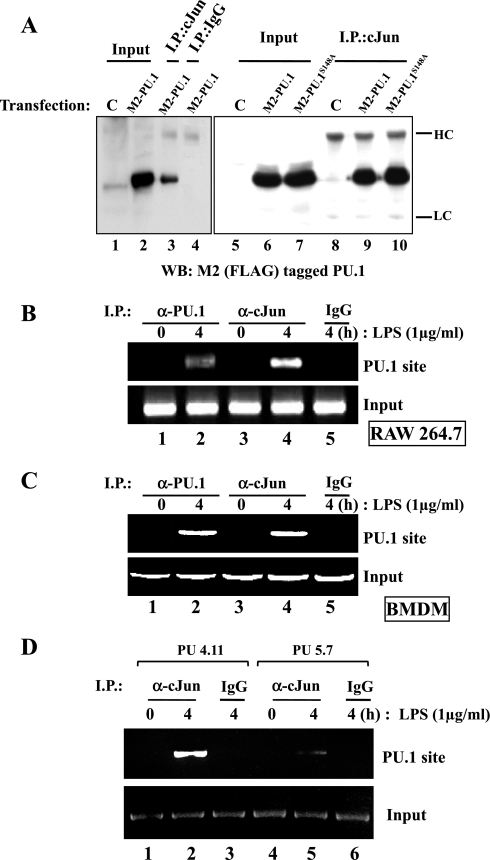
Since cJun, a bZIP DNA binding transcription factor, is involved in L-PGDS expression (25) and PU.1 physically binds to cJun (5), we examined a functional relationship between the two factors. First, we tested whether PU.1 interacts physically with cJun in our experimental setting. HEK-293 cells were transfected with a plasmid encoding FLAG-PU.1 or FLAG-PU.1S148A. At 48 h after transfection, total cell lysate was prepared and added with α-cJun antibody to precipitate cJun, which was analyzed by Western blotting for FLAG-tagged PU.1 to detect coprecipitated PU.1. As shown in Fig. 4A, both PU.1 and PU.1S148A physically associated with cJun. These results suggest that the phosphorylation of Ser148 may not be necessary for a protein-protein interaction between PU.1 and cJun.
Fig. 4.
PU.1 interacts with cJun in the PU.1 binding site of the L-PGDS promoter. A: HEK-293 cells were transfected with pCMV (C) or a plasmid encoding FLAG-tagged PU.1 or PU.1S148A. Total cell lysate was incubated with an α-cJun antibody (lanes 3, 8, 9, and 10) or with an isotypic IgG (lane 4) to exclude nonspecific precipitation. The immune complexes precipitated with the antibodies were analyzed by Western blotting with M2 antibody. One-tenth of total cell lysate was used as input controls (lanes 1, 2, 5, 6, and 7). HC and LC represent heavy and light chains of an immunoglobulin, respectively. A ChIP assay was performed with RAW 264.7 cells (B) or BMDM of C57BL/6 mice (C) treated with LPS for 4 h. The nuclear fractions were equally divided and added with either an α-PU.1 antibody (lanes 1 and 2, top) or an α-cJun antibody (lanes 3 and 4). DNA eluted from each precipitated immune complex was amplified by PCR with the set of primers flanking the PU.1 binding site of the L-PGDS promoter. DNA precipitated by an isotypic IgG (lane 5) and 100 ng of genomic DNA were used as controls (bottom). D: similarly, we performed a ChIP assay with PU5.7, along with the control, PU4.11, after treatment with LPS for 4 h. The nuclear fractions were added with the α-cJun antibody (lanes 1, 2, 4, and 5) or the isotypic IgG (lane 3 and 6) to exclude nonspecific precipitation. Eluted DNA was analyzed by PCR as described above.
Next, to examine whether the interaction between PU.1 and cJun occurs on the PU.1 binding site of the endogenous L-PGDS promoter, we performed a ChIP assay (Fig. 4B). The nuclear fractions of RAW 264.7 cells treated with or without LPS were prepared and divided equally. α-PU.1 antibody was added to one half, and α-cJun antibody to the other half. DNA coprecipitated with each antibody was amplified by PCR with the set of primers flanking the PU.1 site of the murine L-PGDS promoter as indicated in Fig. 1A. As shown in Fig. 4B, LPS treatment induced binding of PU.1 as well as cJun to the PU.1 site, which was confirmed in a similar experiment with BMDM (Fig. 4C). These results suggest that PU.1 and cJun interact with each other on the PU.1 binding site of the endogenous L-PGDS promoter.
Finally, we examined whether or not the interaction on the PU.1 binding site is mediated by PU.1. To this end, we performed a ChIP assay with the PU.1-silenced cell line, PU5.7 cell. The control cell line, PU4.11, and PU5.7 were treated with LPS for 4 h, and the DNA-protein complexes were precipitated by the α-cJun antibody. As shown in Fig. 4D, cJun bound to the PU.1 site in PU5.7 less strongly than in PU4.11, suggesting that the binding of cJun to the PU.1 site is likely mediated by PU.1, not cJun alone. Together, these results suggest that PU.1 provides a platform to form a complex with cJun at the PU.1 binding site of the endogenous L-PGDS promoter in response to LPS treatment.
LPS-induced binding of PU.1 to its cognate site in the endogenous L-PGDS promoter occurs without cJun.
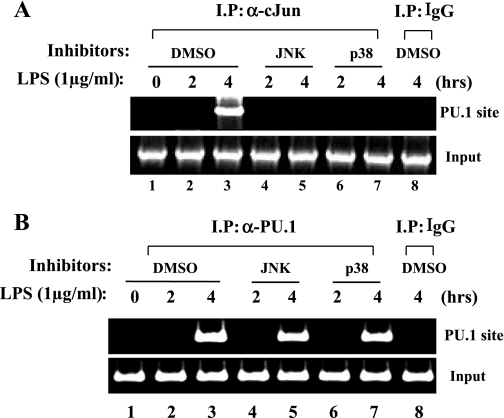
We examined whether the complex formation of PU.1 and cJun is prerequisite for PU.1 to bind to the cognate site in the promoter. First, since both JNK and p38 kinase activate cJun (4), we examined whether JNK and p38 kinase affect the interaction between PU.1 and cJun on the promoter. Before LPS treatment, RAW 264.7 cells were treated with JNK or p38 kinase inhibitor (10 μM each) for 1 h and analyzed by a ChIP assay with the α-cJun antibody. As shown in Fig. 5A, the JNK inhibitor or the p38 inhibitor prevented cJun from binding to the PU.1 site following LPS treatment, suggesting that JNK and p38 kinase regulate the physical association of cJun with PU.1 in response to LPS. Next, to determine whether PU.1 requires cJun for its binding to the cognate site, we performed a similar experiment with α-PU.1 antibody. As shown in Fig. 5B, neither the JNK inhibitor nor the p38 kinase inhibitor interfered with the binding of PU.1 to the PU.1 site. Together, these results demonstrate that PU.1 binds to its cognate site without cJun.
Fig. 5.
PU.1 binding to the cognate site occurs without cJun. A: to examine whether cJun binding to the PU.1 site is affected by either JNK or p38 kinase, we performed a ChIP assay. RAW 264.7 cells were treated with DMSO (lanes 1–3) or 10 μM JNK inhibitor (lanes 4 and 5) or p38 kinase inhibitor (lanes 6 and 7) 1 h before LPS treatment. DNA bound to cJun was precipitated by the α-cJun antibody and amplified by PCR with the set of primers flanking the PU.1 site (top). To exclude nonspecific immunoprecipitation, we added an isotypic IgG to the nuclear fraction of the cells treated with LPS for 4 h (lane 8). Genomic DNA (100 ng) was amplified by PCR as input controls (bottom). B: to measure the possible effect of JNK or p38 kinase on binding of PU.1 to the PU.1 binding site, we performed a ChIP assay with the α-PU.1 antibody (lanes 1–7) or an isotypic IgG (lane 8). Precipitated DNA was amplified by PCR with the set of primers flanking the PU.1 binding site (top). Similarly, genomic DNA was amplified (bottom).
cJun enhances the transcriptional activity of PU.1 and the expression of L-PGDS.
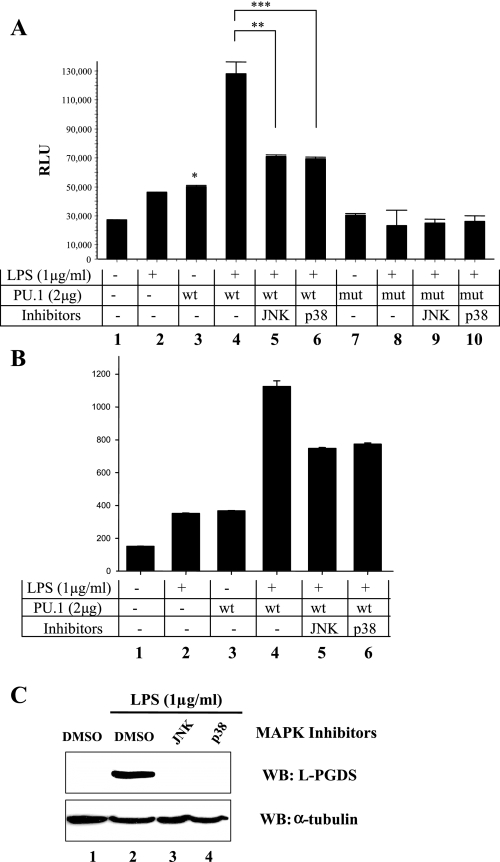
To determine the impact of cJun on PU.1-driven transcription, we transfected RAW 264.7 cells with a plasmid encoding PU.1 or PU.1S148A, along with a PU.1 reporter construct that harbors three PU.1 binding sites but no AP-1 site. At 48 h after transfection, the transfected cells were treated with LPS for 4 h before luciferase assay. As shown in Fig. 6A, overexpression of PU.1 increased luciferase activity. LPS treatment significantly augmented PU.1-driven luciferase activity, which was, however, blunted by the JNK or p38 kinase inhibitor. On the other hand, overexpression of PU.1S148A, even with LPS treatment, failed to increase luciferase activity. It is notable that the expression levels of PU.1 and PU.1S148A were similar (Fig. 4A). To consolidate these results, we performed a similar experiment by using another PU.1 reporter construct that contains three repeats of the PU.1 binding site of the murine L-PGDS promoter but no AP-1 binding site. As shown in Fig. 6B, we obtained similar results. These data indicate that cJun, without anchoring to its binding site, enhances the transcriptional activity of PU.1 in the L-PGDS promoter.
Fig. 6.
Effects of JNK and p38 MAPK on PU.1-driven transcription and L-PGDS expression in macrophages. RAW 264.7 cells were transfected with pCMV (columns 1 and 2) or a plasmid encoding PU.1 (columns 3–6) or PU.1S148A (columns 7–10), along with tk-Renilla luciferase construct and a PU.1 firefly luciferase reporter construct. At 48 h after transfection, the cells were treated with either JNK inhibitor (columns 5 and 9) or p38 kinase inhibitor (columns 6 and 10) 1 h before LPS treatment for 4 h. Luciferase activity was measured from triplicate of the experimental settings, and the experiment was performed 3 times independently. Each bar represents the mean RLU ± SE. *P < 0.05 compared with the control value (lane 1). ** and ***P < 0.05 in comparison of lane 4 with lanes 5 and 6, respectively. B: similar experiment was performed with a PU.1-luciferase reporter that contains 3 repeats of the PU.1 binding site of the murine L-PGDS promoter. C: RAW 264.7 cells were treated with vehicle (lanes 1 and 2), 10 μM JNK inhibitor (lane 3), or 10 μM p38 kinase inhibitor (lane 4) 1 h before LPS treatment for 16 h to induce L-PGDS. Western blot was performed for L-PGDS and tubulin.
To examine whether cJun also affects the expression of L-PGDS protein, we treated the macrophage cells with the JNK or p38 kinase inhibitor, showing that the treatments suppressed L-PGDS expression induced by LPS treatment (Fig. 6C). Together, these results show that the transcriptional activity of PU.1 was enhanced by cJun following LPS treatment and suggest that the signal-dependent, physical interaction between the two proteins at the PU.1 binding site is required for the maximal expression of L-PGDS.
DISCUSSION
L-PGDS is induced in macrophages during inflammation and protects from bacterial pneumonia (25). L-PGDS is also responsible for the majority of PGD2 produced by macrophages, the major effector cell that produces PGD2 during acute inflammation (25). In self-limiting inflammation, PGD2 plays a key role in resolving inflammation (46). Thus, elucidating the mechanism that regulates the induction of L-PGDS expression in a macrophage-specific fashion could be essential for understanding the effect of PGD2 on host immunity.
In this study, we tested the hypothesis that a macrophage-specific factor regulates L-PGDS expression, along with cJun, an ubiquitous transcription factor that is involved in L-PGDS expression in macrophages (25). A promoter analysis that located a putative PU.1 site in the murine L-PGDS promoter led us to investigate the potential role of PU.1, a macrophage-specific transcription factor, in L-PGDS expression. Our results show that PU.1 bound to the cognate site in the endogenous L-PGDS promoter in response to LPS, overexpression of PU.1 increased L-PGDS expression, and epigenetic silencing of PU.1 by siRNA resulted in the suppression of L-PGDS expression and PGD2 production. We also have shown that there was a unique functional interaction between PU.1 and cJun at the PU.1 binding site in the promoter in response to treatment with LPS, which enhanced PU.1-mediated L-PGDS expression. These results suggest that PU.1, through interaction with cJun, provides a macrophage-specific regulatory mechanism for L-PGDS expression in macrophages in response to treatment with LPS.
Both PU.1 and cJun are important for monocyte to macrophage differentiation and macrophage effector function in inflammation (17, 31, 35, 40, 57). In expression of some genes, cJun physically binds to PU.1 and serves as an activator of PU.1 in a JNK-independent fashion (5). Consistent with this, our results show that there were physical and functional interactions between PU.1 and cJun. However, our results clearly demonstrate that the functional interaction between the two proteins is dependent on not only JNK but also p38 kinase. This difference may be due to differential experimental conditions, possibly related to activation by treatment with LPS.
PU.1 regulates TLR4 mRNA expression (48), which raised the possibility that blunted L-PGDS expression is due to deficient TLR4 signaling. We have recently reported that the levels of TLR4 mRNA in the PU.1-silenced cell lines are lower than controls (24). However, in spite of the differences in TLR4 mRNA levels, there were no differences in the levels of cell surface TLR4/MD2 in the PU.1 siRNA cell lines that were treated with LPS compared with control cells. In addition, there was no difference in NF-κB activation, a robust indicative that TLR4 signaling was normal in the PU.1 siRNA cell lines compared with control cells (24). These important control data demonstrate that decreased L-PGDS gene expression was not simply due to impaired TLR4 expression or function in the siRNA cells that were treated with LPS.
It is not clear how TLR4 signaling induced the interaction between PU.1 and cJun on the L-PGDS promoter. Our results show that the Ser148 residue of PU.1, a key residue phosphorylated by CKII or NIK that is activated by LPS, was critical for the transcriptional activity of PU.1 in L-PGDS expression. However, it seems that the phosphorylation of the residue did not participate in the physical interaction with cJun. In addition, binding of PU.1 to its cognate site in the promoter in response to LPS was not affected by JNK or p38 kinase and took place without involvement of cJun. On the other hand, binding of cJun to PU.1 in response to LPS was dependent on a phosphorylation of cJun because treatment of JNK or p38 kinase inhibitors abolished the interaction between the two proteins at the PU.1 binding site and blunted L-PGDS expression. In addition, it seems that cJun was recruited to the PU.1 binding site only when PU.1 occupied the site. Therefore, it is possible that PU.1, activated by CKII or NIK pathway, binds to the PU.1 binding site, which provides a platform for cJun to bind the PU.1 site. In the meantime, cJun becomes phosphorylated and activated by the MAPK and then physically associates with the PU.1 that occupied the PU.1 binding site of the L-PGDS promoter.
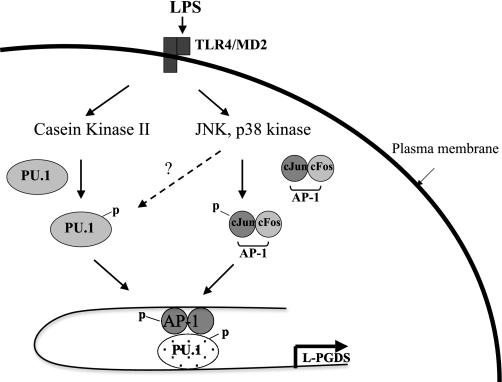
Combined, the current results and our previous report that the L-PGDS promoter harbors an AP-1 binding site, to which cJun binds in a JNK- and p38 kinase-dependent manner (25), we can speculate the mechanism of L-PGDS expression in a macrophage-specific fashion (Fig. 7). LPS-triggered TLR4 signaling activates CKII or NIK that phosphorylates PU.1 at Ser148 residue, resulting in the activation and binding of PU.1 to its cognate site in the promoter. On the other hand, the TLR4 signaling activates JNK and p38 kinases, which phosphorylate and activate cJun. Activated cJun could bind to both the AP-1 binding site directly and the PU.1 binding site indirectly through PU.1 that occupies the site. Although cJun could directly bind to PU.1 without tethering to its cognate AP-1 site, the presence of the AP-1 binding site in the promoter could increase the availability, or the local concentration, of cJun, which could facilitate physical interactions between cJun and PU.1. Since the sites for AP-1 and PU.1 are separated by ∼350 nt, it is conceivable that these physical interactions among proteins and their binding sites could result in sufficient torsion of the DNA to contribute to L-PGDS expression in macrophage.
Fig. 7.
Proposed mechanism for the roles of PU.1 and cJun in L-PGDS expression in macrophages. Scheme depicts a possible mechanism by which PU.1 induces L-PGDS expression in macrophages following LPS treatment. LPS triggers TLR4 signaling, which activates casein kinase II (CKII). CKII phosphorylates PU.1 at Ser148, and PU.1 activated by the phosphorylation binds to its cognate site in the promoter. Meanwhile, the TLR4 signaling also activates JNK and p38 kinase that phosphorylate cJun and make it active. Activated cJun binds to both the AP-1 site of the promoter and PU.1. PU.1 and cJun activated by LPS form a transcriptionally active complex in the L-PGDS promoter, leading to L-PGDS gene expression.
PGD2, the end product of L-PGDS and H-PGDS, has been known to suppress inflammation in animal models for acute lung inflammation that was induced by bleomycin or monosodium urate monohydrate crystal challenges (1, 22, 42). Since PGD2 is detected in resolving exudates and the increased production of PGD2 is associated with resolution of inflammation in a carrageenin-induced self-limiting acute inflammatory model (18), PGD2 is considered as a proresolving lipid molecule (54). Given that PGD2 is mainly produced by macrophages and mast cells (3, 8, 19, 30, 36), it has been presumed that H-PGDS is the major enzyme that produces PGD2 and regulates inflammation. In fact, the importance of H-PGDS in regulation of inflammation was recently demonstrated in a zymosan-induced peritonitis model with H-PGDS knockout mice (46), in which the lack of H-PGDS and thereof the decrease of PGD2 apparently impairs resolution of peritonitis. Given our current results of PU.1-mediated induction of L-PGDS expression in macrophages, in conjunction with our published results (25), it is possible that L-PGDS expression occurs and contributes to the late production of PGD2 during the resolution of peritonitis. In addition, our results may provide an explanation of how the level of PGD2 in exudates is increased during the resolution of pleurisy (18). However, it is interesting that during the resolution of peritonitis the level of PGD2 in exudates was decreased (46). While this could be due to egression of activated macrophages from the peritoneal cavity during the resolution of peritonitis, the differences in the levels of PGD2 during the resolution of inflammation in different models may reflect a differential inflammatory milieu generated by treatment with varying stimuli.
In summary, we investigated how L-PGDS expression is induced in macrophages in the inflammatory milieu. We found that PU.1, a macrophage-specific factor, played a critical role in regulating L-PGDS expression and PGD2 production, in which PU.1 provided a platform for cJun to bind to the PU.1 binding site for induction of L-PGDS in LPS-stimulated macrophages. Therefore, our results highlight the central role of PU.1 in inducing L-PGDS expression in macrophages. We propose that regulation of L-PGDS by PU.1 is the mechanism to produce L-PGDS and PGD2 that play important roles in host immunity.
GRANTS
This work was supported by the Department of Veterans Affairs, National Heart, Lung, and Blood Institute Grants HL-061419, HL-075557, and HL-066196, and Pusan National University Research Grant 2008.
Acknowledgments
We thank Dr. Lawrence Marnett and his laboratory for measuring prostaglandin D2 and Dr. Hye Jeong Lee for technical support.
REFERENCES
- 1.Ando M, Murakami Y, Kojima F, Endo H, Kitasato H, Hashimoto A, Kobayashi H, Majima M, Inoue M, Kondo H, Kawai S, Hayashi I. Retrovirally introduced prostaglandin D2 synthase suppresses lung injury induced by bleomycin. Am J Respir Cell Mol Biol 28: 582–591, 2003. [DOI] [PubMed] [Google Scholar]
- 2.Azim AC, Wang X, Park GY, Sadikot RT, Cao H, Mathew B, Atchison M, van Breemen RB, Joo M, Christman JW. NF-kappaB-inducing kinase regulates cyclooxygenase 2 gene expression in macrophages by phosphorylation of PU.1. J Immunol 179: 7868–7875, 2007. [DOI] [PubMed] [Google Scholar]
- 3.Balter MS, Eschenbacher WL, Peters-Golden M. Arachidonic acid metabolism in cultured alveolar macrophages from normal, atopic, and asthmatic subjects. Am Rev Respir Dis 138: 1134–1142, 1988. [DOI] [PubMed] [Google Scholar]
- 4.Baud V, Karin M. Signal transduction by tumor necrosis factor and its relatives. Trends Cell Biol 11: 372–377, 2001. [DOI] [PubMed] [Google Scholar]
- 5.Behre G, Whitmarsh AJ, Coghlan MP, Hoang T, Carpenter CL, Zhang DE, Davis RJ, Tenen DG. c-Jun is a JNK-independent coactivator of the PU.1 transcription factor. J Biol Chem 274: 4939–4946, 1999. [DOI] [PubMed] [Google Scholar]
- 6.Beutler B, Rietschel ET. Innate immune sensing and its roots: the story of endotoxin. Nat Rev Immunol 3: 169–176, 2003. [DOI] [PubMed] [Google Scholar]
- 7.Bjorkbacka H, Fitzgerald KA, Huet F, Li X, Gregory JA, Lee MA, Ordija CM, Dowley NE, Golenbock DT, Freeman MW. The induction of macrophage gene expression by LPS predominantly utilizes Myd88-independent signaling cascades. Physiol Genomics 19: 319–330, 2004. [DOI] [PubMed] [Google Scholar]
- 8.Brown GP, Monick MM, Hunninghake GW. Human alveolar macrophage arachidonic acid metabolism. Am J Physiol Cell Physiol 254: C809–C815, 1988. [DOI] [PubMed] [Google Scholar]
- 9.Carl VS, Brown-Steinke K, Nicklin MJ, Smith MF Jr. Toll-like receptor 2 and 4 (TLR2 and TLR4) agonists differentially regulate secretory interleukin-1 receptor antagonist gene expression in macrophages. J Biol Chem 277: 17448–17456, 2002. [DOI] [PubMed] [Google Scholar]
- 10.Chen HM, Zhang P, Voso MT, Hohaus S, Gonzalez DA, Glass CK, Zhang DE, Tenen DG. Neutrophils and monocytes express high levels of PU.1 (Spi-1) but not Spi-B. Blood 85: 2918–2928, 1995. [PubMed] [Google Scholar]
- 11.Coutinho A, Forni L, Melchers F, Watanabe T. Genetic defect in responsiveness to the B cell mitogen lipopolysaccharide. Eur J Immunol 7: 325–328, 1977. [DOI] [PubMed] [Google Scholar]
- 12.DeKoter RP, Singh H. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science 288: 1439–1441, 2000. [DOI] [PubMed] [Google Scholar]
- 13.Egan BS, Lane KB, Shepherd VL. PU.1and USF are required for macrophage-specific mannose receptor promoter activity. J Biol Chem 274: 9098–9107, 1999. [DOI] [PubMed] [Google Scholar]
- 14.Feng R, Desbordes SC, Xie H, Tillo ES, Pixley F, Stanley ER, Graf T. PU.1 and C/EBPalpha/beta convert fibroblasts into macrophage-like cells. Proc Natl Acad Sci USA 105: 6057–6062, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujitani Y, Kanaoka Y, Aritake K, Uodome N, Okazaki-Hatake K, Urade Y. Pronounced eosinophilic lung inflammation and Th2 cytokine release in human lipocalin-type prostaglandin D synthase transgenic mice. J Immunol 168: 443–449, 2002. [DOI] [PubMed] [Google Scholar]
- 16.Galson DL, Hensold JO, Bishop TR, Schalling M, D'Andrea AD, Jones C, Auron PE, Housman DE. Mouse beta-globin DNA-binding protein B1 is identical to a proto-oncogene, the transcription factor Spi-1/PU.1, and is restricted in expression to hematopoietic cells and the testis. Mol Cell Biol 13: 2929–2941, 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaynor R, Simon K, Koeffler P. Expression of c-jun during macrophage differentiation of HL-60 cells. Blood 77: 2618–2623, 1991. [PubMed] [Google Scholar]
- 18.Gilroy DW, Colville-Nash PR, Willis D, Chivers J, Paul-Clark MJ, Willoughby DA. Inducible cyclooxygenase may have anti-inflammatory properties. Nat Med 5: 698–701, 1999. [DOI] [PubMed] [Google Scholar]
- 19.Goth A, Johnson AR. Current concepts on the secretory function of mast cells. Life Sci 16: 1201–1213, 1975. [DOI] [PubMed] [Google Scholar]
- 20.Guha M, Mackman N. LPS induction of gene expression in human monocytes. Cell Signal 13: 85–94, 2001. [DOI] [PubMed] [Google Scholar]
- 21.Himes SR, Sester DP, Ravasi T, Cronau SL, Sasmono T, Hume DA. The JNK are important for development and survival of macrophages. J Immunol 176: 2219–2228, 2006. [DOI] [PubMed] [Google Scholar]
- 22.Ianaro A, Ialenti A, Maffia P, Pisano B, Di Rosa M. Role of cyclopentenone prostaglandins in rat carrageenin pleurisy. FEBS Lett 508: 61–66, 2001. [DOI] [PubMed] [Google Scholar]
- 23.Joo M, Hahn YS, Kwon M, Sadikot RT, Blackwell TS, Christman JW. Hepatitis C virus core protein suppresses NF-kappaB activation and cyclooxygenase-2 expression by direct interaction with IkappaB kinase beta. J Virol 79: 7648–7657, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joo M, Kwon M, Azim AC, Sadikot RT, Blackwell TS, Christman JW. Genetic determination of the role of PU.1 in macrophage gene expression. Biochem Biophys Res Commun 372: 97–102, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joo M, Kwon M, Sadikot RT, Kingsley PJ, Marnett LJ, Blackwell TS, Peebles RS Jr, Urade Y, Christman JW. Induction and function of lipocalin prostaglandin D synthase in host immunity. J Immunol 179: 2565–2575, 2007. [DOI] [PubMed] [Google Scholar]
- 26.Joo M, Park GY, Wright JG, Blackwell TS, Atchison ML, Christman JW. Transcriptional regulation of the cyclooxygenase-2 gene in macrophages by PU.1. J Biol Chem 279: 6658–6665, 2004. [DOI] [PubMed] [Google Scholar]
- 27.Kanaoka Y, Ago H, Inagaki E, Nanayama T, Miyano M, Kikuno R, Fujii Y, Eguchi N, Toh H, Urade Y, Hayaishi O. Cloning and crystal structure of hematopoietic prostaglandin D synthase. Cell 90: 1085–1095, 1997. [DOI] [PubMed] [Google Scholar]
- 28.Kawasaki K, Nogawa H, Nishijima M. Identification of mouse MD-2 residues important for forming the cell surface TLR4-MD-2 complex recognized by anti-TLR4-MD-2 antibodies, and for conferring LPS and taxol responsiveness on mouse TLR4 by alanine-scanning mutagenesis. J Immunol 170: 413–420, 2003. [DOI] [PubMed] [Google Scholar]
- 29.Kingsley PJ, Rouzer CA, Saleh S, Marnett LJ. Simultaneous analysis of prostaglandin glyceryl esters and prostaglandins by electrospray tandem mass spectrometry. Anal Biochem 343: 203–211, 2005. [DOI] [PubMed] [Google Scholar]
- 30.Lewis RA, Soter NA, Diamond PT, Austen KF, Oates JA, Roberts LJ. Prostaglandin D2 generation after activation of rat and human mast cells with anti-IgE. J Immunol 129: 1627–1631, 1982. [PubMed] [Google Scholar]
- 31.Li J, King I, Sartorelli AC. Differentiation of WEHI-3B D+ myelomonocytic leukemia cells induced by ectopic expression of the protooncogene c-jun. Cell Growth Differ 5: 743–751, 1994. [PubMed] [Google Scholar]
- 32.Lien E, Means TK, Heine H, Yoshimura A, Kusumoto S, Fukase K, Fenton MJ, Oikawa M, Qureshi N, Monks B, Finberg RW, Ingalls RR, Golenbock DT. Toll-like receptor 4 imparts ligand-specific recognition of bacterial lipopolysaccharide. J Clin Invest 105: 497–504, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lloberas J, Soler C, Celada A. The key role of PU.1/SPI-1 in B cells, myeloid cells and macrophages. Immunol Today 20: 184–189, 1999. [DOI] [PubMed] [Google Scholar]
- 34.Lodie TA, Savedra R Jr, Golenbock DT, Van Beveren CP, Maki RA, Fenton MJ. Stimulation of macrophages by lipopolysaccharide alters the phosphorylation state, conformation, and function of PU.1 via activation of casein kinase II. J Immunol 158: 1848–1856, 1997. [PubMed] [Google Scholar]
- 35.Lord KA, Abdollahi A, Hoffman-Liebermann B, Liebermann DA. Proto-oncogenes of the fos/jun family of transcription factors are positive regulators of myeloid differentiation. Mol Cell Biol 13: 841–851, 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MacDermot J, Kelsey CR, Waddell KA, Richmond R, Knight RK, Cole PJ, Dollery CT, Landon DN, Blair IA. Synthesis of leukotriene B4, and prostanoids by human alveolar macrophages: analysis by gas chromatography/mass spectrometry. Prostaglandins 27: 163–179, 1984. [DOI] [PubMed] [Google Scholar]
- 37.Matsuoka T, Hirata M, Tanaka H, Takahashi Y, Murata T, Kabashima K, Sugimoto Y, Kobayashi T, Ushikubi F, Aze Y, Eguchi N, Urade Y, Yoshida N, Kimura K, Mizoguchi A, Honda Y, Nagai H, Narumiya S. Prostaglandin D2 as a mediator of allergic asthma. Science 287: 2013–2017, 2000. [DOI] [PubMed] [Google Scholar]
- 38.McKercher SR, Torbett BE, Anderson KL, Henkel GW, Vestal DJ, Baribault H, Klemsz M, Feeney AJ, Wu GE, Paige CJ, Maki RA. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J 15: 5647–5658, 1996. [PMC free article] [PubMed] [Google Scholar]
- 39.Miller SI, Ernst RK, Bader MW. LPS, TLR4 and infectious disease diversity. Nat Rev Microbiol 3: 36–46, 2005. [DOI] [PubMed] [Google Scholar]
- 40.Mollinedo F, Gajate C, Tugores A, Flores I, Naranjo JR. Differences in expression of transcription factor AP-1 in human promyelocytic HL-60 cells during differentiation towards macrophages versus granulocytes. Biochem J 294: 137–144, 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morham SG, Langenbach R, Loftin CD, Tiano HF, Vouloumanos N, Jennette JC, Mahler JF, Kluckman KD, Ledford A, Lee CA, Smithies O. Prostaglandin synthase 2 gene disruption causes severe renal pathology in the mouse. Cell 83: 473–482, 1995. [DOI] [PubMed] [Google Scholar]
- 42.Murakami Y, Akahoshi T, Hayashi I, Endo H, Hashimoto A, Kono S, Kondo H, Kawai S, Inoue M, Kitasato H. Inhibition of monosodium urate monohydrate crystal-induced acute inflammation by retrovirally transfected prostaglandin D synthase. Arthritis Rheum 48: 2931–2941, 2003. [DOI] [PubMed] [Google Scholar]
- 43.Nagai Y, Akashi S, Nagafuku M, Ogata M, Iwakura Y, Akira S, Kitamura T, Kosugi A, Kimoto M, Miyake K. Essential role of MD-2 in LPS responsiveness and TLR4 distribution. Nat Immunol 3: 667–672, 2002. [DOI] [PubMed] [Google Scholar]
- 44.Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282: 2085–2088, 1998. [DOI] [PubMed] [Google Scholar]
- 45.Pongubala JM, Van Beveren C, Nagulapalli S, Klemsz MJ, McKercher SR, Maki RA, Atchison ML. Effect of PU.1 phosphorylation on interaction with NF-EM5 and transcriptional activation. Science 259: 1622–1625, 1993. [DOI] [PubMed] [Google Scholar]
- 46.Rajakariar R, Hilliard M, Lawrence T, Trivedi S, Colville-Nash P, Bellingan G, Fitzgerald D, Yaqoob MM, Gilroy DW. Hematopoietic prostaglandin D2 synthase controls the onset and resolution of acute inflammation through PGD2 and 15-deoxyDelta12 14 PGJ2. Proc Natl Acad Sci USA 104: 20979–20984, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Re F, Strominger JL. Monomeric recombinant MD-2 binds toll-like receptor 4 tightly and confers lipopolysaccharide responsiveness. J Biol Chem 277: 23427–23432, 2002. [DOI] [PubMed] [Google Scholar]
- 48.Rehli M, Poltorak A, Schwarzfischer L, Krause SW, Andreesen R, Beutler B. PU.1 and interferon consensus sequence-binding protein regulate the myeloid expression of the human Toll-like receptor 4 gene. J Biol Chem 275: 9773–9781, 2000. [DOI] [PubMed] [Google Scholar]
- 49.Roger T, David J, Glauser MP, Calandra T. MIF regulates innate immune responses through modulation of Toll-like receptor 4. Nature 414: 920–924, 2001. [DOI] [PubMed] [Google Scholar]
- 50.Roger T, Miconnet I, Schiesser AL, Kai H, Miyake K, Calandra T. Critical role for Ets, AP-1 and GATA-like transcription factors in regulating mouse Toll-like receptor 4 (Tlr4) gene expression. Biochem J 387: 355–365, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rothenberg EV, Scripture-Adams DD. Competition and collaboration: GATA-3, PU.1, and Notch signaling in early T-cell fate determination. Semin Immunol 20: 236–246, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schromm AB, Lien E, Henneke P, Chow JC, Yoshimura A, Heine H, Latz E, Monks BG, Schwartz DA, Miyake K, Golenbock DT. Molecular genetic analysis of an endotoxin nonresponder mutant cell line: a point mutation in a conserved region of MD-2 abolishes endotoxin-induced signaling. J Exp Med 194: 79–88, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science 265: 1573–1577, 1994. [DOI] [PubMed] [Google Scholar]
- 54.Serhan CN, Chiang N, Van Dyke TE. Resolving inflammation: dual anti-inflammatory and pro-resolution lipid mediators. Nat Rev Immunol 8: 349–361, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spik I, Brenuchon C, Angeli V, Staumont D, Fleury S, Capron M, Trottein F, Dombrowicz D. Activation of the prostaglandin D2 receptor DP2/CRTH2 increases allergic inflammation in mouse. J Immunol 174: 3703–3708, 2005. [DOI] [PubMed] [Google Scholar]
- 56.Sultzer BM Genetic control of leucocyte responses to endotoxin. Nature 219: 1253–1254, 1968. [DOI] [PubMed] [Google Scholar]
- 57.Szabo E, Preis LH, Birrer MJ. Constitutive cJun expression induces partial macrophage differentiation in U-937 cells. Cell Growth Differ 5: 439–446, 1994. [PubMed] [Google Scholar]
- 58.Urade Y, Eguchi N. Lipocalin-type and hematopoietic prostaglandin D synthases as a novel example of functional convergence. Prostaglandins Other Lipid Mediat 68–69: 375–382, 2002. [DOI] [PubMed] [Google Scholar]
- 59.Urade Y, Hayaishi O. Biochemical, structural, genetic, physiological, and pathophysiological features of lipocalin-type prostaglandin D synthase. Biochim Biophys Acta 1482: 259–271, 2000. [DOI] [PubMed] [Google Scholar]
- 60.Wang JM, Lai MZ, Yang-Yen HF. Interleukin-3 stimulation of mcl-1 gene transcription involves activation of the PU.1 transcription factor through a p38 mitogen-activated protein kinase-dependent pathway. Mol Cell Biol 23: 1896–1909, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]