Abstract
We report the emergence of Salmonella enterica isolates of serotype Concord (and its monophasic variant 6,7:l,v:-) producing the extended-spectrum β-lactamases (ESBLs) SHV-12 and CTX-M-15 in France and Norway between 2001 and 2006 (43 in France and 26 in Norway). The majority of these isolates were from adopted children from Ethiopia, most of whom were healthy carriers. Several symptomatic secondary cases were found in the adoptive families and health care facilities in France. Serotype Concord isolates collected before 2003 produced SHV-12 encoded on a 340-kb conjugative plasmid of replicon IncI1. Isolates collected after 2003 produced CTX-M-15. We detected two conjugative plasmids carrying blaCTX-M-15. One plasmid, approximately 300 kb in size, was positive for the IncHI2 replicon and the plasmid-mediated quinolone resistance gene qnrA1. The other plasmid, from one of the earliest CTX-M-15-producing isolates collected, was a fusion plasmid with IncY and IncA/C2 replicons and was 200 kb in size. However, we showed, using Southern hybridization of I-CeuI-digested chromosomal DNA and S1 nuclease analysis of plasmid DNA, that most isolates had a blaCTX-M-15 gene located on chromosomal DNA. Analysis of the flanking regions of the chromosomally located blaCTX-M-15 gene by cloning revealed an ISEcp1 truncated by an intact IS26 upstream from the blaCTX-M-15 gene and a truncated orf477 gene downstream from blaCTX-M-15. We found regions beyond the IS26 and the orf477 genes that were derived from IncA/C2 plasmids, suggesting the chromosomal integration of part of the blaCTX-M-15-carrying IncY and IncA/C2 fusion plasmid from early CTX-M-15-producing isolates.
Extended-spectrum cephalosporins (ESC) are the drugs of choice in children requiring effective chemotherapy for nontyphoidal salmonellosis. Indeed, fluoroquinolones are contraindicated for use in children, and Salmonella isolates resistant to classical first-line antibiotics, such as aminopenicillins and cotrimoxazole, have emerged over recent decades. The emergence of Salmonella isolates resistant to ESC is a new public health concern (1). Resistance to these drugs is mainly mediated by the bacterial production of β-lactamases that degrade ESC. Two main classes of plasmid β-lactamases that inactivate ESC have been identified in Salmonella: the Ambler class A extended-spectrum β-lactamases (ESBLs), the most prevalent class in the genus, and the Ambler class C cephamycinases. Most ESBLs belong to three families, TEM, SHV, and CTX-M (2). Over the last decade, CTX-Ms have been described in many species (including several Salmonella serotypes) and plasmids from nosocomial and community settings and have become the most prevalent family of ESBLs globally (7). There are currently at least 40 enzymes (a dozen of which have been identified from Salmonella isolates) divided into five major phylogenetic groups (1, 5). CTX-Ms (at least four groups) appear to be derived from Kluyvera chromosomal β-lactamases from the environment, in particular, Kluyvera ascorbata and Kluyvera georgiana (23, 30, 31, 32). Various insertion sequences, such as ISEcp1, or putative transposases, such as CR1 (for common region 1; formerly orf513), are involved in the mobilization of blaCTX-M genes.
The number of records of ESC-resistant human isolates of S. enterica serotype Concord has been increasing in France since 2004 and in Norway since 2001. Concord is a very rare serotype with the antigenic formula 6,7:l,v:1,2. It was identified for the first time in 1944 from four cultures, three of which were isolated from fatal infections in chicks in the United States and one from the stools of a patient affected in a small outbreak of food poisoning in England (14). In the early 1980s, Concord was isolated in chicken-breeding farms in Saudi Arabia (2). Two sporadic ESBL-producing isolates of serotype Concord were reported in recent studies. One, containing blaSHV-12, was isolated in Holland in 2001 (21). The other, a CTX-M-15-producing isolate, was isolated in Ireland in 2005 in a patient originally from Ethiopia (28).
We studied the French and a selection of the Norwegian S. enterica serotype Concord isolates (i) to determine the genomic diversity of these isolates by standardized pulsed-field gel electrophoresis (PFGE), (ii) to determine the genetic basis for antibiotic resistance, and (iii) to briefly describe the epidemiology of infections with the organisms.
MATERIALS AND METHODS
Salmonella isolates.
A total of 146,160 Salmonella clinical isolates were registered at the French National Reference Centre for Salmonella (FNRC-Salm), Institut Pasteur, Paris, France, between January 1996 and December 2006. The FNRC-Salm network is based on approximately 1,400 voluntary hospital or private clinical laboratories (approximately 30% of all French clinical laboratories) and covers a population of approximately 23.7 million people (39.4% of the French population based on the 1999 census). During this 10-year period, 54 serotype Concord isolates (0.04%) isolated from 54 different cases were identified (Table 1). One isolate was not recovered in subculture, and consequently, 53 serotype Concord isolates were included in this study. We also studied seven isolates with the antigenic formula 6,7:l,v:- (probable monophasic variants of serotype Concord) received at the FNRC-Salm between 2004 and 2006.
TABLE 1.
Antimicrobial resistance patterns, bla genes, class 1 integrons, and XbaI PFGE types of S. enterica serotype Concord and monophasic 6,7:l,v:- isolates from France, 1996 to 2006, and of selected S. enterica serotype Concord isolates from Norway, 2001 to 2005
| Yr | No. of isolates | Antimicrobial resistance phenotype (n)a,b | β-Lactam resistance gene (n)b | Class 1 integron size in kb [gene cassette] (n)b | PFGE type (n)b |
|---|---|---|---|---|---|
| France | |||||
| 1996 | 2 | Pansusceptible (2) | − | − | X6 (1), X7 (1) |
| 1997 | 2 | Pansusceptible (2) | − | − | X9 (2) |
| 1998 | 1 | Pansusceptible (1) | − | − | X5 (1) |
| 1999 | 4 | Pansusceptible (4) | − | − | X7 (3), X5 (1) |
| 2000 | 0 | ||||
| 2001 | 1 | Pansusceptible (1) | − | − | X8 (1) |
| 2002 | 1 | − | − | − | − |
| 2003 | 3 | Pansusceptible (3) | − | − | X1 (1), X4 (1), X5 (1) |
| 2004 | 7 | Pansusceptible (2) | − | − | X2 (1), X3 (1) |
| AMX CRO CAZ STR SPT GEN SXT CHL TET (5) | blaCTX-M-15, blaTEM-1 (5) | 0.8 (dfrA7); 1.5 [qacH-aadA1] (1); 1,5 [qacH-aadA1] (4) | X12 (3), X11a (1), X11b (1) | ||
| 2005 | 23 | AMX CRO CAZ STR SPT GEN SXT CHL TET (15) | blaCTX-M-15, blaTEM-1 (15) | 1.5 (15) | X12 (13), X11b (1), X11c (1) |
| AMX CRO CAZ STR SPT GEN SUL CHL TET (3) | blaCTX-M-15, blaTEM-1 (3) | 1.5 (3) | X11b (3) | ||
| AMX CRO CAZ STR SPT GEN SXT CHL (2) | blaCTX-M-15, blaTEM-1 (2) | 1.5 (2) | X15 (2) | ||
| AMX CRO CAZ STR SPT GEN SUL TET (1) | blaCTX-M-15, blaTEM-1 (1) | 1.5 (1) | X11b (1) | ||
| AMX CRO CAZ STR GEN SXT CHL TET NALd (2) | blaCTX-M-15, blaTEM-1 (2) | NDc | X17 (2) | ||
| 2006 | 17 | AMX CRO CAZ STR SPT GEN SXT CHL (7) | blaCTX-M-15, blaTEM-1 (7) | 1.5 (7) | X12 (3), X16 (2), X11c (1), X15 (1) |
| AMX CRO CAZ STR KAN GEN SXT CHL (4) | blaCTX-M-15 (4) | 0.8 (4) | X18 (4) | ||
| Pan-susceptible (2) | − | − | X20 (2) | ||
| AMX CRO CAZ STR SPT GEN SXT CHL TET (1) | blaCTX-M-15, blaTEM-1 (1) | 1.5 (1) | X12 (1) | ||
| AMX CRO CAZ STR SPT GEN SUL CHL (1) | blaCTX-M-15, blaTEM-1 (1) | 1.5 (1) | X11b (1) | ||
| AMX CRO CAZ STR GEN SXT CHL TET NALd (1) | blaCTX-M-15 (1) | NDc | X17 (1) | ||
| AMX CRO CAZ STR GEN SXT TET (1) | blaCTX-M-15, blaTEM-1 (1) | 1.5 (1) | X14 (1) | ||
| Norway | |||||
| 2001 | 3 | AMX SXT CHL (1) | blaTEM-1 (1) | 2.0 [dfrA5-ereA2] (1) | X10c (1) |
| AMX CRO CAZ STR SPT KAN GEN SXT CHL (1) | blaSHV-12, blaTEM-1 (1) | 1.0, 2.0 (1) | X10c (1) | ||
| AMX CRO CAZ SXT CHL (1) | blaSHV-12, blaTEM-1 (1) | 2.0 [dfrA5-ereA2] (1) | X10c (1) | ||
| 2002 | 1 | AMX CRO CAZ SXT CHL (1) | blaSHV-12, blaTEM-1 (1) | 2.0 (1) | X10c (1) |
| 2003 | 2 | AMX CRO CAZ (1) | blaSHV-12 (1) | − | X10b (1) |
| AMX CRO CAZ STR SPT KAN GEN SXT CHL (1) | blaSHV-12, blaTEM-1 (1) | 1.0 [aadA1] (1) | X10a (1) | ||
| 2004 | 1 | AMX CRO CAZ STR SPT GEN SXT CHL TET (1) | blaCTX-M-15, blaTEM-1 (1) | 1.5 [qacH-aadA1] (1) | X11c (1) |
| 2005 | 1 | AMX CRO CAZ STR SPT GEN SXT CHL TET (1) | blaCTX-M-15, blaTEM-1 (1) | 1.8 [dhfrXII-orfF-aadA2] (1) | X13 (1) |
AMX, amoxicillin; STR, streptomycin; SPT, spectinomycin; GEN, gentamicin; CHL, chloramphenicol; SUL, sulfonamides; SXT, sulfamethoxazole-trimethoprim; TET, tetracycline.
−, not done.
ND, none detected.
Presence of a qnrA1 gene.
In Norway, 16,849 cases of Salmonella infections were reported to the Norwegian Surveillance System for Communicable Diseases at the Norwegian Institute of Public Health, Oslo, Norway, between January 1996 and December 2006. About 80% of the case patients had acquired the infection abroad. All clinical Salmonella isolates were submitted to the Norwegian National Reference Laboratory for Bacterial Enteropathogens at the Norwegian Institute of Public Health for verification and characterization. During this period, 27 serotype Concord isolates (0.16%), corresponding to 27 cases, were identified. Eight isolates, selected on the basis of their diversity (year of isolation and resistance phenotype), were sent to the FNRC-Salm and fully characterized (Table 1).
The S. enterica serotype Concord reference strain 156K (isolated from a chick in 1944 in California), obtained from the World Health Organization Collaborative Centre for Reference and Research on Salmonella, Institut Pasteur, was used for molecular typing methods. S. enterica serotype Braenderup H9812 was used as a molecular size marker for PFGE.
Epidemiological investigations.
Demographical, clinical, and epidemiological data were collected for each case identified between 2004 and 2006 in France by contacting physicians by mail. Eight associations certified by the French Government for international adoption from Ethiopia were contacted by e-mail.
In Norway, it is recommended, but not required, that all adopted infants be screened for intestinal parasites and Salmonella infection. However, the extent to which this recommendation is followed is unclear. Clinicians and microbiological laboratories report all laboratory-confirmed Salmonella infections to the Norwegian Surveillance System for Communicable Diseases. Demographical, clinical, and epidemiological information is collected on the clinical notification forms. The Norwegian association responsible for adoption from Ethiopia was informed.
Antimicrobial susceptibility.
Antibiotic susceptibility was determined by the disk diffusion method with 32 antimicrobial drugs (Bio-Rad, Marnes la Coquette, France), as previously described (35). The MICs of ceftriaxone (CRO) and ceftazidime (CAZ) were determined by Etest (AB Biodisk, Solna, Sweden). The ESBL phenotype was detected by using the ESBL detection Etest strips (AB Biodisk) and the double-disk synergy method (25). Isolates were categorized as susceptible, intermediate, or resistant according to Antibiogram Committee of the French Society for Microbiology cutoff values (http://www.sfm.asso.fr/nouv/general.php?pa=2). The cutoff values used for CRO and CAZ are slightly different from those determined by the Clinical and Laboratory Standards Institute (CLSI); susceptible strains were thus defined by MICs of ≤4 μg/ml (CLSI, ≤8 μg/ml) and resistant strains by MIC of >32 μg/ml (CLSI, ≥64 μg/ml for CRO and ≥32 μg/ml for CAZ).
PCR amplification of antimicrobial resistance genes and sequence analysis.
Total DNA was extracted using the InstaGene matrix kit (Bio-Rad) according to the manufacturer's recommendations. The resistance genes blaTEM, blaSHV, blaOXA-1 group, and blaCTX-M and class 1 integron gene cassettes were amplified by PCR as described previously (15, 35).
Sequencing was performed at Genome Express (Meylan, France) or at the Plateforme de Génotypage des Pathogènes et Santé Publique, PF8 (Institut Pasteur). The nucleotide sequences and the deduced protein sequences were analyzed with EditSeq and Megalign software (Dnastar, Madison, WI). The BLASTN program of NCBI was used for database searches (http://www.ncbi.nlm.nih.gov/BLAST/).
PFGE typing.
The genetic diversity of 68 clinical Salmonella isolates of serotype Concord or the monophasic variant 6,7:l,v:- (60 from France and 8 from Norway) and serotype Concord reference strain 156K was assessed by PFGE of genomic DNA digested with XbaI (Roche, Mannheim, Germany), as described previously (34). The running conditions and the molecular size marker were as described in the standardized PulseNet protocol (24). BioNumerics 4.0 (Applied Maths, Sint-Martens-Latem, Belgium) was used for image normalization and construction of similarity matrices. Bands were assigned manually. Clustering was carried out by the unweighted pair-group method with arithmetic averages based on the Dice similarity index, using a 1% optimization parameter and 1% band position tolerance. Each profile that differed by one or more extra bands of >100 kb in size was assigned a type (e.g., type X10). Profiles that differed only by the position of a high-molecular-weight band (400- to 600-kb region) were assigned to subtypes (e.g., subtypes X10a, X10b, and X10c).
Resistance transfer determination.
We carried out a resistance transfer experiment using a subset of 12 (8 French and 4 Norwegian) ESBL-producing isolates on liquid and solid media, using either Escherichia coli K-12 BM14 resistant to sodium azide or E. coli C1a resistant to nalidixic acid (NAL) as the recipient strain (35). Transconjugants were selected on Drigalski agar (Bio-Rad) supplemented with cefotaxime (CTX) (4 μg/ml) and sodium azide (500 μg/ml) or NAL (64 μg/ml). Three E. coli transconjugants were arbitrarily selected for each experiment.
Plasmid DNA from four S. enterica serotype Concord isolates that was unsuccessfully transferred by conjugation was used to transform electrocompetent E. coli DH10B by standard electroporation techniques with a MicroPulser electroporation apparatus (Bio-Rad). Transformants were selected on Mueller-Hinton agar containing CTX (4 μg/ml).
Plasmid analysis.
Plasmids were characterized for a subset of 19 Salmonella isolates (and four E. coli transconjugants) according to the year of isolation, antimicrobial resistance phenotype, and PFGE profile. Two plasmid-profiling methods were used: alkaline lysis and S1 nuclease analysis.
Plasmid DNA extracted by alkaline lysis (33) was analyzed by electrophoresis in 0.8% agarose gels. The molecular sizes of plasmids were estimated by comparison with plasmids of known sizes: pIP173 (125.8 kb), RP4 (56 kb), and Tp116 (210 kb), mixed with supercoiled DNA ladder (Invitrogen, Groningen, The Netherlands).
We used S1 nuclease treatment and PFGE to accurately determine the molecular sizes of large bacterial plasmids. Proteinase K-treated gel plugs prepared for PFGE analysis were cut into 1-mm slices and digested with 1 U of S1 nuclease (Roche) as described previously (3). DNA fragments separated by PFGE were transferred onto a nylon membrane (Hybond N+; Amersham) and hybridized with blaCTX-M, blaTEM, IncA/C, and IncY probes (8, 15). Hybridization, labeling, and detection were performed according to the manufacturers' recommendations, using either an enhanced-chemiluminescence nonradioactive kit (GE Healthcare, United Kingdom) or a radioactive kit (MegaprimeTM DNA-labeling system; GE Healthcare).
PCR-based replicon-typing analysis was performed as described by Carattoli et al. (8). The 18 primer pairs targeting FIA, FIB, FIC, HI1, HI2, I1-Iγ, L/M, N, P, W, T, A/C, K, B/O, X, Y and FII replicons were used in separate PCRs.
Chromosomal localization of the blaCTX-M-15 gene by PFGE-I-CeuI.
To determine the chromosomal location of the blaCTX-M-15 gene, plugs of 11 isolates were prepared as described above and digested with the I-CeuI endonuclease (New England Biolabs). The digested fragments were separated using the Chef DRIII system as described previously (27). The sizes of I-CeuI restriction fragments were determined using known I-CeuI fragment sizes of chromosomal DNA from S. enterica serotype Typhimurium LT2 (27) and XbaI-restricted fragments from the chromosomal DNA of S. enterica serotype Braenderup H9812. The I-CeuI restriction fragments were subjected to Southern hybridization with PCR-generated probes for blaCTX-M, blaTEM, and16S rRNA gene probes (15, 20).
Analysis of the genetic environment of the blaCTX-M-15 gene by cloning.
We used cloning techniques to determine the DNA sequences flanking the blaCTX-M-15 gene in isolate 05-0004 (a representative Concord isolate with a chromosomal blaCTX-M-15 gene, as determined by PFGE I-CeuI and S1 nuclease experiments). DNA prepared using the Promega Wizard kit was partially digested with Sau3aI, purified with Qiaquick PCR purification kit columns (Qiagen, Courtaboeuf, France), and ligated into dephosphorylated, BamHI-restricted phagemid pBK-CMV (Roche, Meylan, France). Recombinant plasmids were introduced into E. coli DH10B by electroporation (Gene Pulser II; Bio-Rad). Antibiotic-resistant colonies were selected on Luria-Bertani agar containing kanamycin (KAN) (30 μg/ml) and CRO (4 μg/ml). Recombinant plasmid DNA was recovered using Qiaprep spin miniprep columns (Qiagen).
The following primers were used for PCR mapping: CP604Av, 5′-GGGTATTTACGAGATGGCAGC-3′ (nucleotides [nt] 157212 to 157232 of the plasmid pSN254 of S. enterica serotype Newport SL254; GenBank accession number CP000604); CP604Am, 5′-GCATTTCTCGGGTGACTTCCT-3′ (nt 12446 to 12466; accession number CP000604); and CP60412000, 5′-GTTGGCAATCTCCTGGGTGAT-3′ (nt 12000 to 12020; accession number CP000604).
Nucleotide sequence accession number.
The nucleotide sequence of the class 1 integron gene cassette qacH-aadA1 was assigned GenBank accession number EU200458.
RESULTS AND DISCUSSION
Epidemiological background.
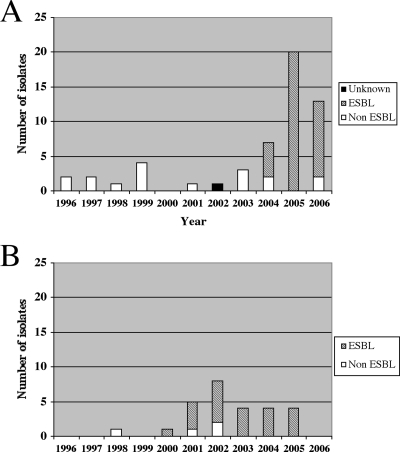
Serotype Concord was rare in Norway before 2000 and in France before 2004 (Fig. 1A and B).
FIG. 1.
(A) Numbers of S. enterica serotype Concord isolates producing or not producing an ESBL in France (A) and in Norway (B) between 1996 and 2006.
In France, of the 14 serotype Concord isolates collected before 2004, 13 were tested (1 was not viable), and all were pansusceptible. In two cases, the subjects had traveled (to Madagascar or Ethiopia) before becoming ill. No epidemiological information was available for the remaining 12 cases. Among isolates collected between 2004 and 2006, 36/40 were multidrug resistant (MDR) with resistance to ESC (Table 1). The seven monophasic isolates were also MDR with resistance to ESC. Most of the 43 ESC-resistant Salmonella (Concord and monophasic) isolates were recovered from children aged <1 year (34/43) or between 1 and 4 years (6/43). Three cases were adults (36, 36, and 93 years old). Epidemiological data were available for 35 of the 43 cases. Of the 35 cases, 28 were children recently adopted from Ethiopia. The seven remaining cases were probable or possible secondary cases in the household or in a health care setting. Clinical data were available for 22 adopted children and six French autochthonous cases. For the adopted children, clinical symptoms were absent in 12, whereas bronchopulmonary infections were seen in 8, pyelonephritis (due to E. coli) in 1, and gastroenteritis in 1. For the French autochthonous cases, five had gastroenteritis and one died.
In Norway, the single serotype Concord isolate collected between 1996 and 2000 (acquired after travel to Eritrea) was susceptible to all antibiotics tested. Between 2000 and 2006, 27 Concord isolates were registered. All but one isolate were from children adopted from Ethiopia. Twenty-three of the 27 Concord isolates were MDR with resistance to ESC (not shown and Table 1). The first ESBL-positive isolate was registered in 2000 in an adopted child from Ethiopia.
We evaluated the prevalence of carriers using data obtained from the systematic initial screening of children arriving in France (clinical examination, stool bacteriology and parasitology, and viral serologies). To this end, a French-certified association contacted all 187 families that adopted children from Ethiopia through the association between January 2005 and August 2006. One hundred and four families participated, and three carriers (2.9%) were found. Most of the adopted infants were from the same orphanage in Addis Ababa. We learned that there were two or three children per bed and that ESC were used extensively for treating life-threatening respiratory infections. The strain may have then been disseminated due to the promiscuity between infants and perhaps through the hands of the staff, thus explaining the long-lasting circulation of the strain. Overcrowding and understaffing are risk factors commonly observed in neonatal units in developing countries, for example, in Tunisia in 1988 (serotype Wien producing SHV-2) (19) and in 2002 (serotype Livingstone producing CTX-M-27) (6). However, the source of this strain in Ethiopia has yet to be investigated. As it was very difficult to prevent transmission in Ethiopia, we focused on preventing dissemination of the strain in France by informing all certified associations about the risk of transmission of resistant Salmonella and the adoptive families about the importance of basic hygiene practice. In 2007, the number of ESBL-producing Salmonella isolates in adopted children from Ethiopia was decreasing in France, with only 13 isolates, and no secondary cases, detected at the FRNC-Salm. In Norway, the adoption agency changed transit home in 2005, and in 2006, no new cases were reported.
Antimicrobial resistance phenotypes and genes and molecular typing.
The antimicrobial resistance phenotypes of the 60 French isolates are described in Table 1. Thirty-six MDR serotype Concord and seven MDR monophasic isolates resistant to ESC contained the blaCTX-M-15 ESBL gene. The MICs of CRO, CAZ, and cefepime (FEP) were >256 μg/ml. Of the 43 ESBL-producing isolates, 38 were positive for the penicillinase blaTEM-1 gene and none were positive for the penicillinase blaOXA-1 group gene. All but three MDR strains harbored a class 1 integron containing a novel qacH-aadA1 gene cassette. One isolate harbored an additional class 1 integron containing a dfrA7 gene cassette. Three CTX-M-15-producing isolates with decreased susceptibility to ciprofloxacin (0.25 to 0.5 μg/ml) were found to contain the plasmid-mediated quinolone resistance gene qnrA1. One of these isolates, 05-3728, was studied previously (11).
The resistance profiles of the eight selected Norwegian isolates are described in Table 1. All of the isolates produced an ESBL, with the exception of one, which was positive for the penicillinase blaTEM-1 gene. We identified blaSHV-12 in five ESBL isolates collected before 2004 and blaCTX-M-15 in the remaining two ESBL isolates collected from 2004 to 2005. The MICs of CRO, CAZ, and FEP were >256 μg/ml for the CTX-M-15-producing isolates. The SHV-12-producing isolates had CAZ MICs of >256 μg/ml, CRO MICs of 64 μg/ml, and FEP MICs of 8 to 16 μg/ml. Several different class 1 integron gene cassettes were identified (Table 1).
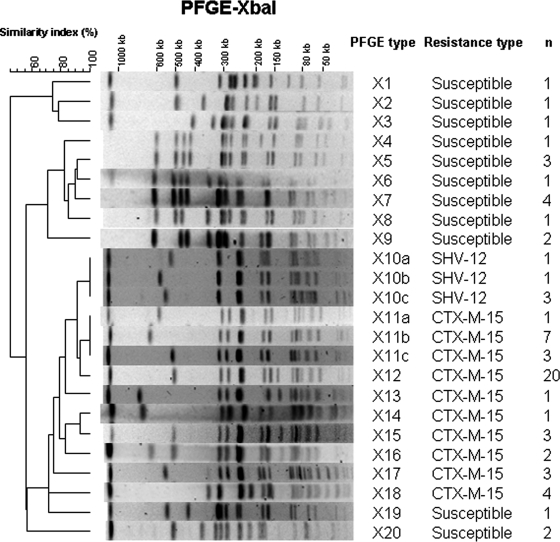
The clonal relatedness of S. enterica serotype Concord and monophasic 6,7:l,v:- isolates was assessed by standard PFGE analysis of XbaI-digested chromosomal DNA (Fig. 2 and Table 1). Twenty different PFGE profiles were obtained from 68 clinical isolates and the reference strain. All ESBL-producing isolates clustered together (Dice correlation coefficient, 72%). SHV-12-producing isolates displayed the same profile, X10 (divided into three subtypes), distinct from the eight obtained from CTX-M-15-producing isolates. Among the CTX-M-15-producing isolates, X12 was predominant (n = 20). This profile was close to other CTX-M-15 isolate profiles, X11 and X13 to X16. However, two profiles, X17 and X18, differed more significantly. Profile X17 was observed in the three isolates containing the qnrA1 gene, and profile X18 was observed in four isolates displaying additional resistance to KAN. The two other clusters contained 15 pansusceptible isolates. The profiles obtained from the seven monophasic isolates were X11c (n = 1), X12 (n = 2), X15 (n = 3), and X16 (n = 1), suggesting that they were derived from the Concord serotype.
FIG. 2.
Representative XbaI-PFGE profiles of S. enterica serotype Concord isolates studied. A dendrogram was generated with Bionumerics software. The PFGE profile, number of isolates, and resistance type are indicated.
Two types of ESBL were identified in the Concord strains from Ethiopia: the blaSHV-12 gene was observed between 2000 and 2003 and the blaCTX-M-15 gene between 2004 and 2006. In France, between 2001 and 2003, there was an outbreak in infants adopted from Mali caused by S. enterica serotypes Babelsberg and Enteritidis producing SHV-12 (35). The shift in the ESBL distribution (i.e., CTX-Ms, in particular CTX-M-15 replacing SHV-12) is consistent with the epidemiology of the ESBLs in the genus Salmonella reported in African countries, such as Senegal (17, 36), Tanzania (4), and South Africa (18, 26).
Transferability and location of the ESBL genes.
Conjugation experiments were carried out on liquid and solid media with a subset of 12 ESBL-producing isolates selected based on the year and country of isolation, the antibiotic resistance phenotype, and the PFGE type. ESC transfer was successful only for the two Norwegian SHV-12-producing isolates tested and two French CTX-M-15-producing isolates (Table 2).
TABLE 2.
Cefotaxime resistance transfer experiments
| Isolate | Yr/country of isolation | Antimicrobial resistance patterna | ESBL gene | PFGE type | Transfer of resistance to ESC by:
|
Transferred resistance traitsa | Plasmids carrying ESBL genes in transconjugants (size [kb], replicon type) | |
|---|---|---|---|---|---|---|---|---|
| Conjugation | Transformation | |||||||
| 04-347 | 2004/France | AMX CRO CAZ STR SPT GEN SXT CHL TET | blaCTX-M-15 | X11a | Yes | AMX CRO CAZ STR SXT CHL TET | 200, IncY/IncA/C2 | |
| 04-7498 | 2004/France | AMX CRO CAZ STR SPT GEN SXT CHL TET | blaCTX-M-15 | X12 | No | |||
| 04-8041 | 2004/France | AMX CRO CAZ STR SPT GEN SXT CHL TET | blaCTX-M-15 | X12 | No | No | ||
| 05-0004 | 2005/France | AMX CRO CAZ STR SPT GEN SUL TET | blaCTX-M-15 | X11b | No | No | ||
| 05-2657 | 2005/France | AMX CRO CAZ STR SPT GEN SUL CHL TET | blaCTX-M-15 | X11b | No | |||
| 05-5343 | 2005/France | AMX CRO CAZ STR GEN SXT CHL TET NAL | blaCTX-M-15 | X17 | Yes | AMX CRO CAZ STR GEN SXT CHL TET NAL | 300, IncHI2 | |
| 06-8636 | 2006/France | AMX CRO CAZ STR KAN GEN SXT CHL | blaCTX-M-15 | X18 | No | |||
| 06-8404 | 2006/France | AMX CRO CAZ STR KAN GEN SXT CHL | blaCTX-M-15 | X18 | No | No | ||
| 07-690 | 2001/Norway | AMX CRO CAZ SXT CHL | blaSHV-12 | X10c | Yes | AMX CRO CAZ | 340, IncI1 | |
| 07-693 | 2002/Norway | AMX CRO CAZ SXT CHL | blaSHV-12 | X10c | Yes | AMX CRO CAZ | 340, IncI1 | |
| 07-698 | 2004/Norway | AMX CRO CAZ STR SPT GEN SXT CHL TET | blaCTX-M-15 | X11c | No | |||
| 07-699 | 2005/Norway | AMX CRO CAZ STR SPT GEN SXT CHL TET | blaCTX-M-15 | X13 | No | No | ||
AMX, amoxicillin; STR, streptomycin; SPT, spectinomycin; GEN, gentamicin; CHL, chloramphenicol; SUL, sulfonamides; SXT, sulfamethoxazole-trimethoprim; TET, tetracycline.
The blaSHV-12 genes were carried by similar plasmids approximately 340 kb in size and belonging to the IncI1 group, as determined by PCR-based replicon typing, whereas the plasmids carrying the blaCTX-M-15 gene were of different types (Table 2). One belonged to IncHI2, while the other was positive for both IncY and IncA/C2 replicons. A positive hybridization of the S1 nuclease-treated plasmid 200-kb band by both IncY and IncA/C2 probes confirmed the double-replicon structure (data not shown).
A large plasmid of approximately 300 kb that did not hybridize to the blaCTX-M probe, but which did hybridize to a blaTEM probe, was present in all CTX-M-15-producing Salmonella isolates, with the exception of isolate 04-347 and the two monophasic isolates tested. This plasmid was also absent in the single susceptible isolate tested (data not shown). Although the Norwegian isolate 07-699 did not undergo transfer of ESC resistance to E. coli, it contained at least two plasmids, one of approximately 250 kb that hybridized to the blaCTX-M-15 probe and one of approximately 140 kb that hybridized to an IncA/C probe (data not shown).
Two French isolates resistant to KAN, 06-8636 and 06-8404, which did not undergo transfer of ESC resistance, were found to contain a large plasmid of approximately 320 kb that did hybridize to the blaCTX-M-15 probe (data not shown).
PCR typing targeting IncA/C and IncY replicons was performed in all other Salmonella isolates. An IncA/C replicon was found only in isolate 07-699, whereas the IncY replicon was found in all isolates except 06-8404 and 06-8636, 07-699, and the susceptible isolates tested. S1 nuclease and Southern blotting showed that the IncY plasmid in Salmonella isolates (except that of 04-347) was approximately 100 kb in size (data not shown). In a previous study, two different replicons were associated with blaCTX-M-15-positive plasmids from Salmonella and E. coli isolates collected between 2001 and 2003 from the United Kingdom, Honduras, and Pakistan (22). Nine of 22 plasmids were positive for the IncI1 replicon, and 12 of 22 plasmids were positive for IncFII (in some strains associated with IncFIA and/or IncFIB replicons in fusion plasmids).
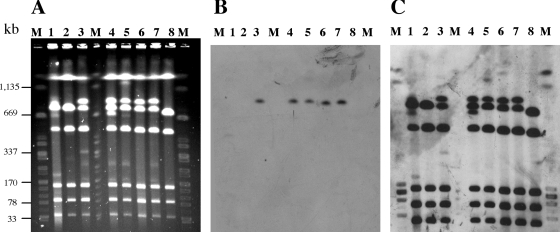
The fact that the ESC resistance transfer experiments were unsuccessful for the majority of the isolates, with no hybridization of their plasmid DNAs to a blaCTX-M-15 gene probe, suggested a chromosomal location of the blaCTX-M-15 gene in these isolates. This hypothesis was confirmed by I-CeuI PFGE, which clearly showed that the blaCTX-M-15 gene was located in a chromosomal fragment of approximately 900 kb (Fig. 3). This ∼900-kb fragment hybridizing to the 16S rRNA gene probe was absent in isolates 05-5343 and 04-347 (carrying blaCTX-M-15 on a plasmid), 06-8636 (an isolate additionally resistant to KAN), and 07-699 (a Norwegian isolate) and in susceptible isolates (Fig. 3 and data not shown). These isolates that did not have the ∼900-kb fragment displayed increased intensity of the band at ∼700 kb, suggesting that two DNA fragments comigrated at this position, as observed for 779-kb and 738-kb fragments of S. enterica serotype Typhimurium strain LT2 (27). Because the endonuclease I-CeuI cleaves only rrn operons and because the number (n = 7) and locations of the rrn genes are highly conserved in Salmonella (27), this ∼900-kb chromosomal fragment was interpreted to derive from one of the two DNA chromosomal fragments contained in the ∼700-kb band by insertion of the blaCTX-M-15 gene and additional DNA. The blaTEM gene was not detected on chromosomal fragments (data not shown). Chromosomal location of ESBL genes in enterobacteria is extremely rare, with only two reports showing chromosomal integration of blaCTX-M-9 or blaCTX-M-15 in E. coli (12, 16).
FIG. 3.
Chromosomal location of the blaCTX-M-15 gene. (A) PFGE separation of I-CeuI-digested DNAs from seven S. enterica serotype Concord and one S. enterica serotype 6,7:l,v:- isolates. (B and C) Southern hybridization with a blaCTX-M-specific internal probe (B) and a 16S rRNA gene probe (C). M, XbaI-digested DNA from S. enterica serotype Braenderup strain H9812; lane 1, isolate 04-347 (Concord, ESBL); lane 2, isolate 05-5343 (Concord, ESBL, qnrA1); lane 3, isolate 04-7498 (Concord, ESBL); lane 4, isolate 05-705 (Concord, ESBL); lane 5, isolate 05-5547 (Concord, ESBL); lane 6, isolate 05-9115 (6,7:l,v:-, ESBL); lane 7, isolate 04-9447 (Concord, ESBL); lane 8, isolate 04-2781 (Concord, pansusceptible).
Genetic context of the blaCTX-M-15 gene.
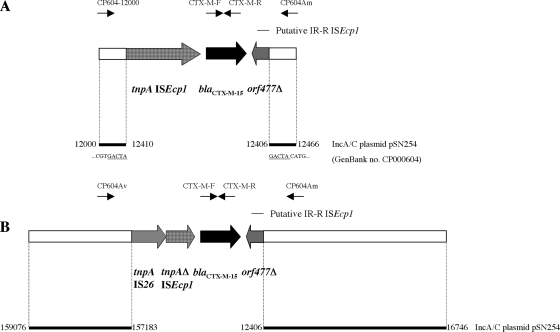
To study the flanking DNA sequences of the chromosomal blaCTX-M-15 gene, we cloned the gene from isolate 05-0004. This representative isolate contained the blaCTX-M-15 gene on the 900-kb chromosomal I-CeuI fragment (data not shown). We obtained 31 E. coli DH10B recombinant clones. Seven of these were tested, and the longest recombinant plasmid, p05-0004-C1 (9 kb), was retained for further analysis. The schematic representation of recombinant plasmid p05-0004-C1 is shown in Fig. 4B. Sequence analysis identified the insertion sequence ISEcp1 48 bp upstream from, and in the same orientation as, the blaCTX-M-15 gene (identical to accession number AY604722). ISEcp1 was truncated at nucleotide position 1102 by an intact IS26, both in the same orientation. orf477 (identical to accession number AY604722) was located downstream from the blaCTX-M-15 gene in the opposite orientation. This open reading frame was truncated at amino acid position 70, corresponding to the end of an 18-bp sequence homologous to the inverted repeat right (IRR) of ISEcp1. We identified 1.9 kb and 4.3 kb of DNA sequence upstream from IS26 and downstream from orf477, respectively, which shared 100% identity with sequences in three MDR IncA/C plasmids (GenBank accession numbers CP000602, CP000603, and CP000604). However, these sequences were located at two distant regions in the plasmids (Fig. 4B). One of these plasmids, pSN254 (accession number CP000604) (37), is the large MDR IncA/C2 plasmid carrying the cephamycinase blaCMY-2 gene that has been increasingly detected in S. enterica serotype Newport in the United States (9, 10, 13).
FIG. 4.
Schematic representations of the various genetic environments of the blaCTX-M-15 gene. (A) The plasmid blaCTX-M-15 gene from S. enterica serotype Concord 04-347. (B) Recombinant clone p05-0004-C1 with the chromosomally located blaCTX-M-15 gene (9-kb insert) from S. enterica serotype Concord 05-0004. Open reading frames and genes surrounding the blaCTX-M-15 gene are shown as boxes, with an arrow indicating the orientation of each coding sequence and the gene name shown under the corresponding box. The putative IRR motif of ISEcp1 is indicated by a horizontal bar. The primers used for PCR mapping are shown with arrows above the boxes. The white boxes denote homology with sequences from MDR IncA/C plasmid pSN254 from S. enterica serotype Newport (GenBank accession number CP000604) (37). Coordinates of the pSN254 sequences are indicated. The nucleotides underlined in panel A correspond to the 5-bp directed repeats.
To verify the flanking structures of the region IS26-ISEcp1-blaCTX-M-15-orf477 on other isolates, we designed primers CP604Av and CP604Am to target pSN254 regions upstream of IS26 and downstream of orf477, respectively (Fig. 4B). PCRs with CP604Av and CTX-M-R and with CP604Am and CTX-M-F were performed on a selection of 15 isolates (comprising 10 tested for resistance transfer and chromosomal or plasmid localization of blaCTX-M-15). PCR products with the expected sizes of 2.2 kb and 1.2 kb were observed in all the isolates with a chromosomally located blaCTX-M-15 gene, whereas no amplification was observed for isolates 05-5343 (IncHI2 plasmid-located blaCTX-M-15 gene), 06-8636 (a representative 2006 French isolate resistant to KAN), and Norwegian isolate 07-699. However, an amplicon of 1.2 kb using CTX-M-F and CP604Am was obtained in the early French isolate 04-347 (IncA/C2-IncY plasmid-located blaCTX-M-15 gene). To analyze the DNA sequence upstream from blaCTX-M-15 of this isolate, we designed a primer (CP604-12000), located at positions 12000 to 12020 of plasmid pSN254, and used it for PCR with CTX-M-R on it and its E. coli transconjugant. We obtained an amplicon of 2.8 kb. Sequencing analysis revealed that the complete ISEcp1 was present and that the ISEcp1- blaCTX-M-15-orf477 region was flanked by 5-bp direct repeats (GACTA) and inserted within IncA/C2 plasmid pSN254 at nucleotide position 12411 (Fig. 4A). PCR with primers CP604-12000 and CP604Am was performed on isolates 06-8636 and 07-699. An amplicon of 450 bp was observed only in isolate 07-699, demonstrating that the blaCTX-M-15 gene was not inserted within the 12000-to-12400 region of pSN254. We did not further characterize the genetic environment of the blaCTX-M-15 genes located on nonconjugative plasmids in these rare isolates.
We found that DNA sequences from the pSN254 IncA/C2 plasmid were in the vicinity of the chromosomal blaCTX-M-15. One of our earliest Concord isolates (04-347), isolated in 2004, contained a blaCTX-M-15-carrying fusion plasmid with replicon elements of types IncY (resident plasmid of MDR Concord) and IncA/C2. The blaCTX-M-15 gene was inserted into this IncA/C2 plasmid at position 12411 by ISEcp1. ISEcp1 was complete (1,656 nt), and 5-bp direct repeats flanked it upstream and downstream from an IRR-like sequence. In isolate 07-699, the blaCTX-M-15 gene was also found in an IncA/C2 plasmid, but in a different position from that identified in isolate 04-347. ESC resistance in isolates 07-699 and 06-8636 did not appear to be transferable, despite the presence of large plasmids; this may have been due to the absence in these isolates of a helper IncY plasmid, which provides functions necessary for mobilization or allows the generation of an autotransferable fusion plasmid (as observed for isolate 04-347). Failure of the transfer of ESC resistance by electroporation might have been due to the loss of integrity of these large plasmid DNAs (250 to 320 kb) after a standard lysis alkaline extraction.
The sequences downstream of the chromosomally located blaCTX-M-15 gene were identical to those of the IncA/C2-IncY plasmid-located blaCTX-M-15 gene of isolate 04-347 (corresponding to nucleotide positions 12400 to 16000 of plasmid pSN254). However, there were rearrangements in the upstream region. The insertion of the IS26 element in ISEcp1 might have been the first event on the original IncA/C2 plasmid (three intact IS26s are present in pSN254). Genetic rearrangements (deletion and inversion) due to IS26 mobilization might have resulted in a structure in which the two distant described parts of pSN254 have been brought in proximity. We can also hypothesize that the whole, or part of, the IncA/C2-IncY fusion plasmid contained in isolate 04-347 was integrated into the chromosome of other Concord-derived isolates collected later; this probably occurred through IS26-mediated cointegration. Naas et al. (29) demonstrated Tn1- or IS26-mediated integration of a plasmid-borne inhibitor-resistant TEM, blaTEM-21, from Proteus mirabilis into E. coli chromosomal DNA. In our study, sequencing the ends of the inserted DNA by genome walking will be necessary in order to elucidate the precise mechanism of integration (homologous recombination, IS- or transposon-mediated transposition, or cointegration).
Acknowledgments
We thank all laboratories of the FNRC-Salm and the Norwegian National Reference Laboratory for Bacterial Enteropathogens networks and Jean Mallet from the association Amis des Enfants du Monde for evaluating the prevalence of healthy carriers.
Footnotes
Published ahead of print on 9 March 2009.
REFERENCES
- 1.Arlet, G., T. J. Barrett, P. Butaye, A. Cloeckaert, M. R. Mulvey, and D. G. White. 2006. Salmonella resistant to extended-spectrum cephalosporins: prevalence and epidemiology. Microbes Infect. 8:1945-1954. [DOI] [PubMed] [Google Scholar]
- 2.Barbour, E. K., and N. H. Nabbut. 1982. Isolation of Salmonella and some other potential pathogens from two chicken breeding farms in Saudi Arabia. Avian Dis. 26:233-244. [PubMed] [Google Scholar]
- 3.Barton, B. M., G. P. Harding, and A. J. Zuccarelli. 1995. A general method for detecting and sizing large plasmids. Anal. Biochem. 226:235-240. [DOI] [PubMed] [Google Scholar]
- 4.Blomberg, B., R. Jureen, K. P. Manji, B. S. Tamim, D. S. M. Mwakagile, W. K. Urassa, M. Fataki, V. Msangi, M. G. Tellevik, S. Y. Maselle, and N. Langeland. 2005. High rate of fatal cases of pediatric septicemia caused by gram-negative bacteria with extended-spectrum beta-lactamases in Dar es Salaam, Tanzania. J. Clin. Microbiol. 43:745-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonnet, R. 2004. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouallegue-Godet, O., Y. Ben Salem, L. Fabre, M. Demartin, P. A. Grimont, R. Mzoughi, and F. X. Weill. 2005. Nosocomial outbreak caused by Salmonella enterica serotype Livingstone producing CTX-M-27 extended-spectrum β-lactamase in a neonatal unit in Sousse, Tunisia. J. Clin. Microbiol. 43:1037-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canton, R., and T. M. Coque. 2006. The CTX-M beta-lactamase pandemic. Curr. Opin. Microbiol. 9:466-475. [DOI] [PubMed] [Google Scholar]
- 8.Carattoli,. A., A. Bertini, L. Villa, V. Falbo, K. L. Hopkins, and E. J. Threlfall. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219-228. [DOI] [PubMed] [Google Scholar]
- 9.Carattoli, A., F. Tosini, W. P. Giles, M. E. Rupp, S. H. Hinrichs, F. J. Angulo, T. J. Barrett, and P. D. Fey. 2002. Characterization of plasmids carrying CMY-2 from expanded-spectrum cephalosporin-resistant Salmonella strains isolated in the United States between 1996 and 1998. Antimicrob. Agents Chemother. 46:1269-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carattoli, A., V. Miriagou, A. Bertini, A. Loli, C. Colinon, L. Villa, J. M. Whichard, and G. M. Rossolini. 2006. Replicon typing of plasmids encoding resistance to newer beta-lactams. Emerg. Infect. Dis. 12:1145-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cattoir, V., F. X. Weill, L. Poirel, L. Fabre, C. J. Soussy, and P. Nordmann. 2007. Prevalence of qnr genes in Salmonella in France. J. Antimicrob. Chemother. 59:751-754. [DOI] [PubMed] [Google Scholar]
- 12.Coque, T. M., A. Novais, A. Carattoli, L. Poirel, J. Pitout, L. Peixe, F. Baquero, R. Cantón, and P. Nordmann. 2008. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum beta-lactamase CTX-M-15. Emerg. Infect. Dis. 14:195-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunne, E. F., P. D. Fey, P. Kludt, R. Reporter, F. Mostashari, P. Shillam, J. Wicklund, C. Miller, B. Holland, K. Stamey, T. J. Barrett, J. K. Rasheed, F. C. Tenover, E. M. Ribot, and F. J. Angulo. 2000. Emergence of domestically acquired ceftriaxone-resistant Salmonella infections associated with AmpC beta-lactamase. JAMA 284:3151-3156. [DOI] [PubMed] [Google Scholar]
- 14.Edwards, P. R., and H. Hughes. 1944. A new Salmonella type isolated from man and fowls. J. Bacteriol. 47:574-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egorova, S., L. Kaftyreva, P. A. D. Grimont, and F. X. Weill. 2007. Prevalence and characterization of extended-spectrum cephalosporin-resistant non-typhoidal Salmonella isolates in adults in Saint-Petersburg, Russia (2002-2005). Microb. Drug Resist. 13:102-107. [DOI] [PubMed] [Google Scholar]
- 16.Garcia, A., F. Navarro, E. Miro, B. Mirelis, S. Campoy, and P. Coll. 2005. Characterization of the highly variable region surrounding the blaCTX-M-9 gene in non-related Escherichia coli from Barcelona. J. Antimicrob. Chemother. 56:819-826. [DOI] [PubMed] [Google Scholar]
- 17.Gassama-Sow, A., A. Aïdara-Kane, N. Raked, F. Denis, and M. C. Ploy. 2004. Integron-associated antibiotic resistance in the newly described Salmonella enterica serovar Keurmassar emerging in Senegal. Emerg. Infect. Dis. 10:1339-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Govinden, U., C. Mocktar, P. Moodley, A. W. Sturm, and S. Y. Essack. 2006. CTX-M-37 in Salmonella enterica serotype Isangi from Durban, South Africa. Int. J. Antimicrob. Agents 28:288-291. [DOI] [PubMed] [Google Scholar]
- 19.Hammami, A., G. Arlet, S. Ben Redjeb, F. Grimont, A. Ben Hassen, A. Rekik, and A. Philippon. 1991. Nosocomial outbreak of acute gastroenteritis in a neonatal intensive care unit in Tunisia caused by multiply drug resistant Salmonella wien producing SHV-2 beta-lactamase. Eur. J. Clin. Microbiol. Infect. Dis. 10:641-646. [DOI] [PubMed] [Google Scholar]
- 20.Harf-Monteil, C., A. Le Fleche, P. Riegel, G. Prevost, D. Bermond, P. A. D. Grimont, and H. Monteil. 2004. Aeromonas simiae sp. nov., isolated from monkey faeces. Int. J. Syst. Evol. Microbiol. 54:481-485. [DOI] [PubMed] [Google Scholar]
- 21.Hasman, H., D. Mevius, K. Veldman, I. Olesen, and F. M. Aarestrup. 2005. Beta-lactamases among extended-spectrum beta-lactamase (ESBL)-resistant Salmonella from poultry, poultry products and human patients in The Netherlands. J. Antimicrob. Chemother. 56:115-121. [DOI] [PubMed] [Google Scholar]
- 22.Hopkins, K. L., E. Liebana, L. Villa, M. Batchelor, E. J. Threlfall, and A. Carattoli. 2006. Replicon typing of plasmids carrying CTX-M or CMY beta-lactamases circulating among Salmonella and Escherichia coli isolates. Antimicrob. Agents Chemother. 50:3203-3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Humeniuk, C., G. Arlet, V. Gautier, P. Grimont, R. Labia, and A. Philippon. 2002. Beta-lactamases of Kluyvera ascorbata, probable progenitors of some plasmid-encoded CTX-M types. Antimicrob. Agents Chemother. 46:3045-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hunter, S. B., P. Vauterin, M. A. Lambert-Fair, M. S. Van Duyne, K. Kubota, L. Graves, D. Wrigley, T. Barrett, and E. Ribot. 2005. Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: converting the national databases to the new size standard. J. Clin. Microbiol. 43:1045-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jarlier, V., M. H. Nicolas, G. Fournier, and A. Philippon. 1988. Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 10:867-878. [DOI] [PubMed] [Google Scholar]
- 26.Kruger, T., D. Szabo, K. H. Keddy, K. Deeley, J. W. Marsh, A. M. Hujer, R. A. Bonomo, and D. L. Paterson. 2004. Infections with nontyphoidal Salmonella species producing TEM-63 or a novel TEM enzyme, TEM-131, in South Africa. Antimicrob. Agents Chemother. 48:4263-4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, S. L., and K. E. Sanderson. 1995. I-CeuI reveals conservation of the genome of independent strains of Salmonella typhimurium. J. Bacteriol. 177:3355-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morris, D., M. Whelan, G. Corbett-Feeney, M. Cormican, P. Hawkey, X. Li, and G. Doran. 2006. First report of extended-spectrum-beta-lactamase-producing Salmonella enterica isolates in Ireland. Antimicrob. Agents Chemother. 50:1608-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naas, T., M. Zerbib, D. Girlich, and P. Nordmann. 2003. Integration of a transposon Tn1-encoded inhibitor-resistant beta-lactamase gene, blaTEM-67 from Proteus mirabilis, into the Escherichia coli chromosome. Antimicrob. Agents Chemother. 47:19-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olson, A. B., M. Silverman, D. A. Boyd, A. McGeer, B. M. Willey, V. Pong-Porter, V. Daneman, and M. R. Mulvey. 2005. Identification of a progenitor of the CTX-M-9 group of extended-spectrum β-lactamases from Kluyvera georgiana isolated in Guyana. Antimicrob. Agents Chemother. 49:2112-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poirel, L., P. Kampfer, and P. Nordmann. 2002. Chromosome-encoded Ambler class A beta-lactamase of Kluyvera georgiana, a probable progenitor of a subgroup of CTX-M extended-spectrum beta-lactamases. Antimicrob. Agents Chemother. 46:4038-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez, M. M., P. Power, M. Radice, C. Vay, A. Famiglietti, M. Galleni, J. A. Ayala, and G. Gutkind. 2004. Chromosome-encoded CTX-M-3 from Kluyvera ascorbata: a possible origin of plasmid-borne CTX-M-1-derived cefotaximases. Antimicrob. Agents Chemother. 48:4895-4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takahashi, S., and Y. Nagano. 1984. Rapid procedure for isolation of plasmid DNA and application to epidemiological analysis. J. Clin. Microbiol. 20:608-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weill, F. X., F. Guesnier, V. Guibert, M. Timinouni, M. Demartin, L. Polomack, and P. A. D. Grimont. 2006. Multidrug resistance in Salmonella enterica serotype Typhimurium from humans in France (1993 to 2003). J. Clin. Microbiol. 44:700-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weill, F. X., M. Demartin, D. Tandé, E. Espié, I. Rakotoarivony, and P. A. D. Grimont. 2004. Extended-spectrum-β-lactamase (SHV-12 like)-producing strains of Salmonella enterica serotypes Babelsberg and Enteritidis isolated in France among infants adopted from Mali. J. Clin. Microbiol. 42:2432-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weill, F. X., J. D. Perrier-Gros-Claude, M. Demartin, S. Coignard, and P. A. Grimont. 2004. Characterization of extended-spectrum β-lactamase (CTX-M-15)-producing strains of Salmonella enterica isolated in France and Senegal. FEMS Microbiol. Lett. 238:353-358. [DOI] [PubMed] [Google Scholar]
- 37.Welch, T. J., W. F. Fricke, P. F. McDermott, D. G. White, M. L. Rosso, D. A. Rasko, M. K. Mammel, M. Eppinger, M. J. Rosovitz, D. Wagner, L. Rahalison, J. E. LeClerc, J. M. Hinshaw, L. E. Lindler, T. A. Cebula, E. Carniel, and J. Ravel. 2007. Multiple antimicrobial resistance in plague: an emerging public health risk. PLoS ONE 2:e309. [DOI] [PMC free article] [PubMed] [Google Scholar]