Abstract
Tandem affinity purification (TAP) has been used to isolate proteins that interact with human hepatic lipase (HL) during its maturation in Chinese hamster ovary cells. Using mass spectrometry and Western blotting, we identified 28 proteins in HL-TAP isolated complexes, 16 of which localized to the endoplasmic reticulum (ER), the site of HL folding and assembly. Of the 12 remaining proteins located outside the ER, five function in protein translation or ER-associated degradation (ERAD). Components of the two major ER chaperone systems were identified, the BiP/Grp94 and the calnexin (CNX)/calreticulin (CRT) systems. All factors involved in CNX/CRT chaperone cycling were identified, including UDP-glucose:glycoprotein glucosyltransferase 1 (UGGT), glucosidase II, and the 57 kDa oxidoreductase (ERp57). We also show that CNX, and not CRT, is the lectin chaperone of choice during HL maturation. Along with the 78 kDa glucose-regulated protein (Grp78; BiP) and the 94 kDa glucose-regulated protein (Grp94), an associated peptidyl-prolyl cis-trans isomerase and protein disulfide isomerase were also detected. Finally, several factors in ERAD were identified, and we provide evidence that terminally misfolded HL is degraded by the ubiquitin-mediated proteasomal pathway. We propose that newly synthesized HL emerging from the translocon first associates with CNX, ERp57, and glucosidase II, followed by repeated posttranslational cycles of CNX binding that is mediated by UGGT. BiP/Grp94 may stabilize misfolded HL during its transition between cycles of CNX binding and may help direct its eventual degradation.
Keywords: protein folding, endoplasmic reticulum, chaperones, calnexin, tandem affinity purification, mass spectrometry
Hepatic lipase (HL) has diverse functions in lipoprotein metabolism and in the etiology of atherosclerosis (1–4). It is synthesized in hepatocytes and macrophages (5, 6) and found in steroidogenic tissues as well (5). It is a secreted glycoprotein, and its site of action is within the subendothelial space or at the lumenal surface of capillary endothelium, where it is bound to heparan-sulfate proteoglycans (2, 3). HL is a lipolytic enzyme that hydrolyzes triacylglycerols and phospholipids in chylomicron remnants, IDL and HDL, thus serving as an important determinant in LDL and HDL remodeling (3). HL is inversely correlated with HDL-cholesterol levels and plays a major role in determining LDL subclass distribution (7, 8). Additionally, HL functions as a ligand facilitating the uptake of lipoproteins by cell surface receptors, a role independent of its catalytic activity (3). While it is unclear whether HL is a pro- or anti-atherogenic factor in lipoprotein metabolism, its expression in macrophages has been assessed as pro-atherogenic. At this site, HL is thought to possibly alter macrophage gene expression and promote retention of VLDL remnants, IDL, and small dense LDL in the region of lesion development (1).
In all its roles, HL function ultimately depends on the success of proper folding and assembly in the endoplasmic reticulum (ER). HL maturation encompasses the processes involved in the formation of a complex three-dimensional structure comprised of a globular N-terminal domain and a β-sheet C-terminal domain using, in all, five disulfide bridges; after folding, monomers must assemble into a functional head-to-tail homodimer (4). While synthesis of a full-length protein requires minutes, protein folding occurs in milliseconds (9). Thus, for most proteins, it is essential to delay folding until synthesis is complete (9). This requires an interaction with chaperones, which in general slow down folding and stabilize unfolded intermediates (9–12). When associated with chaperones, unfolded proteins can also interact with folding factors that facilitate important posttranslational modifications, such as glycan processing, disulfide bond formation, and peptidyl-prolyl cis-trans isomerization (9, 10).
HL maturation necessarily precedes its secretion to the lumenal or subendothelial space where HL function occurs. However, HL maturation is slow, taking hours as opposed to the minutes required by its homologous family member, LPL; thus, HL maturation is the rate-limiting step in its secretion (13). As a consequence, at steady state, most HL molecules in the ER have not yet achieved a functional conformation but are present as folding intermediates or terminally misfolded forms associated with heterogeneously sized protein complexes (13). In addition, HL maturation is inefficient, with nearly half of newly synthesized molecules destined for eventual degradation by an unknown pathway (13).
Although slow maturation kinetics and the presence of HL-protein complexes suggest a robust role for folding factors in HL maturation, only calnexin (CNX), an ER lectin chaperone, has been positively identified as an interacting factor in this process (13, 14). We have recently identified another essential ER factor, Lipase Maturation Factor 1 (Lmf1), although its precise function in maturation has yet to be determined (15). Nevertheless, the absence of functional Lmf1 (15), or limiting access of HL to CNX (13), severely decreases the efficiency of HL folding, indicating that its maturation is dependent on the assistance of at least these ER factors.
Our eventual aim is the identification of all factors that associate with HL and its lipase family members during their sojourn in the ER. This study has begun this analysis by isolating HL complexes using a tandem-affinity tag fused to the C terminus of the protein. The use of such tags as “entry points” permits the isolation of specific proteins under conditions that favor the retention of protein-protein associations (16–20). Indeed, this approach has been successfully applied to isolate many protein interactions occurring in both yeast and mammalian systems (16, 21, 22). In this study, we used such a strategy to isolate proteins that copurified with human HL in Chinese hamster ovary (CHO) cells. After tandem affinity purification, we identified 28 proteins in isolated HL complexes by tandem mass spectrometry (MS/MS) and/or Western blot analysis, many involved in some aspect of protein folding. We discuss the implications of these various proteins in the process of HL maturation and propose the composition of HL/protein complexes occurring both co- and posttranslationally. Overall, our analysis supports the central role played by CNX and associated factors in HL maturation and ER-associated degradation (ERAD).
MATERIALS AND METHODS
Expression constructs and tandem affinity purification synthesis
Human HL and pancreatic lipase (PL) cDNAs were subcloned into the pcDNA6 expression vector (Invitrogen) containing a C-terminal V5 epitope tag as described (13). For both HL and PL pcDNA6/V5 constructs, a tandem affinity purification (TAP) tag was synthesized for in-frame integration into an AgeI site occurring just after the V5 epitope tag. The TAP tag is 170 amino acids long and consists of a single calmodulin binding peptide domain, a tobacco etch virus (TEV) protease site, followed by two adjacent IgG binding domains from Protein A. The TAP tag was synthesized by several rounds of PCR using overlapping amplicons generated from the calmodulin binding peptide domain of the pCAL-c vector (Stratagene) and the Protein A domain from the pEZZ 18 Protein A gene fusion vector (Amersham). The complete protein sequence of HL-TAP is given in Fig. 1. In addition, the human asialoglycoprotein H2b receptor cDNA was subcloned into the pcDNA6/V5 expression vector as described (23).
Fig. 1.
Sequence of the human HL-TAP construct. The boxed sequences represent the relevant domains, including the V5 epitope tag and the tandem affinity tag. The TAP tag is comprised of a calmodulin binding domain, a TEV cleavage site, and two IgG binding domains.
CHO cell experiments
The proline auxotroph derivatives (Pro5) of CHO cells were cultured and maintained as described (13, 23). After calcium phosphate-mediated transfection of CHO cells with the lipase-TAP pcDNA6/V5 constructs, blasticidin-resistant colonies were selected (13, 23). Lipase-expressing colonies were identified by assaying the medium for HL activity (13, 23) after incubating the cells for 16 h in heparin (10 units/ml). For all lipase-TAP experiments, cells were subcultured on 100-mm plates and kept under blasticidin selection until use. Cells were harvested at 70–80% confluency. At the end of all experiments, cells were washed twice with PBS and scraped off in 1 ml of PBS. The resultant cell pellets were stored at −80°C until use.
In TAP experiments using untreated cells, monolayers were incubated for 16 h in fresh medium without blasticidin, containing 10 units/ml heparin. In experiments employing the cross-linker dithiobis-(succinimidylpropionate) (DSP; Pierce), cell monolayers were treated with heparin and then incubated for 10 min on ice with ice-cold PBS containing 20 mM N-ethylmaleimide (Calbiochem). After one wash with ice-cold PBS, DSP in Me2SO was added (4 mM final concentration), and the cell suspension incubated for 1 h on ice with intermittent shaking. Cross-linking was quenched by the addition of glycine (50 mM final concentration) and N-ethylmaleimide (40 mM final concentration). In experiments using DTT, cell monolayers were treated with 4 mM DTT for 2 h prior to harvesting. For experiments using proteasomal inhibitors MG132 or MG262 (Calbiochem), these agents were added in Me2SO at final concentrations of 10 and 1 μM, respectively, along with heparin (10 units/ml). After 6 h incubation, cell monolayers were washed and harvested as described above.
TAP
Eight separate HL-TAP purifications were performed, six using untreated cells, and one each for cells treated with the cross-linker DSP or reducing agent DTT. One PL-TAP purification was also performed. Each purification experiment included an equivalent number of untransfected Pro5 cells to act as a negative control. For each experiment, cell pellets from 10–30 100 mm plates were used. Each pellet was sonicated in 0.5 ml of ice-cold 2% CHAPS, 0.15 M NaCl, and 10 mM Tris-HCl, pH 8.0. The combined lysates were centrifuged at 1,000 g for 15 min at 4°C to remove any cellular debris.
TAP was carried out essentially as described (18). All purification steps used ice-cold solutions, and all incubations were performed at 4–10°C. The first column step consisted of 0.25 ml of IgG-Sepharose 6 Fast Flow resin (Amersham) added to a 0.8 × 4 cm Poly-Prep chromatography column (Bio-Rad), and washed with 30 ml IgG binding buffer (0.5% CHAPS, 0.15 M NaCl, and 10 mM Tris-HCl, pH 8.0) by centrifugation at 100 g for 20 s. Lysate (5–8 ml) was applied to the washed resin, the column capped, and then incubated with constant mixing for 16 h.
After incubation, the resin was washed again with 30 ml IgG binding buffer, and after centrifugation, the resin containing bound complexes was resuspended in 1 ml TEV cleavage buffer (0.5% CHAPS, 0.15 M NaCl, 0.5 mM EDTA, 1 mM DTT, and 10 mM Tris-HCl, pH 8.0) containing 100 units of AcTEV protease (Invitrogen). The column was incubated for 2 h at 10°C with constant mixing, after which the TEV eluate was collected by centrifugation at 500 g for 2 min. Three milliliters of calmodulin binding buffer (0.5% CHAPS, 0.15 M NaCl, 10 mM 2-mercaptoethanol, 1 mM Mg Acetate, 1 mM imidazole, 2 mM CaCl2, and 10 mM Tris-HCl, pH 8.0) was added to the TEV eluate along with 40 μl of 1 M CaCl2. After gentle mixing, the eluate was added to a fresh Poly-Prep column containing 0.20 ml calmodulin affinity resin (Stratagene), washed beforehand with 10 ml calmodulin binding buffer. The column was capped and incubated for 2 h with constant mixing.
After incubation, the column was washed with 30 ml calmodulin binding buffer and eluted with 0.5 ml calmodulin elution buffer (0.5% CHAPS, 0.15 M NaCl, 10 mM 2-mercaptoethanol, 1 mM Mg Acetate, 1 mM imidazole, 20 mM EGTA, and 10 mM Tris-HCl, pH 8.0). Proteins in the resulting eluate were precipitated by adding 0.25 ml of 100% TCA and incubated overnight at 4°C. The protein pellet was washed twice with 0.5 ml ice-cold acetone and allowed to dry under a heat lamp. After drying, SDS-PAGE sample buffer was added (2% SDS, 5% 2-mercaptoethanol, 12% glycerol, 0.0025% bromophenol blue, and 50 mM Tris base, pH 10), and the sample was placed in a boiling water bath for 2 min and then subjected to SDS-PAGE using a 8% SDS Tris/glycine polyacrylamide gel (Invitrogen). Proteins were visualized using the SilverXpress® silver staining kit (Invitrogen) following the manufacturer's instructions. Western blot analysis was carried out using commercial antibodies (StressGen) at the following dilutions: CNX (1:15,000), calreticulin (CRT) (1:15,000), 47 kDa heat shock protein (Hsp47) (1:1,000), BiP (1:1,000), 94 kDa glucose-regulated protein (Grp94) (1:2,000), and α-glucosidase II (1:1,000). Incubations for all primary antibodies were carried out overnight at 4°C. After washing, appropriate HRP-conjugated secondary antibodies were incubated for 1 h at 4°C and the blot developed with the ECL Pico kit (Pierce).
Mass Spectroscopy and protein identification
Lanes from gels following SDS-PAGE of HL, PL, or untransfected TAP isolates were sequentially cut into 20–25 pieces and subjected to trypsin digestion after reduction of cysteines with DTT and alkylation with iodoacetamide. Tryptic peptides were subjected to online LC-MS/MS, performed on an Applied Biosystems QSTAR Pulsar XL (QqTOF) mass spectrometer equipped with a nanoelectrospray interface (Protana) and an LC Packings nano-LC system.
Proteins were identified using the Mascot database search engine (Matrix Science). All searches were performed against the MSDB database restricted to the Rodentia taxon (107,035 sequences). Accession numbers in Tables 1–3 are for the UniProtKB/TrEMBL database (http://ca.expaxy.org) with priority given to Chinese hamster (Cricetulus griseus) or Golden hamster (Mesocricetus auatus) sequence when available. For protein sequence searches, the following variable modifications were set: carbamidomethylation of cysteines and oxidation of methionines. In all searches, one missed tryptic cleavage was allowed, and a mass tolerance of 0.3 Da was set for the precursor and product ions. A Mascot score (combined ions score corrected for low-scoring matches) of ≥50 was considered a significant match. While 20 proteins were identified by multiple assigned peptides, eight proteins were identified by a single assigned peptide, and Table 3 gives Mascot score, coverage, peptide sequence, observed m/z, mass error, and P value for these proteins. The protein (CNX) assigned the lowest score and P value (score = 49, P = 0.012) was confirmed by Western blot analysis, as were four others (Tables 1, 2).
TABLE 1.
Proteins that copurify with HL in untreated CHO cells
| Protein Identification |
||||||||
|---|---|---|---|---|---|---|---|---|
| Location | Protein | MWa | Accession Numberb | Function | WBc | Scored | Peptidese | Coveragef |
| ER | UGGT | 174,049 | Q9JLA3 | Protein folding | 55 | 3 | 2 | |
| GII | 107,300 | Q3UE86 | Protein folding | √ | ND | |||
| Grp94g | 92,476 | P08113 | Protein folding, ERAD | √ | 190 | 3 | 6 | |
| Grp78; BiPh | 72,379 | P07823 | Protein folding, ERAD | √ | 717 | 24 | 31 | |
| CNX | 67,266 | Q8K3H8 | Protein folding, ERAD | √ | ND | |||
| ERp57g | 56,796 | Q91Z81 | Protein folding | 63 | 1 | 2 | ||
| PDIg | 48,161 | P38660 | Protein folding | 174 | 2 | 7 | ||
| Hsp47 | 46,599 | Q9Z1W7 | Protein folding | √ | 80 | 1 | 2 | |
| Reticulocalbin-1 | 38,113 | Q05186 | Ca2+ binding | 146 | 4 | 13 | ||
| PPIaseg | 22,713 | P24369 | Protein folding | 70 | 1 | 6 | ||
| Cytoskeleton | α-Tubulin | 50,020 | Q5VLK2 | Microtubule component | 340 | 11 | 25 | |
| β-Tubulin | 49,742 | Q60454 | Microtubule component | 460 | 10 | 21 | ||
| β-Actin | 41,738 | P48975 | F-actin component | 678 | 15 | 39 | ||
| Cytoplasm | eEF1A | 50,114 | Q540F6 | Translation, ERAD | 285 | 8 | 24 | |
| Glyceraldehyde-3-phosphate dehydrogenase (GAPDH)h | 35,748 | P17244 | Glycolysis | 196 | 4 | 15 | ||
| Mitochondria | Phosphate carrier protein (PTP)h | 39,632 | Q8VEM8 | Solute carrier protein | 184 | 6 | 10 | |
| ADP/ATP translocase 2 (ANT2)h | 32,931 | P51881 | Solute carrier protein | 199 | 11 | 25 | ||
| ADP/ATP translocase 1 (ANT1) | 32,904 | P48962 | Solute carrier protein | 108 | 3 | 10 | ||
Predicted molecular weight (MW) based on an unprocessed precursor protein.
UniProtKB/Swiss-Prot entry numbers (http//ca.expasy.org) for Golden or Chinese Hamster (Mesocricetus auatus; Cricetulus griseus), mouse (Mus musculus), or rat (Rattus norvegicus).
Identified by Western blot analysis.
Score is the combined ions score after correction for low-scoring random matches; ND indicates not detected by MS/MS or detected with a score <50.
The total number of peptides assigned to the protein. For additional information on single peptide-based identifications, see Table 3.
Sequence coverage, expressed as the number of amino acids spanned by the assigned peptides as a percentage of the sequence length.
Detected only when chemical cross-linking was employed.
Also copurified in CHO cells using PL as the entry point.
TABLE 2.
Proteins that copurify with HL in DTT-treated CHO cells
| Protein Identification |
||||||||
|---|---|---|---|---|---|---|---|---|
| Location | Protein | MWa | Accession Numberb | Function | WBc | Scored | Peptidese | Coveragef |
| ER | UGGT1g | 174,049 | Q9JLA3 | Protein folding | 123 | 3 | 2 | |
| Nodal modulator 1h | 133,420 | Q6GQT9 | Carbohydrate binding | 132 | 4 | 2 | ||
| GIIg | 107,300 | Q3UE86 | Protein folding | √ | 84 | 1 | 1 | |
| Grp78; BiPg | 72,379 | P07823 | Protein Folding, ERAD | √ | 717 | 24 | 31 | |
| CNXg | 67,266 | Q8K3H8 | Protein folding, ERAD | √ | ND | |||
| Peptidyl-prolyl cis-trans isomerase (FKBP65RS) | 62,995 | Q9Z247 | Protein folding | 75 | 4 | 5 | ||
| CRT | 46,599 | Q8K3H7 | Protein folding, ERAD | √ | ND | |||
| Hsp47g | 46,599 | Q9Z1W7 | Protein folding | √ | 65 | 1 | 2 | |
| TorsinB | 37,858 | Q2M2S1 | Protein folding | 130 | 2 | 8 | ||
| MSAP | 20,767 | Q9QXT0 | Cytoskeletal effector | 55 | 1 | 5 | ||
| SSRδ | 18,937 | Q3TVJ8 | Signal peptide translocation | 70 | 1 | 6 | ||
| Cytoskeleton | α-Tubuling | 50,020 | Q5VLK2 | Microtubule component | 263 | 5 | 15 | |
| β-Tubuling | 49,742 | Q60454 | Microtubule component | 181 | 6 | 14 | ||
| β-Acting | 41,738 | P48975 | F-actin component | 58 | 2 | 9 | ||
| Cytoplasm | Heat shock 70 kDa protein 8 (Hsc70) | 70,871 | P63017 | Chaperone, ERAD | 190 | 4 | 6 | |
| Ribosomal Rpl11 protein | 19,024 | Q8VC94 | Translation | 78 | 1 | 7 | ||
| Mitochondria | ADP/ATP translocase 2 (ANT2)g | 32,931 | P51881 | Solute carrier protein | 169 | 5 | 16 | |
| ADP/ATP translocase 1 (ANT1)g | 32,904 | P48962 | Solute carrier protein | 147 | 5 | 16 | ||
Predicted molecular weight based on an unprocessed precursor protein.
UniProtKB/Swiss-Prot entry numbers (http//ca.expasy.org) for Golden or Chinese Hamster, mouse, or rat.
Identified by Western blot analysis.
Score is the combined ions score after correction for low-scoring random matches; ND indicates not detected by MS/MS or detected with a score <50.
The total number of peptides assigned to the protein. For additional information on single peptide-based identifications, see Table 3.
Sequence coverage, expressed as the number of amino acids spanned by the assigned peptides as a percentage of the sequence length.
Also detected in untreated cells (Table 1).
Location inferred by the presence of a predicted signal peptide sequence.
TABLE 3.
MS/MS parameters for proteins identified by single assigned peptides
| Protein | Accession Numbera | WBb | Scorec | Coveraged | Peptide Sequence | m/z (Observed) | Mass Errore | P Valuef |
|---|---|---|---|---|---|---|---|---|
| GII | Q3UE86 | √ | 84 | 1 | K.VLLVLELQGLQK.N | 676.9499/2 | 0.0414 | 3.10e-06 |
| CNX | Q8K3H8 | √ | 49 | 2 | K.APVPTGEVYFADSFDR.G | 885.9504/2 | 0.0602 | 0.012 |
| ERp57 | Q91Z81 | 63 | 2 | R.ELNDFINYLQR.E | 712.9000/2 | 0.0759 | 0.0005 | |
| Hsp47 | Q9Z1W7 | √ | 80 | 2 | R.DNQSGSLLFIGR.L | 653.9165/2 | 0.1508 | 8.50e-06 |
| MSAP | Q9QXT0 | 55 | 5 | R.IDSDISGTLK.F | 524.8042/2 | 0.0492 | 0.0026 | |
| PPIase | P24369 | 70 | 6 | K.TVDNFVALATGEK.G | 682.8464/2 | −0.0202 | 8.10e-05 | |
| Ribosomal Rpl11 protein | Q8VC94 | 78 | 7 | K.VLEQLTGTPVFSK.A | 773.9287/2 | 0.0025 | 1.40e-05 | |
| SSRδ | Q3TVJ8 | 70 | 6 | R.FFDEESYSLLR.K | 703.3656/2 | 0.0606 | 8.50e-05 |
UniProtKB/Swiss-Prot entry numbers (http//ca.expasy.org) for Golden or Chinese Hamster, mouse, or rat.
Identified by Western blot analysis.
Score is the combined ions score after correction for low-scoring random matches.
Sequence coverage, expressed as the number of amino acids spanned by the assigned peptides as a percentage of the sequence length.
Difference (error) between the observed and calculated masses.
The number of times it is expected to obtain an equal or higher score purely by chance.
RESULTS
TAP and an overview of HL interacting proteins
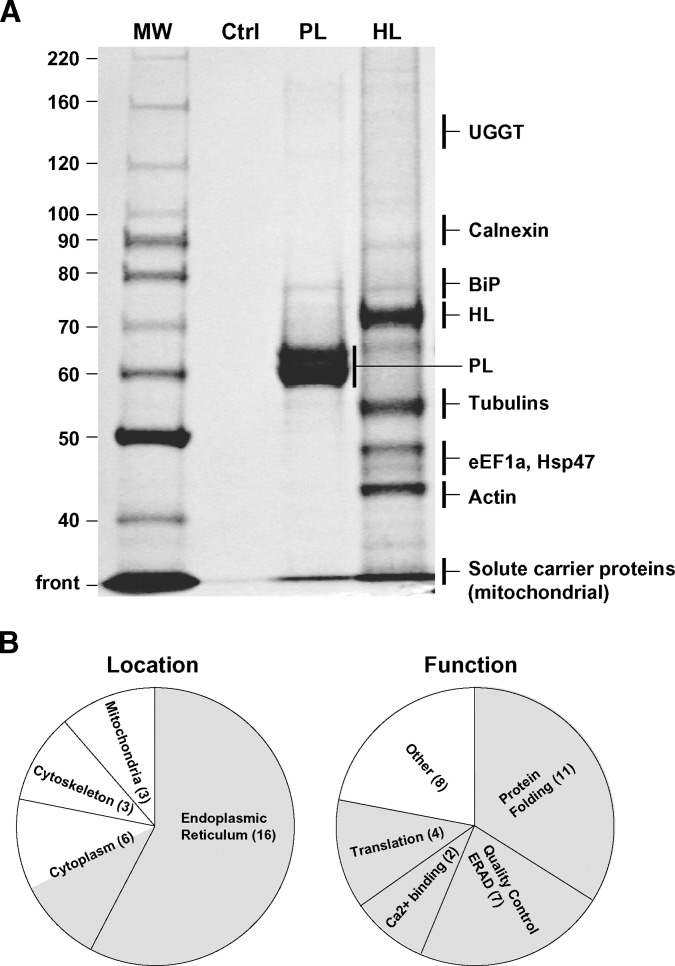
To isolate HL interacting proteins in CHO cells, a human HL-TAP construct was used as the entry point. Importantly, the resulting TAP-tagged HL protein was secreted and exhibited enzyme activity comparable to a nontagged construct (data not shown). Thus, newly synthesized HL-TAP protein in the ER underwent normal maturation by folding and assembling into functionally active homodimers. To isolate proteins that may play a role in its maturation, cell lysates were fractionated by tandem affinity chromatography using, in sequential order, the IgG and calmodulin binding domains present in the C-terminal TAP tag (Fig. 1). Binding and elution were performed under nondenaturing conditions to protect protein-protein interactions (see Materials and Methods). To determine if nonspecific proteins were isolated during tandem affinity chromatography, untransfected cell lysates were used. To test for HL-specific interactions, proteins copurifying with a human PL-TAP fusion protein were used to discern shared proteins from those specific to HL. Indeed, while PL is structurally similar to HL (4, 24, 25), the bulk of PL in the ER is functionally mature and not associated with proteins; this is in stark contrast to HL, which is predominately found in the ER as a nonfunctional protein associated with large, heterogeneous complexes (13). Thus, the fundamental processes directing HL and PL maturation are kinetically and qualitatively dissimilar, providing a basis to identify proteins specific to HL maturation.
An array of proteins copurified with the HL-TAP construct, in comparison with the few isolated when PL was used as an alternative TAP entry point; nonspecific proteins were not detected when untransfected CHO cell lysates were subjected to tandem affinity chromatography (Fig. 2A). In all TAP experiments performed, a total of 28 proteins were identified by MS/MS and Western blot analysis of SDS-PAGE gels (see Tables 1, 2 for a complete listing). Significantly, 57% of the identified proteins localized to the ER, the site of HL maturation and its degradation (Fig. 2B). Many of these proteins have roles in folding. In addition, of the 21% present in the cytoplasm, half function in protein synthesis and/or protein degradation (Fig. 2B). In all, ∼60% of the proteins copurifying with HL-TAP play roles in translation, folding, quality control, and ERAD. Of the remaining proteins, some coisolated with PL-TAP, suggesting functions unrelated or nonspecific to HL maturation, while others may represent isolation artifacts commonly encountered in TAP experiments (17, 22). Indeed, the intracellular location (mitochondria, cytoskeleton) and overall abundance of these proteins suggest that they represent isolation artifacts (see below).
Fig. 2.
A: Representative SDS PAGE gel containing silver-stained proteins isolated by TAP using PL or HL as entry points. The control (Ctrl) lane is a TAP of CHO lysates that have not been transfected with any TAP constructs. Shown on the right are regions of the gel containing some examples of proteins copurifying with an HL-TAP construct (see Tables 1, 2 for a comprehensive list). Only BiP, mitochondrial solute carrier proteins (PTP and ANT2), and GAPDH copurified with the PL-TAP construct. B: All proteins copurifying with HL in all experiments have been combined and presented with regard to location or function. The number of proteins identified in a given class is given in parentheses. The shaded areas represent locations and functions consistent with proteins that may directly or indirectly interact with HL during its co and posttranslational maturation and degradation.
HL interacting proteins in untreated CHO cells
Cells expressing HL-TAP were used to isolate interacting protein in six separate experiments, some using chemical cross-linking to stabilize protein-protein interactions (see Materials and Methods). The resulting HL/protein complexes were fractionated by SDS-PAGE. Protein identification included Western blot analysis or analysis by MS/MS after trypsin cleavage of excised bands. For proteins identified by MS/MS, inclusion criteria were detection in two or more experiments, with a combined ions score of ≥50 after correction for low-scoring random matches. Using these criteria, 16 total proteins were identified by MS/MS, with two additional proteins detected by Western blot analysis alone (Table 1).
Of the 18 total proteins identified, 10 were in the ER, all having roles in protein folding, ERAD and Ca2+ binding (Table 1). CNX, the only protein shown previously to interact with HL (13, 14), was detected along with other proteins promoting the CNX cycle, a well-established ER folding pathway (9, 10, 26, 27). These include UDP-glucose:glycoprotein glycosyltransferase 1 (UGGT), α-glucosidase II (GII), and the oxidoreductase 57 kDa endoplasmic reticulum protein (ERp57) (Table 1). Besides CNX cycle enzymes, five additional factors in protein folding were identified, specifically the 78 kDa glucose-regulated protein (Grp78; BiP), Grp94, Hsp47, protein disulfide isomerase (PDI), and a peptidyl-prolyl cis-trans isomerase (PPIase). Reticulocalbin-1, a Ca2+ binding protein localized to the ER (28), was also identified (Table 1). Calcium storage is a basic function of the ER, and a number of calcium binding proteins, such as CNX and reticulocalbin, serve in this capacity (28 29 30 31). It is unclear whether reticulocalbin participates specifically in HL maturation or is part of a multiprotein complex associating with HL, but with functions unrelated to its maturation.
All but one of these ER proteins were specific to HL-TAP and not present in PL-TAP experiments. The exception, BiP, was identified using both entry points. Thus, BiP may be a general factor required in the maturation/degradation of both lipase proteins. BiP associates with a multiprotein complex of chaperones in the ER, including Grp94, UGGT, PDI, and PPIase, all of which were identified in our analysis (32). The detection of all five proteins with HL-TAP suggests that HL is interacting with the BiP multiprotein complex. However, only BiP was present in PL-TAP experiments, suggesting that PL may interact with BiP alone.
Two cytoplasmic proteins were also detected (Table 1). Only one, Elongation factor 1-α (eEF1A), was specific to HL. In addition, in one HL-TAP experiment, Eukaryotic initiation factor 4A-1 (eLF4A-1) was also detected (MS score 59; two peptides detected covering 6% of the protein). The presence of eEF1A and eLF4A-1 suggests the isolation of HL complexes of cotranslational origin, as these would be expected to include components of the translational machinery (see Discussion). Interestingly, eEF1A has also been implicated in ubiquitin-mediated degradation (33), a pathway used by HL (see below). The elongation factor eEF1A is also known to bind actin and tubulin (34), both of which were detected in HL-TAP experiments (Table 1). It is possible, however, that the association of tubulin/actin with eEF1A occurs only after cell lysis, when cell compartmentalization is disrupted (i.e., a postlysis artifact).
Finally, members of the mitochondrial solute carrier family were also identified, although most were found using PL-TAP as an alternative entry point (Table 1). These proteins localize to the inner mitochondrial membrane and most likely play no role in lipase maturation.
HL interacting proteins in DTT-treated CHO cells
The isolation of proteins that transiently interact with an entry point protein is challenging because of the low level of interactions that would be observed during steady-state conditions. To extend the time of such interactions, and to increase levels of ER chaperones and folding factors in general, we experimentally induced protein unfolding by treating CHO cells with the membrane-permeable reducing agent DTT. DTT treatment causes prolonged association of ER factors with unfolded proteins (such as HL) to reduce protein aggregation and ER stress (23, 35).
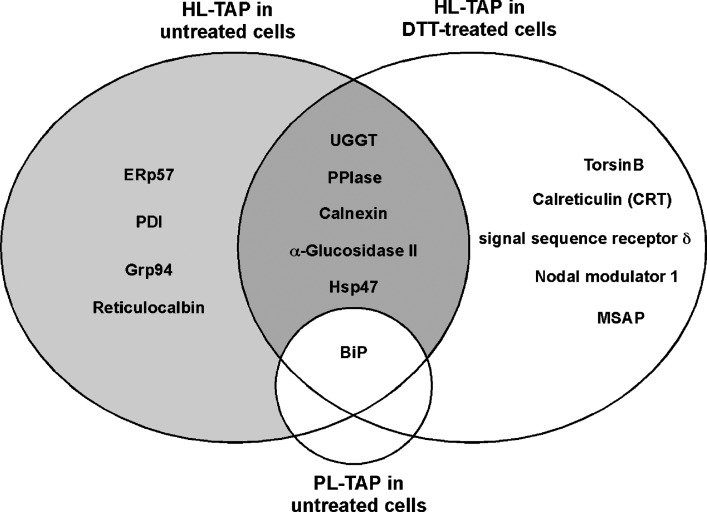
Out of the 18 total proteins identified after DTT treatment (Table 2), 10 were identical to those isolated in untreated cells (Table 1); most were detected with increased MS/MS scores, likely due to prolonged interactions. Thus, a subset of proteins is present regardless of condition, including UGGT, CNX, GII, PPIase, and BiP (Fig. 3). Along with ERp57 and Grp94, we believe that these proteins represent core factors engaged in HL folding and assembly. Hsp47 was also present in this shared group, but its function in HL maturation (if any) remains unclear. Hsp47 is a collagen-specific chaperone, and it binds to Xaa-Arg-Gly repeats in the context of the collagen triple helix (36–39), structural elements not present in HL. In addition, transfection of HL in an Hsp47-deficient cell line (40) showed no obvious HL maturation defects (data not shown). Thus, at this time, there is no evidence to support a functional contribution of Hsp47 to the process of HL maturation.
Fig. 3.
Venn diagram comparing ER proteins copurifying with HL-TAP in untreated CHO cells or cells treated with DTT, a reducing agent that causes protein unfolding. Included is BiP, which also copurified with PL-TAP. The shaded areas represent proteins that are the strongest candidates for roles in HL maturation and degradation.
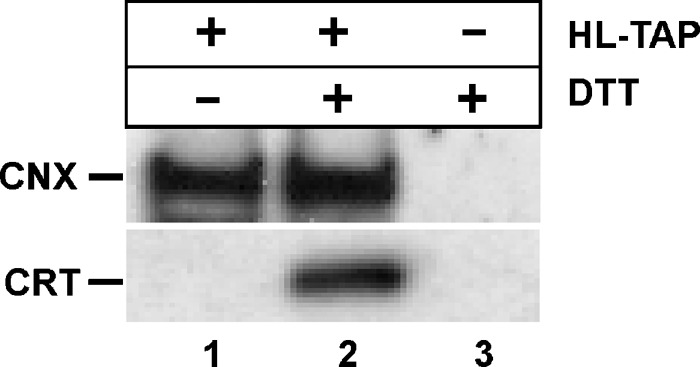
Interestingly, a second lectin chaperone, CRT, was detected only after DTT treatment (Table 2) (Fig. 4, lane 2). CRT has been suggested to be the preferred lectin chaperone during LPL maturation, an assumption based on cotransfection studies in insect cells (41). CNX and CRT share homology and function; both interact with glycoproteins and cycle enzymes in mechanistically identical manners (9, 10, 26, 27, 42). The main difference is in their topology: CNX is a type 1 membrane protein, while CRT is soluble and found in the ER lumen (9, 27, 43). However, while CNX copurified with HL-TAP in both untreated and DTT-treated cells, CRT associated with HL only after DTT treatment (Fig. 4, compare lanes 1, 2). It is likely that CRT association with HL occurs only in the specialized context of DTT-induced protein unfolding. In contrast, membrane-bound CNX consistently interacts with HL, both co- and posttranslationally (see Discussion), an association that is further enhanced in the DTT environment. Thus, CNX appears to be the preferred lectin chaperone involved in HL maturation.
Fig. 4.
Association of the lectin chaperones CNX and CRT with HL in untreated and DTT-treated cells. Cells transfected with (+) or without (−) the HL-TAP construct were treated (+) or not treated (−) with DTT, a reducing agent that causes protein unfolding in the ER. After TAP, the resulting isolates were subjected to Western blot analysis using CNX and CRT antibodies.
Five additional ER proteins, including CRT, were identified only after DTT treatment, including a PPIase family member (FKBP65RS), signal sequence receptor δ (SSRδ), TorsinB, MIR-interacting saposin-like protein (MSAP), and Nodal modulator 1 (Fig. 3) (Table 2). These proteins, like CNX, GII, and UGGT, may normally interact with HL and are enriched in DTT-treated cells due to prolonged associations and increased levels; like CRT, however, they may represent specialized HL associations that do not occur normally. In the former case, the PPIase FKBP65RS associates with BiP-bound substrates (44), as does the PPIase detected in untreated cells (Table 1) (32), and thus may represent a legitimate HL interaction. Both catalyze the cis/trans isomerization of peptidyl-prolyl bonds, a rate-limiting step in protein folding (9, 45). Similarly, SSRδ and the ribosomal Rpl11 protein (Table 2) may be part of a normal cotranslational CNX complex, isolated along with translational factors that were detected in untreated cells (see Discussion). TorsinB is a member of the AAA(+) superfamily of ATPases, but its function is unclear. It is a soluble, N-linked glycoprotein that is localized to the ER (46). It may be involved in protein folding, regulation of protein degradation via the proteasome, and assembly and disassembly of protein complexes, among other functions (46, 47). Thus, in any of these capacities, TorsinB is a candidate for HL maturation; however, without detecting this protein in untreated cells, its participation in normal HL maturation is tentative. MSAP prevents ubiquitination of myosin regulatory light chain, an important component of the cytoskeleton (48). However, a function in HL maturation is unlikely, and its detection may result from the presence of tubulin/actin in HL-TAP isolates. Lastly, nodal modulator 1 has an inferred gene ontology function in carbohydrate binding (GO:0030246; http://www.geneontology.org). At this time, its participation and function in normal HL maturation is tentative and unclear.
HL degradation occurs via the proteasomal pathway
During its maturation, a high percentage of HL never folds successfully (13) and is destined for ERAD. ERAD often occurs after retrotranslocation of the protein into the cytoplasm, where it is ubiquitinated (Ub) by protein-specific E3 ubiquitin-protein ligases (31). The Ub protein is then recognized and degraded by the 26S proteasome. While Ub-mediated degradation is the most common pathway for ERAD (9, 31, 49), LPL degradation was unaffected by proteasomal inhibitors (23). Indeed, other examples indicate that multiple, nonproteasomal pathways of ERAD exist (50, 51). However, in this study, we identified several proteins that suggest HL degradation is Ub mediated: CNX, Grp78, Grp94, and eEF1A are all established players in Ub-mediated degradation (9, 31, 33, 49). In addition, the E3 ubiquitin-protein ligase RNF123 was detected in one HL-TAP experiment in untreated cells (score of 50, three peptides detected with a coverage of 2%; accession number Q5XPI3).
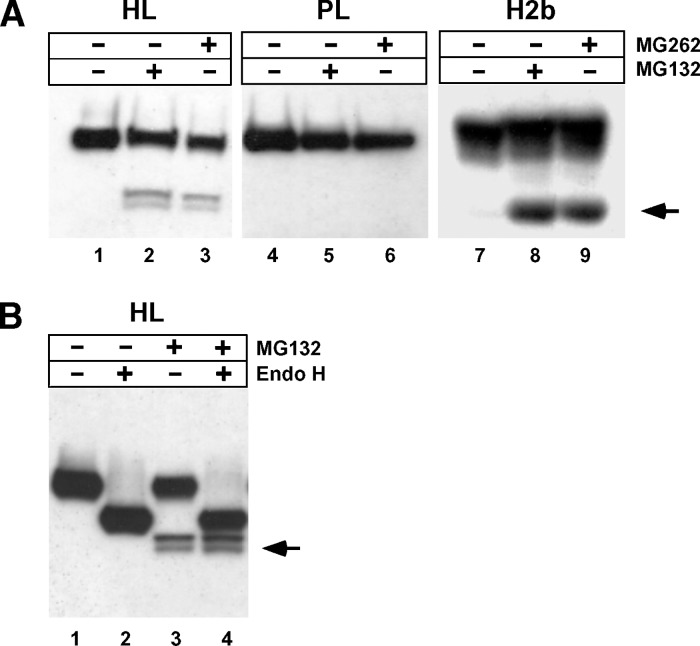
The presence of such proteins suggests that, contrary to LPL, HL may be interacting with factors involved in retrotranslocation and ubiquitination. To test this possibility, the effect of proteasomal inhibitors on HL expressed in CHO cells was investigated. The asialoglycoprotein receptor H2b subunit that undergoes proteasomal degradation (23, 52) was added as a positive control. In addition, PL was included in the analysis, as no candidate proteins involved in ERAD were isolated by PL-TAP. As shown in Fig. 5A, degradation products were clearly seen in cells expressing H2b (lanes 8, 9) and HL (lanes 2, 3) after treatment with proteasomal inhibitors MG132 or MG262 (see arrow); however, such products were absent in PL transfected cells (lanes 5, 6). To evaluate whether HL degradation products were retrotranslocated, their glycosylation status was assessed by endoglycosidase H (endo H) treatment, which removes N-linked high-mannose glycans in vitro. Glycosylation status is a surrogate measure of retrotranslocation, as glycans are removed from ER lumenal proteins emerging into the cytoplasm by a cytosolic-based N-glycanase (9, 53). Fig. 5B shows that that migration of the two degradation products (see arrow) are unaffected after endo H treatment, indicating that they are unglycosylated (compare lanes 3, 4); in contrast, migration of HL in the ER lumen is markedly effected because of the presence of four glycan chains (compare lanes 1, 2). Thus, as suggested by the association of HL with proteins in the proteosomal pathway, misfolded HL is retrotranslocated prior to its degradation by the 26S proteasome.
Fig. 5.
HL ERAD occurs via the proteasomal pathway. A: CHO cells expressing HL, PL, or H2b were incubated with or without the proteasomal inhibitors MG132 and MG262. The accumulation of degradation products is clearly seen in H2b (arrowhead), a well-established example of a secretory protein that undergoes proteasome-mediated degradation. Degradation products also appear when HL is treated with either inhibitor, in contrast to PL, showing no detectable degradation via the proteasomal pathway. B: The HL degradation product is unglycosylated. HL from CHO cells incubated with or without the proteasomal inhibitor MG132 was subjected to endo H cleavage. The arrow points to the HL degradation product (two bands).
DISCUSSION
Glycoprotein maturation occurs through sequential, interdependent stages that rely on groups of chaperones and folding factors in the ER and cytoplasm. Until this study, only two factors were implicated in HL maturation: the lectin chaperone, CNX (13, 14), and the recently identified Lmf1, an ER transmembrane protein affected by the cld mutation (15). This study increases the list substantially and shows that HL uses both multichaperone systems operating in the ER, the BiP/Grp94 system, and the CNX/CRT system (31, 32). Together, these factors presumably assist in HL maturation in a variety of ways. They limit unproductive folding pathways by preventing the inappropriate interaction of nascent N-terminal sequences until HL translation is complete; they catalyze the formation and isomerization of disulfide bonds; they detect and stabilize misfolded forms until productive HL folding occurs; and in the case of terminally misfolded HL, they direct its retrotranslocation and ultimate degradation in the cytoplasm. Our results suggest that HL maturation relies on normal ER function and the integrity of the CNX and BiP/Grp94 chaperone systems; conditions, such as ER stress, that alter these systems would conceivably affect HL folding and assembly.
Cotranslational HL maturation
The tandem affinity tag used in this study was located at the C terminus of the HL protein. Thus, only full-length HL polypeptides were isolated. However, there are compelling reasons to suspect that at least some of the proteins that copurified with the HL-TAP construct associated with HL cotranslationally, when growing nascent chains entered the ER lumen through the translocon complex. For any protein, this is a sensitive time, when inappropriate aggregation and misfolding of nascent chains can occur before the protein is fully synthesized (9 10 11 12). The immediate cotranslational interaction of chaperones with nascent chains stabilizes any hydrophobic constellations that might give rise to aggregation and prevents folding reactions before important modifications can occur, such as disulfide bond formation and peptidyl-prolyl cis/trans isomerization (9).
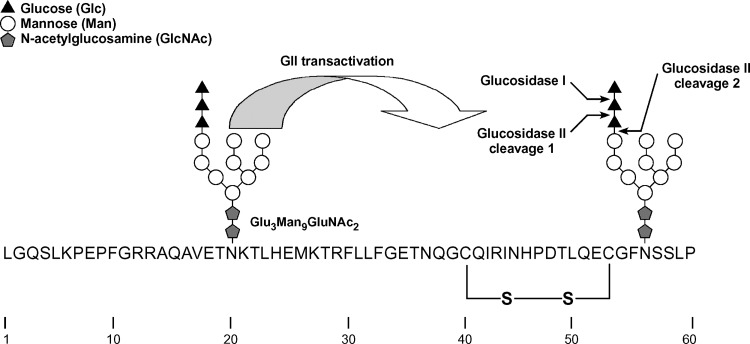
Generally, nascent proteins associate cotranslationally with one of the two major chaperone systems in the ER, the BiP/Grp94 complex, or the CNX/CRT lectin chaperone system (54). For HL, we propose that the latter system is preferred, more specifically, the membrane-bound lectin chaperone, CNX. CNX recognizes and binds nascent and full-length proteins via their N-linked glycan moieties. The lectin domain of CNX recognizes a specific glycan structure, Glc1Man9GluNAc2, termed the monoglucosylated high mannose form (10, 26, 27, 55). Glucosidase I, a membrane-bound enzyme, immediately removes the outermost glucose, while GII, a soluble heterodimer (56), more slowly removes the next glucose to form the monoglucosylated form recognized by CNX (Fig. 6). GII also removes the last glucose residue, causing release of the bound glycoprotein from CNX (10, 26, 27, 55). In the context of HL maturation, CNX and GII were detected as part of the proteome of HL interacting factors, in both untreated and DTT-treated cells (Tables 1, 2) (Fig. 7A).
Fig. 6.
The sequence and glycosylation of the first N-terminal 60 amino acid residues of human HL lacking the signal peptide. Two N-linked glycosylation sites are present at positions 20 and 56. The glycan structure shown is the unprocessed Glc3Man9GlcNAc2 that is transferred by the oligosaccharyl transferase to the growing polypeptide chain. The first two processing events are removal of the outermost glucose residue by glucosidase I (GI), followed by removal of the second glucose residue by GII. The removal of the second glucose residue from the N-linked glycan at Asn56 is greatly facilitated by transactivation of GII by the N-linked glycan at Asn20 (see text). The resulting Glc1Man9GlcNAc2 is a substrate for CNX binding. The removal of the final glucose residue is also catalyzed by GII, and its removal signals the release of HL from CNX. This second cleavage is not affected by transactivation. Notice the close juxtaposition of the first disulfide bond in HL (Cys40—Cys53) with regard to the second N-glycan chain at Asn56.
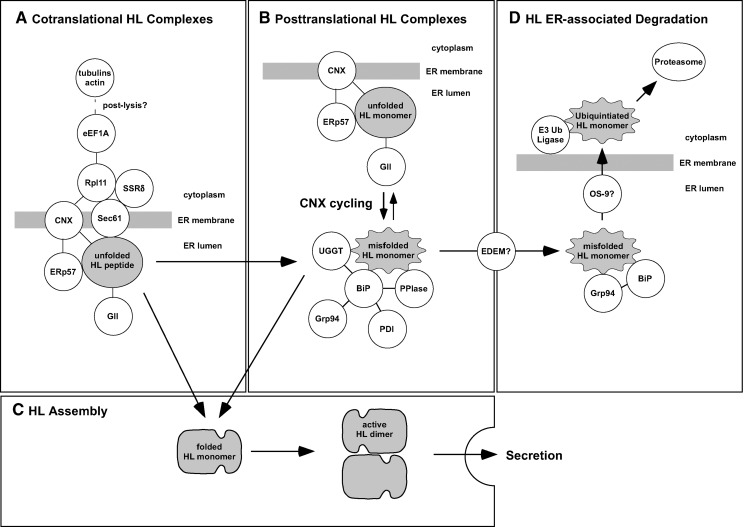
Fig. 7.
A model of protein complexes formed during HL maturation/degradation. HL is depicted in four states of folding: partially or fully translated HL that is unfolded (gray oval); misfolded HL monomer (wavy gray oval); folded HL monomer (gray hammerhead); and assembled HL homodimer (head-to-tail gray hammerhead). Only the HL dimer exhibits enzyme activity, and it is the only form secreted by the cell. All depicted interacting proteins (while circles) were identified in this study with the exceptions of the Sec61 complex, EDEM, and OS-9, which are inferred.
While both CNX and BiP/Grp94 were detected in our study, we believe that HL first uses the CNX chaperone system based on the position of its glycan moieties. Using model viral glycoprotein substrates, it has been shown that the initial choice between the two chaperone systems depends on the location of the first N-linked glycan. Direct interaction with CNX without prior interaction with BiP/Grp94 occurs if the glycans are present within ∼50 amino acids of the N terminus of the protein (54). In the case of HL, two such glycans fall into this category, one at Asn20 and the second at Asn56 (Fig. 6). The glycan at Asn56 is conserved in all species and among the lipase family members LPL and endothelial lipase (4, 25, 57). In HL and LPL, only removal of this early conserved Asn-linked consensus site, and no other N-glycan sites, results in major maturation defects, leading to inactive lipase molecules that are retained in the ER (24, 58). Thus, among the four-glycan chains in human HL, the glycan at Asn56 is the likely target for CNX binding. Moreover, the N-terminal region surrounding Asn56 may be susceptible to misfolding, as Asn56 is situated next to a Phe residue that is part of a hydrophobic constellation of residues conserved in HL and LPL (59); additionally, it is near two Cys residues at positions 40 and 53 that form the first disulfide bridge (Fig. 6). Neither the hydrophobic constellation nor the disulfide bridge is present in PL, which also lacks N-linked glycosylation in this region. Indeed, neither CNX nor its associated factors were detected when PL-TAP was used as the entry point.
While the glycan at Asn56 may be the target for HL/CNX binding, the presence of an additional nearby glycan at Asn20 suggests that the timing of HL/CNX binding is cotranslational rather than posttranslational. While GII-catalyzed cleavage of the second glucose is the critical event in CNX binding, the rate of this cleavage is dependent on the presence of a nearby (proximal) N-linked glycan, as demonstrated using model substrates (55). The noncatalytic β-subunit of GII contains an MRH domain that is homologous to the mannose-6-phosphate receptor (60) that recognizes the mannose structure of glycan residues. Notably, this domain of GII is oriented with respect to the catalytic α-subunit to recognize the mannose structure of a glycan proximal to the one being cleaved (Fig. 6). The ensuing interaction triggers transactivation of the GII catalytic α-subunit, resulting in rapid cleavage of the second glucose from the distal (target) glycan (55). If a proximal glycan is not present, then glucose removal from the target glycan of model substrates is slow and CNX/CRT binding occurs posttranslationally (55). The juxtaposition of Asn20 to Asn56 (35 residues apart; see Fig. 6) is well within an intrapeptide distance that was used to show GII transactivation in a model substrate having two N-linked glycans 46 residues apart (55).
Figure 7A shows proposed interactions occurring during HL cotranslational maturation. Along with CNX and GII, we also detected ERp57, catalyzing the oxidation, reduction, and isomerization of disulfide bonds during protein folding (61). This oxidoreductase binds specifically to a proline-rich region of CNX, called the p-domain (61, 62). Besides ERp57, a second major oxidoreductase, PDI, was also detected; however, unlike ERp57, PDI associates with BiP (32) and not CNX. In the context of a cotranslational CNX/HL association, we also detected the ribosomal protein Rpl11, SSRδ, and associated translation initiation and elongation factors (eLF4A-1 and eEF1A). CNX has been shown to bind directly to the ribosome via its phosphorylated cytosolic domain, an interaction that increases in response to glycoprotein synthesis (63). Thus, both ER lumenal folding factors and cytosolic translational factors would be expected to be present in a CNX/HL complex of cotranslational origin.
Posttranslational HL maturation
The cotranslational use of CNX by HL suggests that its posttranslational folding would proceed according to pathways described for well-studied model substrates (9, 10, 26). Thus, we propose that HL would be released from CNX by the GII-catalyzed cleavage of the last glucose residue from the Asn56 glycan (Fig. 6); after release, the full-length HL polypeptide would undergo folding, presumably free of chaperones (9). Its posttranslational fate would then be determined by the conformation of the folded structure. A misfolded HL monomer would likely associate with UGGT, a glucosyltransferase with a domain that can sense localized regions of polypeptide disorder (64, 65). UGGT would then reglucosylate the glycan residue of the misfolded HL monomer, making it a target once again for CNX binding. Repeated cycles of CNX release and binding would provide a misfolded HL monomer with multiple chances to fold correctly. Our results show that UGGT coisolated with HL-TAP in both untreated and DTT-treated cells, suggesting that HL cycles between the unfolded CNX-bound state and the misfolded UGGT-associated state (Fig. 7B).
Recently, it has been proposed that only a few of the many glycoproteins that bind CNX/CRT require more than a single cycle to fold efficiently (66). However, the long resident time of HL in the ER of CHO cells (a t1/2 of 2 h) (13), along with our finding of UGGT association, suggests that HL probably engages in repeated rounds of CNX cycling. UGGT has been found associated with a large complex of ER proteins, including BiP, PDI, Grp94, and PPIase (32). Thus, it is possible that misfolded HL monomers are first stabilized by association with BiP, followed by UGGT-catalyzed reglucosylation, all occurring within the same complex (Fig. 7B). BiP has a strong affinity for misfolded proteins, and it plays the major role in the quality control system ensuring that misfolded proteins are retained in the ER (11, 31). Once rebound by CNX, disulfide bonds would again be accessible to ERp57 (Fig. 7B). After its release from CNX by GII, if HL folds correctly, it would no longer be detected by BiP or UGGT and would finally assemble into a fully functional head-to-tail homodimer (Fig. 7C). The HL assembly step does not require the formation of disulfide bonds, and thus may be a process that occurs without assistance. However, Grp94, which was detected in our analysis, has been implicated in promoting the assembly of heterodimeric proteins, such as Toll-like receptors (67).
ERAD of misfolded HL
While at least half of unfolded HL becomes a functional homodimer that is rapidly secreted, the remainder is degraded (13). In this study, we show that HL degradation likely occurs via the Ub-mediated proteasomal pathway. While this is a pathway used by most glycoproteins (9, 31, 49), it was found that LPL is degraded via a nonproteasomal, nonlysosomal pathway that is markedly decreased by ATP depletion (23). Why two close family members use different degradation pathways is unclear, except to note that LPL maturation occurs much more rapidly than does HL (13, 23). The persistence of HL/CNX interaction remains well after translation has completed (13), suggesting that HL is involved in cycles of CNX binding. Indeed, our identification of UGGT supports HL/CNX cycling (see above). For cycling glycoproteins, a glycan “timer” determines when it is time to depart CNX cycling and enter into the degradation pathway (10). Specifically, this timer is ER α-mannosidase I, which catalyzes the removal of the mannose residue from the second (B) branch of the glycan chain to form the Man8GlcNAc2 isomer Man8B (9, 10). The Man8B isomer is a poor substrate for UGGT and GII but a good substrate for the EDEM receptors that initiate targeting of glycoproteins for retrotranslocation and Ub-mediated degradation (9, 10). Eventually, further mannose trimming causes EDEM-bound substrates to associate with mannose binding proteins OS-9/XTP3-B, which along with Grp94/BiP, facilitate retrotranslocation and ubiquitination by the Hrd1-SEL1L complex (68, 69).
While we did not detect any EDEM receptors or downstream effectors in our HL-TAP experiments, we did detect the Grp94/BiP complex and the E3 ubiquitin ligase RNF123 (Fig. 7D). Thus, based on our HL-TAP experiments, it is not possible to determine if HL is degraded through the EDEM/OS-9 pathway. However, based on other CNX-cycling glycoproteins (10), this pathway appears to be the best candidate for HL ERAD.
CONCLUDING REMARKS
While the identification of interacting proteins has been enhanced by effective strategies for the isolation of protein-protein complexes, it is limited when interactions occur in a transient manner. The detection of transient associations depends on how long an association lasts, particularly if it occurs at a single site with low affinity. Since interactions involving HL maturation are transient in nature, we believe the HL-TAP approach used in this study has the resolution to detect interactions occurring at relatively high rates with more abundant proteins in the ER. Thus, our approach undoubtedly fails to detect less abundant proteins that interact with HL transiently during its maturation. We believe the evolution of new tandem affinity tags, particularly those designed for work in mammalian systems and for the detection of transient interactions (16, 17, 21), offer the promise of expanding the current list of ER factors having important roles in lipase maturation.
Acknowledgments
The authors thank Hui Zhen Mao for help in the preparation of the lipase TAP constructs.
Abbreviations
CHO, Chinese hamster ovary
CNX, calnexin
CRT, calreticulin
DSP, dithiobis-(succinimidylpropionate)
eEF1A, elongation factor 1-α
eLF4A-1, Eukaryotic initiation factor 4A-1
endo H, endoglycosidase H
ER, endoplasmic reticulum
ERAD, endoplasmic reticulum–associated degradation
ERp57, 57 kDa endoplasmic reticulum protein
GII, glucosidase II
Grp78/BiP, 78 kDa glucose-regulated protein
Grp94, 94 kDa glucose-regulated protein
HL, hepatic lipase
Hsp47, 47 kDa heat shock protein
Lmf1, Lipase Maturation Factor 1
MS/MS, tandem mass spectrometry
MSAP, MIR-interacting saposin-like protein
PDI, protein disulfide isomerase
PL, pancreatic lipase
SSRδ, signal sequence receptor δ
TAP, tandem affinity purification
Ub, ubiquitin
UGGT, UDP-glucose:glycoprotein glucosyltransferase 1
This work was supported by the Cedars-Sinai Medical Center, the Department of Veterans Affairs, and Grant HL-24841 from the National Institutes of Health.
Published, JLR Papers in Press, January 8, 2009.
References
- 1.Hasham S. N., and S. Pillarisetti. 2006. Vascular lipases, inflammation and atherosclerosis. Clin. Chim. Acta. 372 179–183. [DOI] [PubMed] [Google Scholar]
- 2.Perret B., L. Mabile, L. Martinez, F. Terce, R. Barbaras, and X. Collet. 2002. Hepatic lipase: structure/function relationship, synthesis, and regulation. J. Lipid Res. 43 1163–1169. [PubMed] [Google Scholar]
- 3.Santamarina-Fojo S., H. Gonzalez-Navarro, L. Freeman, E. Wagner, and Z. Nong. 2004. Hepatic lipase, lipoprotein metabolism, and atherogenesis. Arterioscler. Thromb. Vasc. Biol. 24 1750–1754. [DOI] [PubMed] [Google Scholar]
- 4.Wong H., and M. C. Schotz. 2002. The lipase gene family. J. Lipid Res. 43 993–999. [DOI] [PubMed] [Google Scholar]
- 5.Doolittle M. H., H. Wong, R. C. Davis, and M. C. Schotz. 1987. Synthesis of hepatic lipase in liver and extrahepatic tissues. J. Lipid Res. 28 1326–1334. [PubMed] [Google Scholar]
- 6.Gonzalez-Navarro H., Z. Nong, L. Freeman, A. Bensadoun, K. Peterson, and S. Santamarina-Fojo. 2002. Identification of mouse and human macrophages as a site of synthesis of hepatic lipase. J. Lipid Res. 43 671–675. [PubMed] [Google Scholar]
- 7.Cohen J. C., G. L. Vega, and S. M. Grundy. 1999. Hepatic lipase: new insights from genetic and metabolic studies. Curr. Opin. Lipidol. 10 259–267. [DOI] [PubMed] [Google Scholar]
- 8.Deeb S. S., A. Zambon, M. C. Carr, A. F. Ayyobi, and J. D. Brunzell. 2003. Hepatic lipase and dyslipidemia: interactions among genetic variants, obesity, gender, and diet. J. Lipid Res. 44 1279–1286. [DOI] [PubMed] [Google Scholar]
- 9.Hebert D. N., and M. Molinari. 2007. In and out of the ER: protein folding, quality control, degradation, and related human diseases. Physiol. Rev. 87 1377–1408. [DOI] [PubMed] [Google Scholar]
- 10.Hebert D. N., S. C. Garman, and M. Molinari. 2005. The glycan code of the endoplasmic reticulum: asparagine-linked carbohydrates as protein maturation and quality-control tags. Trends Cell Biol. 15 364–370. [DOI] [PubMed] [Google Scholar]
- 11.Sitia R., and I. Braakman. 2003. Quality control in the endoplasmic reticulum protein factory. Nature. 426 891–894. [DOI] [PubMed] [Google Scholar]
- 12.Trombetta E. S., and A. J. Parodi. 2003. Quality control and protein folding in the secretory pathway. Annu. Rev. Cell Dev. Biol. 19 649–676. [DOI] [PubMed] [Google Scholar]
- 13.Ben-Zeev O., and M. H. Doolittle. 2004. Maturation of hepatic lipase. Formation of functional enzyme in the endoplasmic reticulum is the rate-limiting step in its secretion. J. Biol. Chem. 279 6171–6181. [DOI] [PubMed] [Google Scholar]
- 14.Boedeker J. C., M. H. Doolittle, and A. L. White. 2001. Differential effect of combined lipase deficiency (cld/cld) on human hepatic lipase and lipoprotein lipase secretion. J. Lipid Res. 42 1858–1864. [PubMed] [Google Scholar]
- 15.Peterfy M., O. Ben-Zeev, H. Z. Mao, D. Weissglas-Volkov, B. E. Aouizerat, C. R. Pullinger, P. H. Frost, J. P. Kane, M. J. Malloy, K. Reue, et al. 2007. Mutations in LMF1 cause combined lipase deficiency and severe hypertriglyceridemia. Nat. Genet. 39 1483–1487. [DOI] [PubMed] [Google Scholar]
- 16.Burckstummer T., K. L. Bennett, A. Preradovic, G. Schutze, O. Hantschel, G. Superti-Furga, and A. Bauch. 2006. An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat. Methods. 3 1013–1019. [DOI] [PubMed] [Google Scholar]
- 17.Collins M. O., and J. S. Choudhary. 2008. Mapping multiprotein complexes by affinity purification and mass spectrometry. Curr. Opin. Biotechnol. 19 324–330. [DOI] [PubMed] [Google Scholar]
- 18.Puig O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret, E. Bragado-Nilsson, M. Wilm, and B. Seraphin. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 24 218–229. [DOI] [PubMed] [Google Scholar]
- 19.Rigaut G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17 1030–1032. [DOI] [PubMed] [Google Scholar]
- 20.Tagwerker C., K. Flick, M. Cui, C. Guerrero, Y. Dou, B. Auer, P. Baldi, L. Huang, and P. Kaiser. 2006. A tandem affinity tag for two-step purification under fully denaturing conditions: application in ubiquitin profiling and protein complex identification combined with in vivo cross-linking. Mol. Cell. Proteomics. 5 737–748. [DOI] [PubMed] [Google Scholar]
- 21.Guerrero C., C. Tagwerker, P. Kaiser, and L. Huang. 2006. An integrated mass spectrometry-based proteomic approach: quantitative analysis of tandem affinity-purified in vivo cross-linked protein complexes (QTAX) to decipher the 26 S proteasome-interacting network. Mol. Cell. Proteomics. 5 366–378. [DOI] [PubMed] [Google Scholar]
- 22.Gavin A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, et al. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 415 141–147. [DOI] [PubMed] [Google Scholar]
- 23.Ben-Zeev O., H. Z. Mao, and M. H. Doolittle. 2002. Maturation of lipoprotein lipase in the endoplasmic reticulum. Concurrent formation of functional dimers and inactive aggregates. J. Biol. Chem. 277 10727–10738. [DOI] [PubMed] [Google Scholar]
- 24.Ben-Zeev O., G. Stahnke, G. Liu, R. C. Davis, and M. H. Doolittle. 1994. Lipoprotein lipase and hepatic lipase: the role of asparagine-linked glycosylation in the expression of a functional enzyme. J. Lipid Res. 35 1511–1523. [PubMed] [Google Scholar]
- 25.Hide W. A., L. Chan, and W. H. Li. 1992. Structure and evolution of the lipase superfamily. J. Lipid Res. 33 167–178. [PubMed] [Google Scholar]
- 26.Helenius A., and M. Aebi. 2004. Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 73 1019–1049. [DOI] [PubMed] [Google Scholar]
- 27.Ruddock L. W., and M. Molinari. 2006. N-glycan processing in ER quality control. J. Cell Sci. 119 4373–4380. [DOI] [PubMed] [Google Scholar]
- 28.Ozawa M., and T. Muramatsu. 1993. Reticulocalbin, a novel endoplasmic reticulum resident Ca(2+)-binding protein with multiple EF-hand motifs and a carboxyl-terminal HDEL sequence. J. Biol. Chem. 268 699–705. [PubMed] [Google Scholar]
- 29.Gorlach A., P. Klappa, and T. Kietzmann. 2006. The endoplasmic reticulum: folding, calcium homeostasis, signaling, and redox control. Antioxid. Redox Signal. 8 1391–1418. [DOI] [PubMed] [Google Scholar]
- 30.Honore B., and H. Vorum. 2000. The CREC family, a novel family of multiple EF-hand, low-affinity Ca(2+)-binding proteins localised to the secretory pathway of mammalian cells. FEBS Lett. 466 11–18. [DOI] [PubMed] [Google Scholar]
- 31.Ma Y., and L. M. Hendershot. 2004. ER chaperone functions during normal and stress conditions. J. Chem. Neuroanat. 28 51–65. [DOI] [PubMed] [Google Scholar]
- 32.Meunier L., Y. K. Usherwood, K. T. Chung, and L. M. Hendershot. 2002. A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol. Biol. Cell. 13 4456–4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lamberti A., M. Caraglia, O. Longo, M. Marra, A. Abbruzzese, and P. Arcari. 2004. The translation elongation factor 1A in tumorigenesis, signal transduction and apoptosis: review article. Amino Acids. 26 443–448. [DOI] [PubMed] [Google Scholar]
- 34.Condeelis J. 1995. Elongation factor 1 alpha, translation and the cytoskeleton. Trends Biochem. Sci. 20 169–170. [DOI] [PubMed] [Google Scholar]
- 35.Braakman I., J. Helenius, and A. Helenius. 1992. Manipulating disulfide bond formation and protein folding in the endoplasmic reticulum. EMBO J. 11 1717–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koide T., S. Asada, Y. Takahara, Y. Nishikawa, K. Nagata, and K. Kitagawa. 2006. Specific recognition of the collagen triple helix by chaperone HSP47: minimal structural requirement and spatial molecular orientation. J. Biol. Chem. 281 3432–3438. [DOI] [PubMed] [Google Scholar]
- 37.Koide T., Y. Nishikawa, S. Asada, C. M. Yamazaki, Y. Takahara, D. L. Homma, A. Otaka, K. Ohtani, N. Wakamiya, K. Nagata, et al. 2006. Specific recognition of the collagen triple helix by chaperone HSP47. II. The HSP47-binding structural motif in collagens and related proteins. J. Biol. Chem. 281 11177–11185. [DOI] [PubMed] [Google Scholar]
- 38.Koide T., Y. Takahara, S. Asada, and K. Nagata. 2002. Xaa-Arg-Gly triplets in the collagen triple helix are dominant binding sites for the molecular chaperone HSP47. J. Biol. Chem. 277 6178–6182. [DOI] [PubMed] [Google Scholar]
- 39.Makareeva E., and S. Leikin. 2007. Procollagen triple helix assembly: an unconventional chaperone-assisted folding paradigm. PLoS One. 2 e1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ishida Y., H. Kubota, A. Yamamoto, A. Kitamura, H. P. Bachinger, and K. Nagata. 2006. Type I collagen in Hsp47-null cells is aggregated in endoplasmic reticulum and deficient in N-propeptide processing and fibrillogenesis. Mol. Biol. Cell. 17 2346–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang L., G. Wu, C. G. Tate, A. Lookene, and G. Olivecrona. 2003. Calreticulin promotes folding/dimerization of human lipoprotein lipase expressed in insect cells (sf21). J. Biol. Chem. 278 29344–29351. [DOI] [PubMed] [Google Scholar]
- 42.Caramelo J. J., and A. J. Parodi. 2008. Getting in and out from calnexin/calreticulin cycles. J. Biol. Chem. 283 10221–10225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Molinari M., K. K. Eriksson, V. Calanca, C. Galli, P. Cresswell, M. Michalak, and A. Helenius. 2004. Contrasting functions of calreticulin and calnexin in glycoprotein folding and ER quality control. Mol. Cell. 13 125–135. [DOI] [PubMed] [Google Scholar]
- 44.Davis E. C., T. J. Broekelmann, Y. Ozawa, and R. P. Mecham. 1998. Identification of tropoelastin as a ligand for the 65-kD FK506-binding protein, FKBP65, in the secretory pathway. J. Cell Biol. 140 295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kiefhaber T., R. Quaas, U. Hahn, and F. X. Schmid. 1990. Folding of ribonuclease T1. 2. Kinetic models for the folding and unfolding reactions. Biochemistry. 29 3061–3070. [DOI] [PubMed] [Google Scholar]
- 46.O'Farrell C., P. J. Lockhart, S. Lincoln, M. De Lucia, A. B. Singleton, D. W. Dickson, and M. R. Cookson. 2004. Biochemical characterization of torsinB. Brain Res. Mol. Brain Res. 127 1–9. [DOI] [PubMed] [Google Scholar]
- 47.Ogura T., and A. J. Wilkinson. 2001. AAA+ superfamily ATPases: common structure–diverse function. Genes Cells. 6 575–597. [DOI] [PubMed] [Google Scholar]
- 48.Bornhauser B. C., P. A. Olsson, and D. Lindholm. 2003. MSAP is a novel MIR-interacting protein that enhances neurite outgrowth and increases myosin regulatory light chain. J. Biol. Chem. 278 35412–35420. [DOI] [PubMed] [Google Scholar]
- 49.Nakatsukasa K., and J. L. Brodsky. 2008. The recognition and retrotranslocation of misfolded proteins from the endoplasmic reticulum. Traffic. 9 861–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Geier E., G. Pfeifer, M. Wilm, M. Lucchiari-Hartz, W. Baumeister, K. Eichmann, and G. Niedermann. 1999. A giant protease with potential to substitute for some functions of the proteasome. Science. 283 978–981. [DOI] [PubMed] [Google Scholar]
- 51.Mancini R., M. Aebi, and A. Helenius. 2003. Multiple endoplasmic reticulum-associated pathways degrade mutant yeast carboxypeptidase Y in mammalian cells. J. Biol. Chem. 278 46895–46905. [DOI] [PubMed] [Google Scholar]
- 52.Kamhi-Nesher S., M. Shenkman, S. Tolchinsky, S. V. Fromm, R. Ehrlich, and G. Z. Lederkremer. 2001. A novel quality control compartment derived from the endoplasmic reticulum. Mol. Biol. Cell. 12 1711–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hirsch C., D. Blom, and H. L. Ploegh. 2003. A role for N-glycanase in the cytosolic turnover of glycoproteins. EMBO J. 22 1036–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Molinari M., and A. Helenius. 2000. Chaperone selection during glycoprotein translocation into the endoplasmic reticulum. Science. 288 331–333. [DOI] [PubMed] [Google Scholar]
- 55.Deprez P., M. Gautschi, and A. Helenius. 2005. More than one glycan is needed for ER glucosidase II to allow entry of glycoproteins into the calnexin/calreticulin cycle. Mol. Cell. 19 183–195. [DOI] [PubMed] [Google Scholar]
- 56.Trombetta E. S., J. F. Simons, and A. Helenius. 1996. Endoplasmic reticulum glucosidase II is composed of a catalytic subunit, conserved from yeast to mammals, and a tightly bound noncatalytic HDEL-containing subunit. J. Biol. Chem. 271 27509–27516. [DOI] [PubMed] [Google Scholar]
- 57.Rader D. J., and M. Jaye. 2000. Endothelial lipase: a new member of the triglyceride lipase gene family. Curr. Opin. Lipidol. 11 141–147. [DOI] [PubMed] [Google Scholar]
- 58.Wolle J., H. Jansen, L. C. Smith, and L. Chan. 1993. Functional role of N-linked glycosylation in human hepatic lipase: asparagine-56 is important for both enzyme activity and secretion. J. Lipid Res. 34 2169–2176. [PubMed] [Google Scholar]
- 59.Derewenda Z. S., and C. Cambillau. 1991. Effects of gene mutations in lipoprotein and hepatic lipases as interpreted by a molecular model of the pancreatic triglyceride lipase. J. Biol. Chem. 266 23112–23119. [PubMed] [Google Scholar]
- 60.Munro S. 2001. The MRH domain suggests a shared ancestry for the mannose 6-phosphate receptors and other N-glycan-recognising proteins. Curr. Biol. 11 R499–R501. [DOI] [PubMed] [Google Scholar]
- 61.Maattanen P., G. Kozlov, K. Gehring, and D. Y. Thomas. 2006. ERp57 and PDI: multifunctional protein disulfide isomerases with similar domain architectures but differing substrate-partner associations. Biochem. Cell Biol. 84 881–889. [DOI] [PubMed] [Google Scholar]
- 62.Oliver J. D., H. L. Roderick, D. H. Llewellyn, and S. High. 1999. ERp57 functions as a subunit of specific complexes formed with the ER lectins calreticulin and calnexin. Mol. Biol. Cell. 10 2573–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chevet E., H. N. Wong, D. Gerber, C. Cochet, A. Fazel, P. H. Cameron, J. N. Gushue, D. Y. Thomas, and J. J. Bergeron. 1999. Phosphorylation by CK2 and MAPK enhances calnexin association with ribosomes. EMBO J. 18 3655–3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ritter C., K. Quirin, M. Kowarik, and A. Helenius. 2005. Minor folding defects trigger local modification of glycoproteins by the ER folding sensor GT. EMBO J. 24 1730–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Taylor S. C., A. D. Ferguson, J. J. Bergeron, and D. Y. Thomas. 2004. The ER protein folding sensor UDP-glucose glycoprotein-glucosyltransferase modifies substrates distant to local changes in glycoprotein conformation. Nat. Struct. Mol. Biol. 11 128–134. [DOI] [PubMed] [Google Scholar]
- 66.Molinari M., C. Galli, O. Vanoni, S. M. Arnold, and R. J. Kaufman. 2005. Persistent glycoprotein misfolding activates the glucosidase II/UGT1-driven calnexin cycle to delay aggregation and loss of folding competence. Mol. Cell. 20 503–512. [DOI] [PubMed] [Google Scholar]
- 67.Yang Y., B. Liu, J. Dai, P. K. Srivastava, D. J. Zammit, L. Lefrancois, and Z. Li. 2007. Heat shock protein gp96 is a master chaperone for toll-like receptors and is important in the innate function of macrophages. Immunity. 26 215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Christianson J. C., T. A. Shaler, R. E. Tyler, and R. R. Kopito. 2008. OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1–SEL1L ubiquitin ligase complex for ERAD. Nat. Cell Biol. 10 272–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tamura T., J. H. Cormier, and D. N. Hebert. 2008. Sweet bays of ERAD. Trends Biochem. Sci. 33 298–300. [DOI] [PubMed] [Google Scholar]