Abstract
Subtypes of breast cancer that represent the two major types of epithelial cells in the breast (luminal and basal) carry distinct histopathological profiles. Breast cancers of the basal-like subtype, which include the majority of hereditary breast cancers due to mutations in the Breast Cancer Susceptibility Gene 1 (BRCA1), frequently assume triple-negative status, i.e. they lack expression of estrogen receptor-α and progesterone receptor and lack overexpression or amplification of the HER2/NEU oncogene. Defects in DNA damage response pathways result in genome instability and lead to carcinogenesis, but may also be exploited for therapeutic purposes. We analyzed repair of oxidative DNA damage (ODD) by the base-excision repair (BER) pathway, which when aberrant leads to genomic instability and breast carcinogenesis, in cell lines that represent the different subtypes of breast cancer and in the presence of BRCA1 deficiency. We found that basal-like and BRCA1-mutated breast cancer cells were defective in BER of ODD, and that this defect conferred sensitivity to inhibition of poly(ADP-ribose) polymerase, a DNA repair enzyme. The defect may be attributed, at least in part, to a novel role for BRCA1 in the BER pathway. Overall, these data offer preventive, prognostic, and therapeutic usefulness.
Keywords: DNA base excision repair, Oxidative DNA damage, Poly(ADP-ribose) Polymerase, Basal-like breast cancer, Breast cancer susceptibility gene 1
INTRODUCTION
Breast cancer is a heterogeneous disease that consists of different molecular and pathologic subtypes. The subtypes represent the two major types of epithelial cells in the breast (luminal and basal) and were first identified by gene expression profiling (1). Each subtype carries a distinct histopathological profile. Breast cancers of the basal-like subtype frequently assume triple-negative status, i.e. they lack expression of estrogen receptor-α (ER-α) and progesterone receptor (PR) and lack overexpression or amplification of the HER2/NEU oncogene (1-4). Furthermore, they express genes linked to proliferation and often associate with a more aggressive clinical course, including poor clinical outcome, recurrence, and metastasis (1, 2, 4-11).
Hereditary breast cancers due to mutations in the Breast Cancer Susceptibility Gene 1 (BRCA1) are primarily of the basal-like subtype and also tend to be triple-negative (11-13). They share similar gene expression patterns, morphological characteristics, immunohistochemical profiles, and pathological features with sporadic basal-like breast cancers, including high histological grade, high mitotic index, overexpression of EGFR, mutations in p53, and cytogenetic abnormalities (14).
The triple-negative status of many basal-like and BRCA1-mutated breast cancers make these tumors particularly challenging to treat, since targeted agents against ER and HER2/NEU are ineffective in this setting. Therefore, cytotoxic chemotherapy and radiation remain the only therapeutic options. Unfortunately, despite response to conventional regimens, these cancers retain a poor prognosis (15), suggesting the need for improved systemic treatments.
Defects in DNA damage response pathways result in genome instability and lead to carcinogenesis, but may be exploited for therapeutic purposes. For example, BRCA1 encodes a tumor suppressor that functions in multiple DNA damage response pathways, including DNA double-strand break repair (i.e. homologous recombination (HR) and non-homologous end-joining (NHEJ)) and nucleotide excision repair (NER) (16, 17), and thereby maintains genomic stability and prevents carcinogenesis. Likewise, BRCA1-deficient cells, due to defects in DNA repair, are highly sensitive to drugs, such as mitomycin C and cisplatin, that induce interstrand and intrastrand DNA crosslinks, stalled replication forks, and in turn, DNA double-strand breaks. Recently, BRCA1- and BRCA2-deficient cells have been shown to exhibit sensitivity to poly(ADP-ribose) polymerase (PARP) inhibitors, also thought to be due to their DNA repair defects (18, 19). PARP-1 is one of a family of enzymes that synthesize poly(ADP-ribose) and is known to function in BER by recruiting essential mediators to single-strand break intermediates (20). A better understanding of DNA damage response pathways in subtypes of breast cancer may result in therapeutic benefit.
Oxidative DNA damage (ODD) constitutes the majority of DNA damage in human cells due to reactive oxygen species, which are genotoxic agents generated endogenously by metabolism and other biological processes. ODD typically occurs as single-base alterations and undergoes repair by the base-excision repair (BER) pathway. When left unrepaired, ODD results in mutagenesis. For example, the most common ODD lesion, 8-oxoguanine, can mispair and result in GC®TA transversions. Alternatively, ODD converts to single- or double-strand breaks and results in genomic instability. Overall, these events contribute to the initiation, progression, and maintenance of breast cancer.
Given the many phenotypic similarities between basal-like and BRCA1-mutated breast cancers, we hypothesized they may also share defects in maintaining genomic stability via aberrant regulation of ODD. Little is known about the regulation of ODD among the different subtypes of breast cancer, but understanding these mechanisms may be important for developing more targeted treatment strategies, such as inhibition of PARP or other DNA repair proteins. In this study, we analyzed the ability to repair ODD by BER in the different subtypes of breast cancer and in the presence of BRCA1 deficiency and discovered that basal-like and BRCA1-mutated breast cancer cells were both defective in repairing ODD. We also found that selective attenuation of BER by knockdown of 8-oxoguanine DNA glycosylase (hOGG1) conferred sensitivity to inhibition of PARP, an important DNA repair enzyme. Taken together, our results showed that both BRCA1-mutated and basal-like breast cancer cells were defective in BER, and that this defect conferred sensitivity to PARP inhibition, suggesting novel approaches for treatment of triple-negative breast cancer.
MATERIALS AND METHODS
Cell Lines
BRCA1-mutated cell lines included: HCC1937, MDAMB436, SUM149PT, and SUM1315MO2. Basal-like cell lines included: BT549, HCC38, HCC1143, HCC1500, HCC1806, hs578T, MDAMB231, and MDAMB468. Luminal cell lines included: BT474, MCF7, MDAMB361, and T47D. Normal-like cell lines included: MCF10A and MCF12A. SUM149PT and SUM1315MO2 were obtained from and cultured as recommended by Asterand® plc. All other cell lines were cultured as recommended by ATCC®. BRCA1+/+ and BRCA1-/- murine mammary epithelial cells (MMECs) have been previously described (21). To generate stable isogenic cell lines with altered expression of hOGG1, MCF7 cells were transduced with hOGG1 shRNA or non-targeting control shRNA and selected with 1μg/ml puromycin. All cell lines were maintained at 37°C and 5%CO2.
Chemicals and Reagents
Chemicals included: hydrogen peroxide (H2O2, Fisher Scientific), methyl methanesulfonate (MMS, Sigma-Aldrich), and 1,5-isoquinolinediol (IQD, Calbiochem). The wild-type BRCA1 construct was kindly provided by Simon Powell and has previously been described (22). shRNA to BRCA1 (5′-TGCCAAAGTAGCTAATGTA-3′), two different shRNA to hOGG1 (A, 5′-GTATGGACACTGACTCAGA-3′; B, 5′-GTACTTCCAGCTAGATGTT-3′), and non-targeting control shRNA (5′-GGCTAGACCTCCAAGATCA-3′ and 5′-GGAGATCAGCCATTAATAT-3′) were cloned into pSUPER.retro.puro (Oligoengine) according to the manufacturer’s instructions.
RTqPCR
Total RNA was extracted using the RNeasy® mini kit (Qiagen). Reverse transcription was carried out using the First-strand cDNA synthesis kit (GE Healthcare). The real-time PCR reaction mix contained 10ng template cDNA, Taqman Universal PCR master mix (Applied Biosystems), 900nM hOGG1 primer (forward, 5′-AATTCCAAGGTGTGCGACTG-3′; reverse, 5′-CGATGTTGTTGTTGGAGGAAC-3′), 250nM Taqman hOGG1 Probe (5′-CGACAAGACCCCATCGAATGCCTTTTC-3′), and 50nM Taqman® ribosomal RNA Control Reagents/VIC™ Probe (GE Healthcare). Each reaction was dispensed in quadruplicate. The cycling conditions included 50°C for 2 minutes, 95°C for 10 minutes, and 40 cycles of 95°C for 30 seconds and 60°C for 1 minute. The comparative threshold method (23) determined relative RNA expression.
Western blot
Nuclear lysates were prepared using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific) according to the manufacturer’s instructions, resolved by SDS-PAGE, transferred to PVDF membrane, blocked, and probed with the appropriate antibody. Antibodies included anti-BRCA1 (Calbiochem) and anti-tata binding protein (Abcam). Protein was detected by horseradish peroxidase-conjugated secondary antibody and ECL™ detection system (GE Healthcare).
MTT Assay
Cells were seeded in 96-well plates, allowed to adhere, and treated in quadruplicate with vehicle-control or increasing concentrations of drug. After the indicated period of time, MTT (Sigma-Aldrich) was added to each well and incubated until formation of formazan crystals by the mitochondria of living cells; formazan crystals were then made soluble with DMSO. Optical density was determined at 570 nm by spectrophotometry. Sensitivity was expressed as a percentage of vehicle-control treated cells by dividing the optical density of each treated well by the mean of the vehicle-control wells. IC50 defines the concentration at which 50% of the cells were inhibited following treatment.
Alkaline comet assay
The alkaline comet assay was modified for detection of oxidized bases using a bacterial repair endonuclease and carried out as previously described (24, 25). Briefly, following cell lysis of logarithmically-growing cells, DNA was subjected to FPG enzyme (i.e. an ODD-specific endonuclease), denatured under alkaline conditions, electrophoresed to resolve fragmented DNA from intact DNA, and then visualized with SYBR® green by fluorescent microscopy under 20X objective as a comet in shape. Comet heads indicate intact DNA; FPG-induced comet tails indicate ODD. ODD was calculated as the % DNA in tail for FPG-treated (+FPG) less control (-FPG) samples using CometScore™ software (TriTek Corporation). Greater than 200 cells for each sample were scored.
BER Assay
Human-specific adenovirus containing the GFP coding sequence (ad-GFP, Clontech) was incubated with 5μM methylene blue and exposed to 2mW/cm2 visible light for zero (undamaged control) or one minute to induce ODD using the photodynamic (PDT) setup (Supp. Fig. 1A). Cells were then infected with undamaged-control or damaged ad-GFP. PDT and infection was carried out in the dark under blue light. For fluorescent microscopy, cells were visualized under 10X objective. For fluorescent plate reading, cells were collected, washed, resuspended in PBS, and counted. Equal cell numbers were dispensed in quadruplicate to black 96-well plates and read at Ex475/Em505 using a fluorescent plate reader. GFP expression was calculated following log transformation of the fluorescent readings as a ratio of damaged to undamaged control. The confidence interval was computed based on a t-distribution using the logged values.
Statistical Analysis
Unless otherwise noted, data were expressed as mean ± s.e.m. P-values were determined using the two-tailed student t-test. Relationships between variables were determined using linear regression analysis and evaluated using the coefficient of determination (r2). Correlations were determined by calculating the Pearson correlation coefficient (r).
RESULTS
Sensitivity to Oxidative DNA Damage among Subtypes of Breast Cancer
To evaluate the response to ODD among the different subtypes of breast cancer and in BRCA1-mutated breast cancers, we first identified a panel of breast cell lines characteristic of the normal breast (i.e. normal-like; MCF10A and MCF12A), as well as luminal (BT474, MCF7, MDAMB361, and T47D), basal-like (BT549, HCC38, HCC1143, HCC1500, HCC1806, hs578T, MDAMB231, and MDAMB468), and BRCA1-mutated (HCC1937, MDAMB436, SUM149PT, and SUM1315MO2) breast cancers. These cell lines were chosen based on previous reports of their breast cancer subtype, gene expression profile, triple-negative status, and BRCA1 genotype (26-29). The basal-like breast cancer cell lines used in this study have all been reported to be triple-negative without BRCA1 mutations. We confirmed triple-negative and BRCA1 status by immunoblotting for ER, PR, and HER2 expression and for BRCA1 nuclear expression, respectively (data not shown). We then analyzed this panel of human breast cell lines for H2O2 sensitivity as an indicator of response to ODD. Cells were treated with increasing concentrations of H2O2 for 72 hours and analyzed for sensitivity by MTT assay. Based on the average IC50 value for each cell line within a subtype (Table 1), the basal-like and BRCA1-mutated breast cancer cell lines were significantly more sensitive to H2O2 than the normal-like breast cell lines (p=0.0003 and p=0.009, respectively), whereas the luminal breast cancer cell lines were not significantly more sensitive to H2O2 than the normal-like breast cell lines (p=0.09) (Fig. 1A). Likewise, the basal-like and BRCA1-mutated cell lines were similarly sensitive to H2O2 (p=0.6), and were both significantly more sensitive to H2O2 than the luminal breast cancer cell lines (p=0.0003 and p=0.006, respectively) (Fig. 1A).
Table 1. H2O2 sensitivity in breast cell lines.
Breast cell lines of various subtypes as indicated were analyzed for H2O2 sensitivity by MTT assay. The average IC50 values determined from the dose-response curve of at least three independent experiments are shown as an average ± s.e.m. p-value was determined by student t-test relative to the normal-like cell lines
| Cell Line | H2O2 IC50 (μM) | p-Value |
|---|---|---|
| Normal-like | ||
| MCF10A | 1008 ± 14 | |
| MCF12A | 700 ± 265 | |
| Luminal | 0.0981 | |
| MCF7 | 550 ± 58 | |
| BT474 | 750 ± 129 | |
| T47D | 500 ± 100 | |
| MDAMB361 | 483 ± 126 | |
| BRCA1 | 0.0094** | |
| SUM149PT | 95 ± 30 | |
| SUM1315MO2 | 192 ± 63 | |
| HCC1937 | 23 ± 12 | |
| MDAMB436 | 367 ± 58 | |
| Basal-like | 0.0003** | |
| BT549 | 13 ± 6 | |
| hs578T | 150 ± 87 | |
| MDAMB231 | 333 ± 76 | |
| HCC38 | 20 ± 5 | |
| HCC1143 | 20 ± 10 | |
| HCC1806 | 50 ± 29 | |
| HCC1500 | 38 ± 37 | |
| MDAMB468 | 333 ± 58 | |
p<0.01.
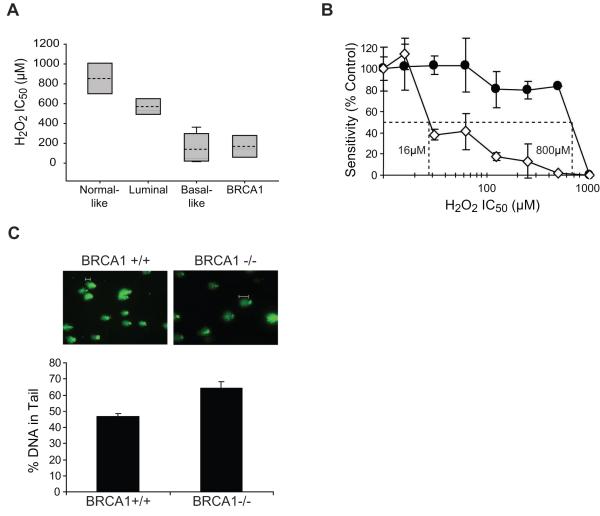
Figure 1. Increased ODD in basal-like and BRCA1-mutated breast cancer cell lines and in the presence of BRCA1 deficiency.
(A) Analysis of H2O2 sensitivity, an indicator of ODD, by MTT assay in cell lines representing the normal breast and each subtype of breast cancer. Each box plot summarizes the data described in Table 1. For each subtype, the grey shaded area indicates the range of IC50 values, the dashed line indicates the average IC50 value, and the solid line indicates the median IC50 value. (B) Analysis of H2O2 sensitivity by MTT assay in BRCA1+/+ (-●-) and BRCA1-/- (-◇-) MMECs. The graph illustrates sensitivity following treatment with increasing concentration of oxidizing agent. The IC50 concentrations as determined by interpolation from the dose-response curves are indicated for each cell line. (C) Basal levels of ODD in BRCA1+/+ and BRCA1-/- MMECs. ODD was measured by the alkaline comet assay modified for detection of oxidized bases using logarithmically growing cells. ODD was visualized by fluorescent microscopy (top), and quantified by measuring the percentage of DNA in comet tails using comet software (bottom). White bars indicate typical comet tails.
Sensitivity to Oxidative DNA Damage in the Presence of BRCA1 Deficiency
To further examine the role of BRCA1 in the response to ODD, we analyzed BRCA1+/+ and BRCA1-/- MMECs for H2O2 sensitivity by MTT assay. These cells have previously been shown to have lost BRCA1 expression by RT-PCR, Northern, and Western blotting (21). We found BRCA1-/- MMECs to be more sensitive to H2O2 than BRCA1+/+ MMECs. Figure 1B illustrates that BRCA1-/- MMECs (IC50=16μM) were 50-fold more sensitive to H2O2 than BRCA1+/+ MMECs (IC50=800μM). Given that H2O2 sensitivity is an indicator of ODD, we reasoned that BRCA1-deficient cells may harbor greater levels of ODD due to endogenous cellular activity that increases generation and/or decreases removal of damaged lesions. Therefore, we determined the basal levels of ODD in BRCA1+/+ and BRCA1-/- MMECs using the alkaline comet assay modified for detection of oxidized bases as described in Materials and Methods. Following visualization of DNA comets by fluorescent microscopy and calculation of the percentage of DNA in the comet tail (i.e. levels of ODD) using comet software, we found that BRCA1-/- MMECs harbored significantly greater DNA in the comet tails (64±4%) compared to BRCA1+/+ MMECs (47±2%) (p=0.03) (Fig. 1C). These data indicate an association between H2O2 sensitivity and elevated levels of ODD in BRCA1-deficient cells.
BER Activity among Subtypes of Breast Cancer
Given that ODD is typically repaired by BER, we examined BER activity in breast cancer cell lines of different subtypes. To do so, we developed a cell-based BER assay (Supp. Fig. 1) that maintains an intact cellular environment and eliminates the need for exogenous agents that often induce nonspecific damage to DNA, protein, and lipids. The BER assay consists of three basic steps: 1) oxidatively damaging a GFP-reporter gene, 2) adenoviral-mediated gene transfer for delivery of the damaged GFP-reporter gene into living cells, and 3) host-cell reactivation, which allows for repair of the oxidatively-damaged reporter gene and expression of GFP. The GFP reporter gene was damaged ex vivo using PDT, a known method for inducing ODD (30) to levels that preclude transcription of the GFP gene and thereby prevent expression of the GFP protein. Adenovirus was chosen due to its robust infection efficiency, ability to infect dividing and non-dividing cells, and ease of delivery. Following sufficient time for repair and expression of the ODD-induced GFP reporter gene by the host-cell, we measured fluorescence. A green fluorescent signal indicated expression of the GFP reporter gene, which occurred if the GFP coding sequence that contained ODD was repaired by BER.
We analyzed the BER activity of several breast cancer cell lines using this novel cell-based assay. The cell lines were chosen to represent the different subtypes of breast cancer (luminal: BT474, MCF7; basal-like: HCC38, HCC1143, and MDAMB468; BRCA1-mutated: SUM149PT) and exhibited efficient viral infectivity, i.e. a requirement for the BER assay. The basal-like and BRCA1-mutated cell lines similarly showed a decrease in expression of GFP at 24 hours following infection with the oxidatively-damaged GFP reporter gene compared to the undamaged control, whereas the luminal breast cancer cell lines showed little decrease in GFP expression (Fig. 2A). Therefore, basal-like and BRCA1-mutated cell lines are defective in BER.
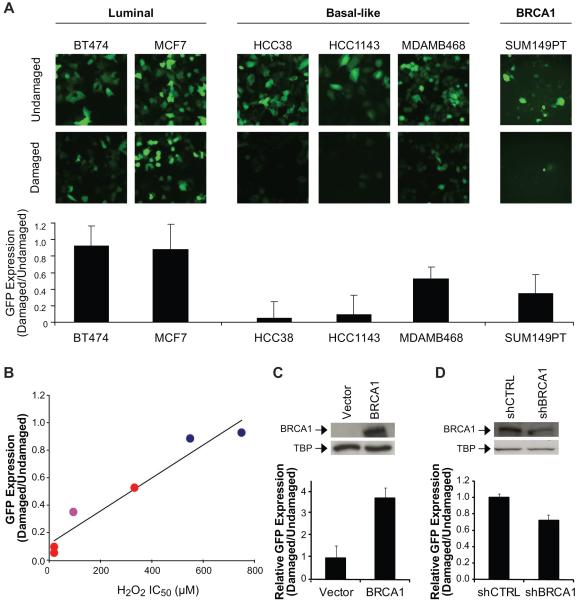
Figure 2. Decreased BER activity in basal-like and BRCA1-mutated or deficient breast cancer cell lines.
(A) BER activity in cell lines representing each subtype of breast cancer using a cell-based BER assay. GFP expression, i.e. an indicator of BER activity, was observed by fluorescent microscopy (top), and quantified by plate reading (bottom). Images were collected under 10X objective and are representative. (B) Relationship between H2O2 sensitivity and BER in breast cancer cell lines. Cell lines included: luminal  , basal-like
, basal-like  , and BRCA1-mutated
, and BRCA1-mutated  . The plot depicts linear regression of the average IC50 value obtained from MTT analysis of H2O2 sensitivity and GFP expression quantified following the BER assay. (C) BER activity in the SUM149PT breast cancer cell line (mutant BRCA1) transfected with wild-type BRCA1. In the top panel, Western blot illustrates nuclear expression of wild-type BRCA1. Tata-binding protein (TBP) was used as a nuclear loading control. In the bottom panel, each bar represents the expression of GFP as determined by the BER assay, which was quantified using a fluorescent plate reader and calculated relative to the vector control. (D) BER activity in the MDAMB361 breast cancer cell line (wild-type BRCA1) transfected with shRNA to BRCA1. Expression of BRCA1 (top) and GFP (bottom) were determined as in (C).
. The plot depicts linear regression of the average IC50 value obtained from MTT analysis of H2O2 sensitivity and GFP expression quantified following the BER assay. (C) BER activity in the SUM149PT breast cancer cell line (mutant BRCA1) transfected with wild-type BRCA1. In the top panel, Western blot illustrates nuclear expression of wild-type BRCA1. Tata-binding protein (TBP) was used as a nuclear loading control. In the bottom panel, each bar represents the expression of GFP as determined by the BER assay, which was quantified using a fluorescent plate reader and calculated relative to the vector control. (D) BER activity in the MDAMB361 breast cancer cell line (wild-type BRCA1) transfected with shRNA to BRCA1. Expression of BRCA1 (top) and GFP (bottom) were determined as in (C).
We next asked whether the sensitivity to H2O2 correlated with GFP expression in the BER assay. When the average IC50 value for H2O2 sensitivity was plotted against the difference in GFP expression between the damaged and undamaged conditions for each of the representative cell lines, we found by regression analysis a near linear relationship between the two parameters (r2=0.94) (Fig. 2B), and by calculation of the Pearson coefficient a positive correlation between the average H2O2 IC50 value and GFP expression (r=0.96), indicating that greater sensitivity to H2O2 corresponds to decreased BER.
BER Activity in the Presence of Altered BRCA1
To further examine the role of BRCA1 in BER, we transfected the BRCA1-mutated SUM149PT breast cancer cell line with wild-type BRCA1, and found using the BER assay that cells expressing wild-type BRCA1 demonstrated greater host-cell reactivation of the GFP reporter gene than the cells containing mutant BRCA1. Figure 2C illustrates an approximate 4-fold increase in BER activity. We also transfected the BRCA1-wild-type MDAMB361 breast cancer cell line with shRNA to BRCA1, and found that shRNA knocked down expression of BRCA1 and decreased host-cell reactivation relative to cells containing non-targeting control shRNA. Knockdown of BRCA1 corresponded to an approximate 28% decrease in BER activity (Fig. 2D). Taken together, BRCA1 functions in BER.
Effect of a BER Defect on Oxidative DNA Damage
To evaluate the response to oxidative stress in the presence of a known BER defect, we generated isogenic cell lines stably expressing shRNA to hOGG1, which is the initiating enzyme for the repair of ODD (8-oxoguanine lesions) by BER. We selected two different cell lines (MCF7shOGG1-A, MCF7shOGG1-B) that knocked down the expression of hOGG1 to 0.21±0.09 and 0.51±0.5 relative to that of a shRNA control cell line (MCF7shCTRL), respectively (A, p=0.00009; B, p=0.008) (Fig. 3A).
Figure 3. Decreased repair of ODD by BER in the presence of shRNA to hOGG1.
(A) mRNA expression of hOGG1 by RTqPCR in isogenic cell lines stably expressing (two different) shRNA to hOGG1 or a non-targeting control shRNA. mRNA expression was calculated by ΔΔCT method. (B) H2O2 and (C) MMS sensitivity in shOGG1-expressing cell lines. Following treatment with increasing concentrations of DNA damaging agent, sensitivity was determined by MTT assay. Each bar represents the average IC50 value ± s.e.m. from at least three independent experiments. (D) BER activity in shOGG1-expressing cell lines. Each bar represents the average GFP expression ± s.e.m. from at least three independent experiments as determined by the BER assay. **, p<0.01; *, p<0.05.
We then analyzed H2O2 sensitivity by MTT assay. By comparing IC50 values, MCF7shOGG1-A and MCF7shOGG1-B cell lines were 3- and 2.4-fold more sensitive to H2O2 compared to MCF7shCTRL, respectively (A, p=0.01; B, p=0.04) (Fig. 3B). MMS methylating agent leads to DNA base alterations that are also repaired by BER, but MMS-mediated BER does not utilize the ODD-specific glycosylase hOGG1. Therefore, to determine specificity for ODD, we analyzed hOGG1-knockdown and control cell lines for MMS sensitivity by MTT assay. MCF7shOGG1-A, MCF7shOGG1-B, and MCF7shCTRL cell lines were similarly sensitive to MMS (A, p=0.5; B, p=0.3) (Fig. 3C).
Finally, we analyzed repair activity using the BER assay. MCF7shOGG1-A and MCF7shOGG1-B decreased host-cell reactivation of the GFP reporter gene to 0.36±0.04 and 0.65±0.13 relative to that of MCF7shCTRL, respectively (A, p=0.00001; B, p=0.01) (Fig. 3D). These results show that the expression of hOGG1 affects H2O2 sensitivity and BER activity, and indicate an ODD-specific response.
PARP Inhibitor Sensitivity in BER-compromised Cells
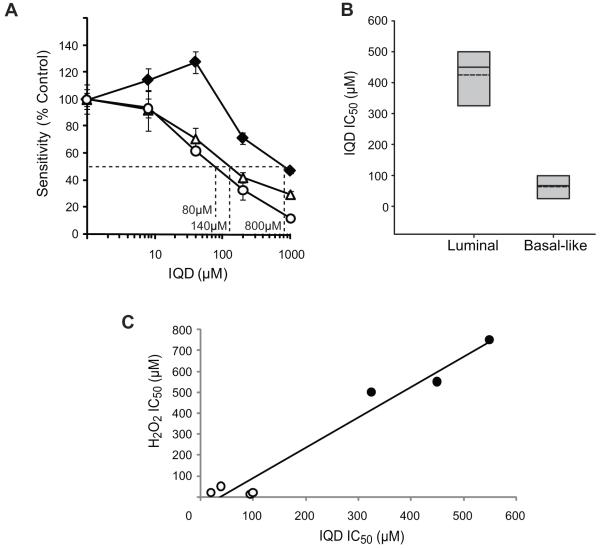
To assess whether the sensitivity to PARP inhibitors observed in BRCA1-mutant cells may be due to their defective BER activity, we subjected the hOGG1-knockdown and control cell lines to increasing concentrations of a PARP inhibitor, IQD, and tested their sensitivity after 96 hours by MTT assay. MCF7shOGG1-A (140μM) and MCF7shOGG1-B (80μM) cell lines were 5 to 10-fold more sensitive to IQD compared to the MCF7shCTRL cell line (800 μM) (Fig. 4A). We also examined sensitivity to IQD in BRCA1+/+ and BRCA1-/- MMECs. The average IC50 values from three independent experiments for BRCA1+/+ and BRCA1-/- were 745±196 μM and 280±67 μM, respectively. Therefore, loss of BRCA1 was associated with a 2.7-fold increase in sensitivity to IQD relative to that of BRCA1 wild-type cells (p=0.02). Due to the similar histopathological profiles (14) and response to ODD (Figs. 1A, 2A) between basal-like and BRCA1-mutated breast cancer cells, we next tested whether sensitivity to PARP inhibition translated to basal-like breast cancer cell lines with defective BER. Luminal (MCF7, BT474, and T47D) and basal-like breast cancer cell lines that were sensitive to H2O2 and/or defective for BER (BT549, HCC38, HCC1143, and HCC1806) were treated with IQD for 96 hours and analyzed for sensitivity by MTT assay. According to the average IC50 value for each subtype, we found that the basal-like cell lines were 6.7-fold more sensitive to IQD compared to the luminal breast cancer cell lines (p= 0.0008) (Fig. 4B), though a considerable range in response was seen between the subtypes with an up to 27.5-fold difference in sensitivity between individual cell lines (Table 2). To examine the relationship between sensitivity to PARP inhibition and H2O2, we plotted the average IC50 value for IQD versus that for H2O2. Regression analysis of the data showed a near linear relationship between the two parameters (r2=0.96) with a clear distinction between the data points of the basal-like and luminal cell lines (Fig. 4C), and calculation of the Pearson coefficient identified a positive correlation between sensitivity to PARP inhibitor and H2O2 (r=0.98). These results demonstrate that the deficiency in hOGG1-mediated BER conferred cellular sensitivity to inhibition of PARP, and that a deficiency in BER contributed at least in part to a selective response of the basal-like subtype of breast cancer cells to treatment with a PARP inhibitor.
Figure 4. Increased sensitivity of BER-compromised cells to PARP inhibition.
(A) Sensitivity of cell lines expressing hOGG1 shRNA to IQD, an inhibitor of PARP. The graph illustrates sensitivity as determined by MTT assay following treatment with increasing concentration of drug. The IC50 concentration for each cell line was interpolated from the dose-response curve as indicated. MCF7CTRL (-◆-) is the control cell line containing non-targeting shRNA. MCF7shOGG1-A (-△-) and MCF7shOGG1-B (-○-) are two different isogenic cell lines stably expressing shRNA to hOGG1. Data are representative of three independent experiments. (B) Sensitivity of luminal and basal-like breast cancer cell lines to IQD. Each box plot summarizes the data described in Table 2. For each subtype, the grey shaded area indicates the range of IC50 values, the dashed line indicates the average IC50 value, and the solid line indicates the median IC50 value. (C) Relationship between PARP inhibitor and H2O2 sensitivity in breast cancer cell lines. Cell lines included: luminal (●) and basal-like (○). The plot depicts linear regression of the average IC50 values obtained from MTT analysis of IQD and H2O2 sensitivity.
Table 2. IQD sensitivity in breast cancer cell lines.
Breast cell lines of the luminal or basal-like subtype as indicated were analyzed for IQD sensitivity by MTT assay. The average IC50 values determined from the dose-response curve of at least two independent experiments are shown as an average ± s.e.m.
| Cell Line | IQD IC50 (μM) | p-Value |
|---|---|---|
| Luminal | ||
| MCF7 | 450 ± 58 | |
| BT474 | 550 ± 71 | |
| T47D | 325 ± 177 | |
| Basal-like | 0.0007** | |
| BT549 | 95 ± 95 | |
| HCC38 | 100 ± 0 | |
| HCC1143 | 20 ± 0 | |
| HCC1806 | 39 ± 49 | |
p<0.01.
DISCUSSION
Currently, limited information exists regarding the specific mechanisms that initiate and maintain BRCA1-mutated cancers, and even less is known for the similarly aggressive but more common basal-like subtype of breast cancer. Defects in response to DNA damage and in DNA repair are central to the pathogenesis of human malignancies (31). The BRCA1 cancer susceptibility gene is known to affect multiple DNA repair processes, including HR, NHEJ, and NER (16, 17, 32). However, whether defects in these processes alone are causative for carcinogenesis remains unclear, and whether deficiencies in DNA repair pathways exist in basal-like breast cancers with a similar phenotype but without mutations in BRCA1 are unknown. We found that repair of ODD by BER, which when aberrant leads to breast carcinogenesis, is defective in triple-negative breast cancer cells and (at least for BRCA1-mutated breast cancers) involves a function by BRCA1. Furthermore, defective repair of ODD by BER conferred sensitivity to inhibition of PARP, a DNA repair enzyme. Therefore, defects in DNA damage response pathways involved in breast carcinogenesis of triple-negative breast cancers, such as that for BER, may serve as targets for therapy and thus be exploited for therapeutic purposes.
We identified for the first time a defect in the repair of ODD by BER in triple-negative breast cancer cells. Basal-like and BRCA1-mutated cell lines showed greater sensitivity to H2O2 and decreased repair of an ODD-induced GFP reporter gene using a novel cell-based BER assay compared to the normal-like and luminal breast cancer cell lines (Table 1 and Figs. 1A, 2A), but showed no statistically significant difference in H2O2 sensitivity or in BER activity between each other (Table 1, Figs. 1A, 2A). Given that ODD has been implicated in the initiation and progression of breast cancer (33-36), basal-like and BRCA1-mutated breast cancers may share similar mechanisms of carcinogenesis. We observed some variability in sensitivity to ODD among the basal-like breast cancer cell lines (Table 1 and Fig. 1A). This variability was expected due to the use of a set of heterogeneous cell lines needed to represent the complicated and variable genetic backgrounds of basal-like breast tumors, but was inconsequential as evidenced by the highly statistically significant difference in H2O2 sensitivity when compared to the normal-like (p=0.0003) and luminal (p=0.0003) cell lines. The variability may also be due to redundancy within the BER pathway or crosstalk among the different repair pathways that exist to preserve genomic stability. For example, the MDAMB468 cell line (H2O2 IC50=333μM) expresses wild-type BRCA1 at higher levels than the HCC38 cell line (H2O2 IC50=20μM) (data not shown), and thus may be able to better manage H2O2-induced ODD. Similar to basal-like and BRCA1-mutated breast cancer cells, BER-proficient cells expressing two different shRNA to hOGG1, the initiating glycosylase for 8-oxoguanine DNA lesions in mammalian cells, showed an increased sensitivity to H2O2 and reduced repair of the ODD-induced GFP reporter gene compared to control cells (Fig. 3A-B, D). Our finding that hOGG1-knockdown and control cells were similarly sensitive to methylating agent MMS (Fig. 3C) supports a defect specific for repair of ODD in hOGG1-knockdown cells. Taken together, the triple-negative breast cancer cell lines respond to oxidative stress as do cells deficient in an important enzyme involved in the repair of ODD by BER. The 2-3 fold difference in H2O2 sensitivity between hOGG1-knockdown and control cells (Fig. 3B) compared to the up to 75-fold difference between the basal-like and normal-like or luminal cells (Table 1) may also be due to redundancy and crosstalk of the different repair pathways, such that the parental cell line of the hOGG1-knockdown cells is a luminal cell line with wild-type BRCA1 and thus may better respond to ODD.
We also showed that BRCA1 plays a role in the repair of ODD by BER and this role likely relates to its function as a tumor suppressor. Consistent with greater H2O2 sensitivity and diminished BER activity in human breast cancer cells with mutant BRCA1, MMECs null for BRCA1 were also more sensitive to H2O2 and contained greater basal levels of ODD than the isogenic wild-type MMECs (Fig. 1B-C). Furthermore, expressing wild-type BRCA1 in a mutant background (Fig. 2C) resulted in greater repair of the ODD-induced GFP reporter gene, and decreasing expression of BRCA1 in a wild-type background using shRNA (Fig. 2D) resulted in less repair. Therefore, we found that elevated levels of ODD are due at least in part to defective BER in the presence of aberrant BRCA1. Rodriguez et al. observed a greater number of ODD lesions induced by ionizing radiation in lymphoblast-derived cell lines from BRCA1 mutation carriers diagnosed with breast cancer compared to non-mutation carriers without cancer (37). These data support our finding that BRCA1 plays a role in BER. The exact mechanism by which BRCA1 regulates BER is yet to be determined, but may be similar to that observed for other types of repair, including transcriptional regulation of repair genes or interaction with other repair proteins (16, 17, 32). It is possible that BRCA1 and p53 act together to regulate ODD since mutations in BRCA1 are often accompanied by mutations in p53 (38-40) and all of the observed H2O2-sensitive and BER-defective cells in this study were mutant or deficient for wild-type p53. Furthermore, p53 regulates hOGG1 expression and enhances BER activity (41-45).
We demonstrated that defective BER confers sensitivity to inhibition of PARP. Basal-like, BRCA1-deficient, and hOGG1- knockdown cells were all found to be compromised for BER and to be sensitive to the IQD PARP inhibitor (Figs. 2, 4A-B). Sensitivity to PARP inhibition is thus not limited to BRCA1 and BRCA2-mutated cancers, but is characteristic of the larger subgroup of basal-like breast cancers. Sensitivity of BRCA1- and BRCA2-mutated tumors to PARP inhibitors has been assumed to be due to a synthetic-lethal mechanism based on the function of BRCA1 and BRCA2 in HR (46). In this scenario, inhibition of PARP precludes repair of single-strand break intermediates of the BER pathway, which upon replication convert to double-strand breaks, thereby sensitizing cells with compromised double-strand break repair (i.e. BRCA1-deficient cells). However, our finding that cells deficient in hOGG1 were more sensitive to IQD compared to the isogenic control (Fig. 4A) indicates a broader mechanism in determining sensitivity to PARP inhibition. Basal-like or BRCA1-deficient cells may also be more sensitive to these agents due to further inhibition of already compromised BER activity. In addition, BRCA1-deficient cells may be more sensitive to PARP inhibition due to an inability to perform NER, which sometimes compensates for BER in the presence of excessive ODD (17, 47). Therefore, the effect of PARP inhibition in triple-negative breast cancer cells likely encompasses the attenuation of multiple DNA repair pathways, resulting in increased susceptibility to DNA damage.
We found that H2O2 sensitivity associated with elevated levels of ODD in BRCA1-mutated cells (Fig. 1C), and that H2O2 sensitivity correlated with BER activity in the different subtypes of breast cancer cell lines (Fig. 2B). Taken together with our data showing defective repair of ODD by BER in the presence of deficient or mutated BRCA1 and in basal-like breast cancer cells (Table 1, Figs 1A-B, 2A, 2C-D), our findings indicate that BRCA1-mutated or the similar basal-like breast cancer cells are more sensitive to H2O2 due to greater levels of ODD that are left unrepaired as a result of compromised BER. This defect in repair of ODD by BER may be causative for carcinogenesis, and may also be exploited for therapeutic benefit. First, our finding that selective inhibition of BER by knockdown of hOGG1 conferred sensitivity to PARP inhibition identified the BER pathway as a potential therapeutic target. Basal-like or BRCA1-mutated breast cancers currently need novel approaches for targeted therapy. Therefore, our data warrant future studies that analyze the use of other inhibitors of the BER pathway for treatment of these triple-negative cancers. Alternatively, combination therapies that target PARP and other BER proteins may be effective in the treatment of luminal breast cancers. Second, we found that H2O2 sensitivity correlated with IQD sensitivity (Fig. 4C). Comparison of these sensitivities revealed two distinct populations, an H2O2/IQD sensitive population representing the basal-like cell lines and a relatively insensitive H2O2/IQD population representing the luminal cell lines (Fig. 4C). Therefore, response to ODD, including H2O2 sensitivity and/or BER, may predict sensitivity to PARP inhibition, specifically for the basal-like subtype of breast cancer.
In summary, we found using a novel assay for in vivo BER activity that triple-negative breast cancer cells are deficient in BER of ODD, and that this defect may be attributed, at least in part, to a novel role for BRCA1 in the BER pathway. We also found that the BER defect contributes to a newly-described mechanism for sensitivity to PARP inhibition. The current study describes a common carcinogenic event for basal-like and BRCA1-mutated cancers with therapeutic potential, including the proposition of novel targets for future therapeutic strategies and the ability to select triple-negative cancers for treatment with PARP inhibitor. Our data have additional clinical implications. First, our finding that BRCA1 may function in ODD-mediated carcinogenesis will be useful for determining novel strategies for cancer prevention, which is particularly important for carriers of mutations in BRCA1 who have an increased risk for developing breast and ovarian cancer (48, 49). Second, our in vivo BER assay may be useful for predicting therapeutic effectiveness of PARP inhibitors in other tumor types. Currently, we are adapting this methodology for use in primary cancer cells obtained from patients on clinical trials.
Supplementary Material
Acknowledgements
We thank Simon Powell for the BRCA1 construct, Ken Cowan for BRCA1+/+ and BRCA1-/- MMECs, and Graciela Spivak for helpful discussion on the BER assay.
Financial Support: U.S. National Institute of Health grant R01 CA108794, U.S. Department of Defense grant DOD S81XWH-04-1-0576, Mary Kay Ash Charitable Foundation 078-07, Breast Cancer Research Foundation, Susan G. Komen for the Cure Postdoctoral Fellowship KG080695
Footnotes
Conflicts of Interest: None.
REFERENCES
- 1.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Hu Z, Fan C, Oh DS, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nielsen TO, Hsu FD, Jensen K, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10:5367–74. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 4.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauer KR, Brown M, Cress RD, Parise CA, Caggiano V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer. 2007;109:1721–8. doi: 10.1002/cncr.22618. [DOI] [PubMed] [Google Scholar]
- 6.Carey LA, Dees EC, Sawyer L, et al. The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res. 2007;13:2329–34. doi: 10.1158/1078-0432.CCR-06-1109. [DOI] [PubMed] [Google Scholar]
- 7.Dent R, Trudeau M, Pritchard KI, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13:4429–34. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- 8.Haffty BG, Yang Q, Reiss M, et al. Locoregional relapse and distant metastasis in conservatively managed triple negative early-stage breast cancer. J Clin Oncol. 2006;24:5652–7. doi: 10.1200/JCO.2006.06.5664. [DOI] [PubMed] [Google Scholar]
- 9.Liedtke C, Mazouni C, Hess KR, et al. Response to neoadjuvant therapy and longterm survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;26:1275–81. doi: 10.1200/JCO.2007.14.4147. [DOI] [PubMed] [Google Scholar]
- 10.Tischkowitz M, Brunet JS, Begin LR, et al. Use of immunohistochemical markers can refine prognosis in triple negative breast cancer. BMC Cancer. 2007;7:134. doi: 10.1186/1471-2407-7-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sorlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–23. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foulkes WD, Stefansson IM, Chappuis PO, et al. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J Natl Cancer Inst. 2003;95:1482–5. doi: 10.1093/jnci/djg050. [DOI] [PubMed] [Google Scholar]
- 13.Lakhani SR, Reis-Filho JS, Fulford L, et al. Prediction of BRCA1 status in patients with breast cancer using estrogen receptor and basal phenotype. Clin Cancer Res. 2005;11:5175–80. doi: 10.1158/1078-0432.CCR-04-2424. [DOI] [PubMed] [Google Scholar]
- 14.Turner NC, Reis-Filho JS. Basal-like breast cancer and the BRCA1 phenotype. Oncogene. 2006;25:5846–53. doi: 10.1038/sj.onc.1209876. [DOI] [PubMed] [Google Scholar]
- 15.Rakha EA, Reis-Filho JS, Ellis IO. Basal-like breast cancer: a critical review. J Clin Oncol. 2008;26:2568–81. doi: 10.1200/JCO.2007.13.1748. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J, Powell SN. The role of the BRCA1 tumor suppressor in DNA double-strand break repair. Mol Cancer Res. 2005;3:531–9. doi: 10.1158/1541-7786.MCR-05-0192. [DOI] [PubMed] [Google Scholar]
- 17.Hartman AR, Ford JM. BRCA1 induces DNA damage recognition factors and enhances nucleotide excision repair. Nat Genet. 2002;32:180–4. doi: 10.1038/ng953. [DOI] [PubMed] [Google Scholar]
- 18.Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 19.Bryant HE, Schultz N, Thomas HD, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–7. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 20.Mortusewicz O, Ame JC, Schreiber V, Leonhardt H. Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells. Nucleic Acids Res. 2007;35:7665–75. doi: 10.1093/nar/gkm933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sgagias MK, Wagner KU, Hamik B, et al. Brca1-deficient murine mammary epithelial cells have increased sensitivity to CDDP and MMS. Cell Cycle. 2004;3:1451–6. doi: 10.4161/cc.3.11.1211. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J, Willers H, Feng Z, et al. Chk2 phosphorylation of BRCA1 regulates DNA double-strand break repair. Mol Cell Biol. 2004;24:708–18. doi: 10.1128/MCB.24.2.708-718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Collins A, Dusinska M, Franklin M, et al. Comet assay in human biomonitoring studies: reliability, validation, and applications. Environ Mol Mutagen. 1997;30:139–46. doi: 10.1002/(sici)1098-2280(1997)30:2<139::aid-em6>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 25.Collins AR, Dusinska M, Gedik CM, Stetina R. Oxidative damage to DNA: do we have a reliable biomarker? Environ Health Perspect. 1996;104(Suppl 3):465–9. doi: 10.1289/ehp.96104s3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elstrodt F, Hollestelle A, Nagel JH, et al. BRCA1 mutation analysis of 41 human breast cancer cell lines reveals three new deleterious mutants. Cancer Res. 2006;66:41–5. doi: 10.1158/0008-5472.CAN-05-2853. [DOI] [PubMed] [Google Scholar]
- 27.Gazdar AF, Kurvari V, Virmani A, et al. Characterization of paired tumor and non-tumor cell lines established from patients with breast cancer. Int J Cancer. 1998;78:766–74. doi: 10.1002/(sici)1097-0215(19981209)78:6<766::aid-ijc15>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 28.Neve RM, Chin K, Fridlyand J, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tomlinson GE, Chen TT, Stastny VA, et al. Characterization of a breast cancer cell line derived from a germ-line BRCA1 mutation carrier. Cancer Res. 1998;58:3237–42. [PubMed] [Google Scholar]
- 30.Schagen FH, Moor AC, Cheong SC, et al. Photodynamic treatment of adenoviral vectors with visible light: an easy and convenient method for viral inactivation. Gene Ther. 1999;6:873–81. doi: 10.1038/sj.gt.3300897. [DOI] [PubMed] [Google Scholar]
- 31.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–45. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 32.Hartman AR, Ford JM. BRCA1 and p53: compensatory roles in DNA repair. J Mol Med. 2003;81:700–7. doi: 10.1007/s00109-003-0477-0. [DOI] [PubMed] [Google Scholar]
- 33.Jackson AL, Loeb LA. The contribution of endogenous sources of DNA damage to the multiple mutations in cancer. Mutat Res. 2001;477:7–21. doi: 10.1016/s0027-5107(01)00091-4. [DOI] [PubMed] [Google Scholar]
- 34.Klaunig JE, Kamendulis LM. The role of oxidative stress in carcinogenesis. Annu Rev Pharmacol Toxicol. 2004;44:239–67. doi: 10.1146/annurev.pharmtox.44.101802.121851. [DOI] [PubMed] [Google Scholar]
- 35.Malins DC, Polissar NL, Gunselman SJ. Progression of human breast cancers to the metastatic state is linked to hydroxyl radical-induced DNA damage. Proc Natl Acad Sci U S A. 1996;93:2557–63. doi: 10.1073/pnas.93.6.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Storz P. Reactive oxygen species in tumor progression. Front Biosci. 2005;10:1881–96. doi: 10.2741/1667. [DOI] [PubMed] [Google Scholar]
- 37.Rodriguez H, Jaruga P, Leber D, Nyaga SG, Evans MK, Dizdaroglu M. Lymphoblasts of women with BRCA1 mutations are deficient in cellular repair of 8,5′-Cyclopurine-2′-deoxynucleosides and 8-hydroxy-2′-deoxyguanosine. Biochemistry. 2007;46:2488–96. doi: 10.1021/bi062022p. [DOI] [PubMed] [Google Scholar]
- 38.Crook T, Brooks LA, Crossland S, et al. p53 mutation with frequent novel condons but not a mutator phenotype in BRCA1- and BRCA2-associated breast tumours. Oncogene. 1998;17:1681–9. doi: 10.1038/sj.onc.1202106. [DOI] [PubMed] [Google Scholar]
- 39.Lakhani SR, Van De Vijver MJ, Jacquemier J, et al. The pathology of familial breast cancer: predictive value of immunohistochemical markers estrogen receptor, progesterone receptor, HER-2, and p53 in patients with mutations in BRCA1 and BRCA2. J Clin Oncol. 2002;20:2310–8. doi: 10.1200/JCO.2002.09.023. [DOI] [PubMed] [Google Scholar]
- 40.Schuyer M, Berns EM. Is TP53 dysfunction required for BRCA1-associated carcinogenesis? Mol Cell Endocrinol. 1999;155:143–52. doi: 10.1016/s0303-7207(99)00117-3. [DOI] [PubMed] [Google Scholar]
- 41.Achanta G, Huang P. Role of p53 in sensing oxidative DNA damage in response to reactive oxygen species-generating agents. Cancer Res. 2004;64:6233–9. doi: 10.1158/0008-5472.CAN-04-0494. [DOI] [PubMed] [Google Scholar]
- 42.Chatterjee A, Mambo E, Osada M, Upadhyay S, Sidransky D. The effect of p53-RNAi and p53 knockout on human 8-oxoguanine DNA glycosylase (hOgg1) activity. Faseb J. 2006;20:112–4. doi: 10.1096/fj.04-3423fje. [DOI] [PubMed] [Google Scholar]
- 43.Offer H, Wolkowicz R, Matas D, Blumenstein S, Livneh Z, Rotter V. Direct involvement of p53 in the base excision repair pathway of the DNA repair machinery. FEBS Lett. 1999;450:197–204. doi: 10.1016/s0014-5793(99)00505-0. [DOI] [PubMed] [Google Scholar]
- 44.Seo YR, Fishel ML, Amundson S, Kelley MR, Smith ML. Implication of p53 in base excision DNA repair: in vivo evidence. Oncogene. 2002;21:731–7. doi: 10.1038/sj.onc.1205129. [DOI] [PubMed] [Google Scholar]
- 45.Zhou J, Ahn J, Wilson SH, Prives C. A role for p53 in base excision repair. Embo J. 2001;20:914–23. doi: 10.1093/emboj/20.4.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ashworth A. A synthetic lethal therapeutic approach: poly(ADP) ribose polymerase inhibitors for the treatment of cancers deficient in DNA double-strand break repair. J Clin Oncol. 2008;26:3785–90. doi: 10.1200/JCO.2008.16.0812. [DOI] [PubMed] [Google Scholar]
- 47.Reardon JT, Bessho T, Kung HC, Bolton PH, Sancar A. In vitro repair of oxidative DNA damage by human nucleotide excision repair system: possible explanation for neurodegeneration in xeroderma pigmentosum patients. Proc Natl Acad Sci U S A. 1997;94:9463–8. doi: 10.1073/pnas.94.17.9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Easton DF, Ford D, Bishop DT, Breast Cancer Linkage Consortium Breast and ovarian cancer incidence in BRCA1-mutation carriers. Am J Hum Genet. 1995;56:265–71. [PMC free article] [PubMed] [Google Scholar]
- 49.Ford D, Easton DF, Bishop DT, Narod SA, Goldgar DE, Breast Cancer Linkage Consortium Risks of cancer in BRCA1-mutation carriers. Lancet. 1994;343:692–5. doi: 10.1016/s0140-6736(94)91578-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.