Abstract
The activity of fenticonazole was studied against 260 West and Southeast European vulvovaginal candidiasis isolates, and low MICs were displayed. Fenticonazole was assessed by European Committee on Antimicrobial Susceptibility Testing and CLSI microdilution methods for the first time, and the results showed excellent agreement (97%) and significant interclass correlation coefficient (P < 0.0001). Also, the levels of agreement for the results for itraconazole, fluconazole, and ketoconazole were 84%, 90%, and 98% (P < 0.0001), respectively. Multilocus typing by PCR fingerprinting and subsequent cluster analysis delineated geographically associated alignments for Candida albicans and fluconazole resistance-related clusters for Candida glabrata.
Uncomplicated vulvovaginal candidiasis (VVC) affects approximately 75% of women at reproductive age (13, 17, 22); Candida albicans is a major cause and Candida glabrata accounts for approximately 5% of cases worldwide (30). The recommended first-line therapy for uncomplicated VVC is topical azoles (4, 7, 25, 27, 28), unless resistance of the isolate is substantiated or azole hypersensitivity is diagnosed (4, 8). Identifying antifungal resistance in vitro is clinically important, but variable host responses to treatment and unpredictable fungal load in the vulvovaginal mucosa (in loco) invariably weaken in vitro with in vivo correlations. However, standardized susceptibility testing of isolates to local antifungals could provide data on the in vitro activity of newer topical antifungals.
Recording agreement of the results of the CLSI (24) and European Committee on Antimicrobial Susceptibility Testing (EUCAST) (10) reference methods in determining the susceptibility of VVC isolates from Belgian and Greek patients to fenticonazole, a topical imidazole (8, 12), forms the basis of this report. Subsequently, PCR fingerprinting was used to investigate whether distinct geographical and azole-resistant clinical isolate subpopulations can be recognized.
A total of 260 baseline C. albicans and C. glabrata isolates from pregnant, nonpregnant, and diabetic women were tested (Table 1). Isolates were identified in Chromagar medium (Chromagar, Paris, France) and identified with the API ID 32 C system (bioMerieux, Marcy l'Etoile, France). All C. albicans isolates were screened for Candida dubliniensis (3, 18, 31) and C. glabrata strains were screened for Candida nivariensis and Candida bracarensis (1, 19) to ensure that susceptibility testing and PCR fingerprints corresponded only to C. albicans and C. glabrata isolates.
TABLE 1.
Origin of isolates from 260 patients with uncomplicated vulvovaginitis
| Patient group (no.) | Mean age (yr) | No. of patients of indicated national origin with isolates of indicated species
|
Total no. of isolates | |||
|---|---|---|---|---|---|---|
| Belgium
|
Greece
|
|||||
| C. albicans | C. glabrata | C. albicans | C. glabrata | |||
| Pregnant (65) | 30 | 61 | 4 | NIa | NI | 65 |
| Nonpregnant (152) | 40 | 132 | 2 | 17 | 1 | 152 |
| Diabetic (43) | 42 | NI | NI | 39 | 4 | 43 |
| All (total no. of patients) | 193 | 6 | 56 | 5 | 260 | |
NI, patient group not included.
Stock fluconazole (Pfizer Inc., Sandwich, Kent, United Kingdom) solutions and a range of concentrations of itraconazole and ketoconazole (Janssen, Beerse, Belgium) were prepared as described for each reference method. Fenticonazole compound (Recordati S.A, Milan, Italy, and Galenica, Athens, Greece) was prepared as a 100× stock in dimethyl sulfoxide (Merck, Darmstadt, Germany) at a final concentration range of 0.0312 μg/ml to 32 μg/ml. Test medium, inoculum preparations, and reading of results were as described in the respective guidelines (10, 24). Candida parapsilosis ATCC 22019 and Candida krusei ATCC 6258 were used as control strains for both methods (Table 2). No CLSI or EUCAST out-of-range MICs were observed for itraconazole, fluconazole, or ketoconazole.
TABLE 2.
Susceptibilities of 260 VVC isolates and quality control strains determined by the CLSI M27-A2 and EUCAST broth microdilution methods
| Candida isolate (no.) or QC straina | Method | Itraconazole
|
Fenticonazole
|
Fluconazole
|
Ketoconazole
|
||||
|---|---|---|---|---|---|---|---|---|---|
| MIC50 range (μg/ml) | GMb (μg/ml) | MIC50 range (μg/ml) | GM (μg/ml) | MIC50 range (μg/ml) | GM (μg/ml) | MIC50 range (μg/ml) | GM (μg/ml) | ||
| C. albicans (249) | CLSI | 0.03-0.5 | 0.07 | 0.03-0.5 | 0.14 | 0.12-16 | 1.86 | 0.12-4.0 | 0.78 |
| EUCAST | 0.03-0.5 | 0.06 | 0.03-0.25 | 0.10 | 0.12-32 | 1.84 | 0.12-4.0 | 0.54 | |
| C. glabrata (11) | CLSI | 0.03-0.5 | 0.10 | 0.03-1 | 0.34 | 2.0-≥64 | 7.51 | 0.5-4.0 | 1.86 |
| EUCAST | 0.03-0.5 | 0.10 | 0.03-0.5 | 0.28 | 2.0-≥64 | 7.51 | 2.0-8.0 | 2.13 | |
| C. parapsilosis ATCC 22019 | CLSI | 0.06-0.25 | 0.14 | 0.03-1c | 0.16 | 2-8 | 3.73 | 0.12-0.25 | 0.17 |
| EUCAST | 0.06-0.12 | 0.09 | 0.03-0.25d | 0.13 | 0.5-4 | 2.14 | 0.12-0.5 | 0.21 | |
| C. krusei ATCC 6258 | CLSI | 0.12-0.5 | 0.10 | 0.06-2e | 0.23 | 16-64 | 45.25 | 0.25-0.5 | 0.33 |
| EUCAST | 0.03-0.12 | 0.08 | 0.06-1f | 0.20 | 32-64 | 45.25 | 0.12-1 | 0.26 | |
Results were obtained for 20 independent tests. QC, quality control.
GM, geometric mean.
One of 20 test results (MIC, 1 μg/ml) was out of the observed range of 0.03 to 0.5 μg/ml.
No test result was out of range.
One of 20 test results (MIC, 2 μg/ml) was out of the observed range of 0.06 to 0.5 μg/ml.
One of 20 test results (MIC, 1 μg/ml) was out of the observed range of 0.06 to 0.25 μg/ml.
No differences in susceptibilities among isolates from the three patient groups were observed, but in contrast to previous reports (21), no geographical associations in susceptibility were recorded for isolates from the two European regions. Fluconazole resistance (Table 2) in C. albicans was rare (6.9%), whereas 45% of the C. glabrata isolates were resistant (6, 11). Fluconazole and ketoconazole cross-resistance was inferred for 20/249 (8.03%) C. albicans isolates and for 3/11 (27.2%) C. glabrata isolates. Generally, lower MICs were recorded for fenticonazole than for the other drugs (Table 2), but their clinical relevance cannot be assessed without correlating the in vitro responses and in loco fenticonazole pharmacokinetics and pharmacodynamics with the in vivo response. Topical ketoconazole efficacy and drug levels have thus far been assessed ex vivo in human skin specimens and have successfully supported standardized susceptibility testing and clinical investigations (2). However, bioassay systems to complement in vitro studies have not been assessed with topical VVC agents.
Agreement between the CLSI and EUCAST results (29) within ±1 dilution was 84 to 98% (Table 3), and interclass correlation coefficients were statistically very significant (P < 0.0001), suggesting that fenticonazole testing with both reference methods gives concordant MICs.
TABLE 3.
Agreement and intraclass correlation coefficients for log2-transformed dataa obtained by CLSI and EUCAST reference methods for azole drugs against vulvovaginitis isolates
| Antifungal drug | Agreement (%)b | ICCb | P |
|---|---|---|---|
| Itraconazole | 84 | 0.64 | <10−4 |
| Fenticonazole | 97 | 0.88 | <10−4 |
| Fluconazole | 90 | 0.90 | <10−4 |
| Ketoconazole | 98 | 0.88 | <10−4 |
SPSS version 10.0 (SPSS Inc., Chicago, IL, 1999) was used to determine these data.
A value of 85% was selected to validate the results. Agreement values and intraclass correlation coefficients (ICCs) were calculated from the results obtained for all C. albicans and C. glabrata isolates.
A possible susceptibility-associated relatedness of strains and the population structures of the C. albicans and C. glabrata isolates from the two geographic regions was studied by PCR fingerprinting using the minisatellite specific oligonucleotide [5′-GAGGGTGGCGGTTCT-3′] M13 (23, 35) as described before (15, 34). All Greek VVC isolates originated exclusively from Greek Caucasians, whereas Belgian strains were isolated from patients of mixed ethnic origin, including African immigrants residing in Belgium.
Each strain was tested on five independent occasions to ensure the reproducibility of the results. Cluster analysis was performed using Bionumerics version 4 (Bio-Maths, Kortrijk, Belgium; analysis done at the National Centre for Meningococcal Disease, Athens School of Public Health, Athens, Greece) and the Dice coefficient of similarity and cluster analysis with the unweighted-pair group method with arithmetic averages, with 1.00% position tolerance and no optimization, to obtain the greatest variation in similarity.
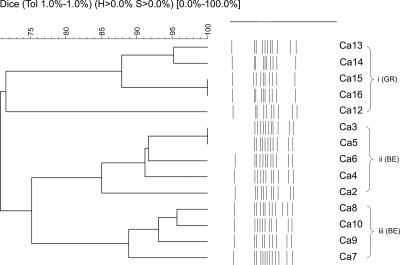
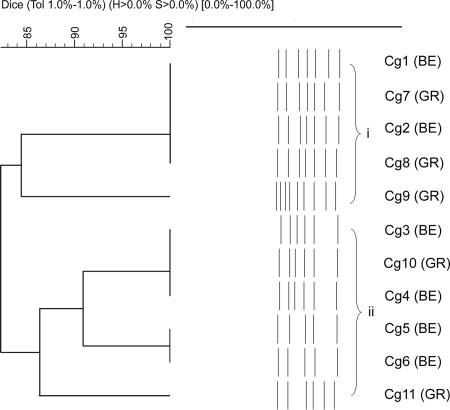
Discrete non-nosocomial and epidemiologically unrelated C. albicans subpopulations in the two European regions were identified (Fig. 1). Despite a microsatellite fingerprinting inference to the contrary (5, 20), our minisatellite typing did not associate fluconazole-resistant C. albicans isolates with any particular cluster. Similarly, multilocus sequence typing (MLST) did not significantly connect isolates with specific azole susceptibility profiles to particular clades (26). At a global level, MLST analysis of C. albicans isolates with different geographical and anatomical origins has shown clades with geographical enrichment (32, 33). Also, microsatellite analysis has even separated German from Austrian C. albicans clades in Central Europe (14), though with no reference to the ethnic origin of the population studied. Our minisatellite assay assembled all strains from Greek Caucasians in a single group (Fig. 1), but irrespective of the geographic origin of the patients, fluconazole-resistant C. glabrata isolates grouped in a single cluster (Fig. 2). An association of fluconazole-resistant strains with specific clades has been also shown by MLST analysis (9). M13 typing is not equivalent to MLST, as each method assays different elements of the genome. However, the acute discrimination of the fluconazole-resistant C. glabrata subpopulation among only 11 isolates adds confidence that M13 typing may be dependably used in discriminating C. glabrata fluconazole-resistant strains. Notably, C. albicans and C. glabrata isolates from pregnant, nonpregnant, and diabetic women did not associate with specific clusters.
FIG. 1.
Summary of results for 249 Candida albicans VVC isolates. A dendrogram representing the three C. albicans clusters of VVC isolates is shown. Groups were defined by 75% similarity. Of the 194 Belgian isolates, 137 (70.6%) grouped in cluster ii and 57 (29.40%) grouped in cluster iii. All 55 Greek isolates grouped in a single cluster, cluster i.
FIG. 2.
Candida glabrata clusters. Groups were defined by 86% similarity. C. glabrata (Cg) isolates 3, 4, 5, 6, 10, and 11, with fluconazole resistance, clustered in group ii.
This study showed excellent agreement between the EUCAST and CLSI methods (97%) in testing fenticonazole against C. albicans and C. glabrata from patients with uncomplicated VVC and limited C. albicans fluconazole resistance. Comparative multilocus typing by PCR fingerprinting has clustered fluconazole-resistant C. glabrata isolates in a separate group irrespective of their geographic origins, whereas C. albicans isolates clustered in geographically distinct groups with no susceptibility associations. The possibility that marker choice (16) and sample size influence the C. albicans geographic distinction patterns cannot be excluded. However, assuming that there are no deviations from the Hardy-Weinberg principle, the observed clustering of VVC strains from Greek Caucasian patients may reflect an ad hoc geographically restricted event that nonetheless requires further investigation.
Acknowledgments
This work was partly supported by the Hellenic Centre for Diseases Prevention and Control (Ministry of Health and Welfare) and was concluded using Mycology Laboratory funds (SARG K.A 70/3/6915 and K.A 70/3/5905) from the National and Kapodistrian University of Athens and the Bodosakis Foundation.
We thank the following individuals for technical assistance: N. Nolard and D. Swinne, Scientific Institute, Public Health, Section Mycology, Brussels, Belgium; S. Gantois, Scientific Institute, Public Health, Section Mycology, and Institute Jules Bordet, Brussels, Belgium; M. Husson, Medical Mycology, Institute Jules Bordet, ULB, Brussels, Belgium; and I. Ilia, Mycology Laboratory, Medical School, University of Athens, Athens, Greece.
A.V. has received unrestricted research grants from Gilead Sciences, Pfizer, and the Schering Plough Research Institute, Kenilworth, NJ.
We declare no conflicts of interest.
Footnotes
Published ahead of print on 17 February 2009.
REFERENCES
- 1.Alcoba-Flórez, J., M. del Pilar Arévalo, F. J. González-Paredes, J. Cano, J. Guarro, E. Pérez-Roth, and S. Méndez-Álvarez. 2005. PCR protocol for specific identification of Candida nivariensis, a recently described pathogenic yeast. J. Clin. Microbiol. 43:6194-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arrese, J. E., L. Fogouang, C. Piérard-Franchimont, and G. E. Piérard. 2002. Euclidean and fractal computer-assisted corneofungimetry: a comparison of 2% ketoconazole and 1% terbinafine topical formulations. Dermatology 204:222-227. [DOI] [PubMed] [Google Scholar]
- 3.Bikandi, J., R. San Millán, M. D. Moragues, G. Cebas, M. Clarke, D. C. Coleman, D. J. Sullivan, G. Quindós, and J. Pontón. 1998. Rapid identification of Candida dubliniensis by indirect immunofluorescence based on differential localization of antigens on C. dubliniensis blastospores and Candida albicans germ tubes. J. Clin. Microbiol. 36:2428-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Diseases Control and Prevention, K. A. Workowski, and S. M. Berman. 2006. Sexually transmitted diseases treatment guidelines, 2006. MMWR Recommend. Rep. 55:1-94. [PubMed] [Google Scholar]
- 5.Chong, P. P., P. Sampaio, L. Gusmão, C. Alves, C. Pina-Vaz, A. Amorim, and C. Pais. 2003. Highly polymorphic microsatellite for identification of Candida albicans strains. J. Clin. Microbiol. 41:552-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts; third informational supplement. Document M27-S3. CLSI, Wayne, PA.
- 7.das Neves, J., E. Pinto, B. Teixeira, G. Dias, P. Rocha, T. Cunha, B. Santos, M. H. Amaral, and M. F. Bahia. 2008. Local treatment of vulvovaginal candidosis: general and practical considerations. Drugs 68:1787-1802. [DOI] [PubMed] [Google Scholar]
- 8.De Bernardis, F., and A. Cassone. 1996. Comparison of the effects of fenticonazole and econazole on the aspartic proteinase secreted by Candida albicans. Contracept. Fertil. Sex. 24:163-165. [PubMed] [Google Scholar]
- 9.Dodgson, A. R., C. Pujol, D. W. Denning, D. R. Soll, and A. J. Fox. 2003. Multilocus sequence typing of Candida glabrata reveals geographically enriched clades. J. Clin. Microbiol. 41:5709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.European Committee for Antimicrobial Susceptibility Testing (EUCAST) Subcommittee on Antifungal Susceptibility Testing (AFST). 2008. EUCAST definitive document EDef 7.1: method for the determination of broth dilution MICs of antifungal agents for fermentative yeasts. 2008. Clin. Microbiol. Infect. 14:398-405. [DOI] [PubMed] [Google Scholar]
- 11.European Committee for Antimicrobial Susceptibility Testing Subcommittee on Antifungal Susceptibility Testing (EUCAST-AFST). 2008. Technical note on fluconazole. Clin. Microbiol. Infect. 14:193-195.18070130 [Google Scholar]
- 12.Fernández-Alba, J., A. Valle-Gay, M. Dibildox, J. A. Vargas, J. González, M. García, L. H. López, and the Fentimex Mexican Study Group. 2004. Fenticonazole nitrate for treatment of vulvovaginitis: efficacy, safety, and tolerability of 1-gram ovules, administered as ultra-short 2-day regimen. J. Chemother. 16:179-186. [DOI] [PubMed] [Google Scholar]
- 13.Fidel, P. L., Jr. 2005. Immunity in vaginal candidiasis. Curr. Opin. Infect. Dis. 18:107-111. [DOI] [PubMed] [Google Scholar]
- 14.Fundyga, R. E., T. J. Lott, and J. Arnold. 2002. Population structure of Candida albicans, a member of the human flora, as determined by microsatellite loci. Infect. Genet. Evol. 2:57-68. [DOI] [PubMed] [Google Scholar]
- 15.Gaitanis, G., A. Velegraki, E. C. Alexopoulos, E. Kapsanaki-Gotsi, L. Zisova, Y. Ran, H. Zhang, G. Arsenis, I. D. Bassukas, and J. Faergemann. 20 November 2008, posting date. Malassezia furfur fingerprints as possible markers for human phylogeography. ISME J. doi: 10.1038/ismej.2008.112. [DOI] [PubMed]
- 16.Gräser, Y., M. Volosek, J. Arrington, G. Schonian, W. Presber, T. G. Mitchell, and R. Vilgalys. 1996. Molecular markers reveal that population structure of the human pathogen Candida albicans exhibits both clonality and recombination. Proc. Natl. Acad. Sci. USA 93:12473-12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kent, H. L. 1991. Epidemiology of vaginitis. Am. J. Obstet. Gynecol. 165:1168-1175. [DOI] [PubMed] [Google Scholar]
- 18.Kurzai, O., W. J. Heinz, D. J. Sullivan, D. C. Coleman, M. Frosch, and F. A. Mühlschlegel. 1999. Rapid PCR test for discriminating between Candida albicans and Candida dubliniensis isolates using primers derived from the pH-regulated PHR1 and PHR2 genes of C. albicans. J. Clin. Microbiol. 37:1587-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Linton, C. J., A. M. Borman, G. Cheung, A. D. Holmes, A. Szekely, M. D. Palmer, P. D. Bridge, C. K. Campbell, and E. M. Johnson. 2007. Molecular identification of unusual pathogenic yeast isolates by large ribosomal subunit gene sequencing: 2 years of experience at the United Kingdom Mycology Reference Laboratory. J. Clin. Microbiol. 45:1152-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, X. P., S. R. Fan, F. Y. Bai, J. Li, and Q. P. Liao. 2009. Antifungal susceptibility and genotypes of Candida albicans strains from patients with vulvovaginal candidiasis. Mycoses 52:24-28. [DOI] [PubMed] [Google Scholar]
- 21.Manferdi, M., M. J. McCullough, L. Polonelli, S. Conti, Z. M. Al-Karaawi, P. Vescovi, and S. R. Porter. 2006. In vitro antifungal susceptibility to six antifungal agents of 229 Candida isolates from patients with diabetes mellitus. Oral Microbiol. Immunol. 21:177-182. [DOI] [PubMed] [Google Scholar]
- 22.Marrazzo, J. 2002. Vulvovaginal candidiasis. Clin. Evid. 7:1784-1796. [PubMed] [Google Scholar]
- 23.Meyer, W., and T. G. Mitchell. 1995. Polymerase chain reaction fingerprinting in fungi using single primers specific to minisatellites and simple repetitive DNA sequences: strain variation in Cryptococcus neoformans. Electrophoresis 16:1648-1656. [DOI] [PubMed] [Google Scholar]
- 24.National Committee for Clinical Laboratory Standards. 2002. Reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard M27-A2. NCCLS, Wayne, PA.
- 25.Nurbhai, M., J. Grimshaw, M. Watson, C. Bond, J. Mollison, and A. Ludbrook. 2007. Oral versus intra-vaginal imidazole and triazole anti-fungal treatment of uncomplicated vulvovaginal candidiasis (thrush). Cochrane Database Syst. Rev. 4:CD002845. [DOI] [PubMed] [Google Scholar]
- 26.Odds, F. C., M. E. Bougnoux, D. J. Shaw, J. M. Bain, A. D. Davidson, D. Diogo, M. D. Jacobsen, M. Lecomte, S. Y. Li, A. Tavanti, M. C. Maiden, N. A. Gow, and C. d'Enfert. 2007. Molecular phylogenetics of Candida albicans. Eukaryot. Cell 6:1041-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quaglia, M. G., E. Donati, N. Desideri, S. Fanali, F. D. D'auria, and M. Tecca. 2002. Chiral discrimination by HPLC and CE and antifungal activity of racemic fenticonazole and its enantiomers. Chirality 5:449-454. [DOI] [PubMed] [Google Scholar]
- 28.Rex, J. H., T. J. Walsh, J. D. Sobel, S. G. Filler, P. G. Pappas, W. E. Dismukes, and J. E. Edwards. 2000. Practice guidelines for the treatment of candidiasis. Clin. Infect. Dis. 30:662-678. [DOI] [PubMed] [Google Scholar]
- 29.Rodriguez-Tudela, J. L., J. P. Donnelly, M. A. Pfaller, E. Chryssantou, P. Warn, D. W. Denning, A. Espinel-Ingroff, F. Barchiesi, and M. Cuenca-Estrella. 2007. Statistical analyses of correlation between fluconazole MICs for Candida spp. assessed by standard methods set forth by the European Committee on Antimicrobial Susceptibility Testing (E.Dis. 7.1) and CLSI (M27-A2). J. Clin. Microbiol. 45:109-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sobel, J. D. 2007. Vulvovaginal candidosis. Lancet 369:1961-1971. [DOI] [PubMed] [Google Scholar]
- 31.Staib, P., and J. Morschhaüser. 1999. Chlamydospore formation on Staib agar as a species-specific characteristic of Candida dubliniensis. Mycoses 42:521-524. [DOI] [PubMed] [Google Scholar]
- 32.Takakura, S., S. Ichiyama, J. M. Bain, A. D. Davidson, M. D. Jacobsen, D. J. Shaw, N. A. Gow, and F. C. Odds. 2008. Comparison of Candida albicans strain types among isolates from three countries. Int. J. Med. Microbiol. 298:663-668. [DOI] [PubMed] [Google Scholar]
- 33.Tavanti, A., A. D. Davidson, M. J. Fordyce, N. A. R. Gow, M. C. J. Maiden, and F. C. Odds. 2005. Population structure and properties of Candida albicans, as determined by multilocus sequence typing. J. Clin. Microbiol. 43:5601-5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Velegraki, A., M. Kambouris, A. Kostourou, G. Chalevelakis, and N. J. Legakis. 1999. Rapid extraction of fungal DNA from clinical samples for PCR amplification. Med. Mycol. 37:69-73. [PubMed] [Google Scholar]
- 35.Xu, J., C. M. Boyd, E. Livingston, W. Meyer, J. F. Madden, and T. G. Mitchell. 1999. Species and genotypic diversities and similarities of pathogenic yeasts colonizing women. J. Clin. Microbiol. 37:3835-3843. [DOI] [PMC free article] [PubMed] [Google Scholar]