Abstract
Metronidazole resistance in Helicobacter pylori has been attributed to mutations in rdxA or frxA. Insufficient data correlating RdxA and/or FrxA with the resistant phenotype, and the emergence of resistant strains with no mutations in either rdxA or frxA, indicated that the molecular basis of H. pylori resistance to metronidazole required further characterization. The rdxA and frxA genes of four matched pairs of metronidazole-susceptible and -resistant strains were sequenced. The resistant strains had mutations in either rdxA, frxA, neither gene, or both genes. The reduction rates of five substrates suggested that metabolic differences between susceptible and resistant strains cannot be explained only by mutations in rdxA and/or frxA. A more global approach to understanding the resistance phenotype was taken by employing two-dimensional gel electrophoresis combined with tandem mass spectrometry analyses to identify proteins differentially expressed by the matched pair of strains with no mutations in rdxA or frxA. Proteins involved in the oxireduction of ferredoxin were downregulated in the resistant strain. Other redox enzymes, such as thioredoxin reductase, alkyl hydroperoxide reductase, and superoxide dismutase, showed a pI change in the resistant strain. The data suggested that metronidazole resistance involved more complex metabolic changes than specific gene mutations, and they provided evidence of a role for the intracellular redox potential in the development of resistance.
Metronidazole (Mtr) is an important component of therapeutic regimens that currently are used to treat many microbial infections. Metronidazole is considered a prodrug whose uptake and activation requires intracellular reduction, resulting in the production of cytotoxic short-lived radicals and other reactive species (29). 5-Nitroimidazole is activated via interactions with redox systems capable of reducing the low-potential (−415 mV) nitro group in position 5 of the imidazole ring (29). This property makes metronidazole effective against organisms in a low-intracellular-redox state, such as anaerobic bacteria and protozoa, as well as some microaerophiles, such as Campylobacter spp. and Helicobacter pylori (16).
Helicobacter pylori is found in the gastric mucous layer or adhering to the epithelial lining of the human stomach and is one of the most prevalent infections in humans (1, 19, 39). The frequent use of metronidazole has resulted in increased resistance to the antibiotic by H. pylori. The emergence of resistant isolates that do not respond to the drug fostered an interest in understanding the primary causes of resistance to metronidazole in this bacterium. Extensive investigations of H. pylori established that the main causes of metronidazole resistance are mutations in the gene rdxA or frxA (6, 7, 14, 15). However, insufficient data correlating the oxygen-insensitive nitroreductase RdxA and/or the NAD(P)H flavin oxidoreductase FrxA with the resistant phenotype and the fact that a small percentage of resistant strains do not have mutations in either rdxA or frxA indicated that the molecular basis of H. pylori resistance to Mtr has not been characterized completely.
Early studies showed that the oxygen tension has a large impact on the resistance of H. pylori to Mtr (23, 31-33), and several investigations have linked the activities of specific oxidoreductases to the Mtr-susceptible phenotype of the bacterium (9, 23, 36). Three disulfide reduction activities, which use dithiobis-2-nitrobenzoic acid (DTNB), oxidized glutathione (GSSG) and NADH, or ferredoxin (Fdx) and NADH as substrates, were identified in H. pylori (12). Metronidazole inhibited disulfide reduction competitively in each of the three activities, and the measured inhibition constants of Mtr for the reduction of different substrates indicated that the effects of Mtr were stronger in susceptible strains than in resistant ones (12). The presence of DTNB, GSSG, or Fdx inhibited Mtr reduction in situ, indicating that these substrates competed with Mtr as acceptors in redox reactions catalyzed by the corresponding disulfide reductases and suggesting that the enzymes participated in the reduction of Mtr (12). These results indicated a possible role in Mtr activation by enzymes catalyzing redox reactions that modulate the intracellular redox status, and that in metronidazole-susceptible strains, the cellular machinery regulating the redox status maintains an intracellular potential sufficiently low to activate metronidazole-reducing pathways (12).
In this study, four matched pairs of susceptible and resistant strains with different mutations in their rdxA and frxA genes were investigated. The redox state of the cells and the metabolic reduction of five substrates were measured. A more global approach was required to understand the resistant phenotype; thus, changes in the proteome of a matched pair of strains with no mutations in rdxA or frxA and changes induced in the proteome of the Mtr-resistant strain subjected to Mtr were investigated using two-dimensional gel electrophoresis and mass spectrometry (MS).
MATERIALS AND METHODS
Materials.
Blood agar base no. 2, brain heart infusion medium, defibrinated horse blood, and horse serum were from Oxoid (Heidelberg West, Victoria, Australia). Amphotericin B (Fungizone), bicinchoninic acid, bovine serum albumin, chloramphenicol, copper II sulfate, dithiothreitol (DTT), flavin adenine dinucleotide (FAD), spinach Fdx, GSSG, iodoacetamide (IA), metronidazole, mineral oil, ß-NAD (NADH), polymyxin B, trimethoprim, and sodium 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide (XTT) were from Sigma (Castle Hill, New South Wales, Australia). Vancomycin was from Eli Lilly (North Ryde, New South Wales, Australia). Deuterium oxide was from Cambridge Isotope Laboratories (Cambridge, England, United Kingdom). Tris base and sodium dodecyl sulfate (SDS) were from Amersham Biosciences (Melbourne, Victoria, Australia). All other reagents were of analytical grade.
Bacterial strains and growth conditions.
Helicobacter pylori strains 10593/2 and 10593a/2, from the University of New South Wales culture collection, and CAS 015 J0, CAS 015 J56, HER 126 V1, HER 126 V4, RIG 117 J0, and RIG 117 J56, from the Université Victor Segalen Bordeaux II collection, were grown at 37°C under an aerobic atmosphere of 5% CO2 in air in regular CO2 incubators or under a microaerobic atmosphere of 5% CO2, 5% O2, and 90% N2 in a Sony Tri-Carb incubator. The strains were matched pairs isolated pretreatment (susceptible) and posttreatment (resistant) from patients with gastritis whose treatment with metronidazole failed (18). Their susceptibilities to metronidazole were confirmed by Etest (AB Biodisk, Solna, Sweden). H. pylori identification was made by positive urease and catalase tests, and motility and morphology were observed under phase-contrast microscopy. The Mtr phenotype and the status of the rdxA and frxA genes of all of these strains are presented in Table 1.
TABLE 1.
Status of rdxA and frxA genes in four H. pylori genetic backgrounds with susceptible and resistant matched pairsa
| H. pylori strain | Mtr MIC (mg liter−1) | Mtr-resistant counterpart | Mtr MIC (mg liter−1) | Mutation in rdxA | Mutation in frxA |
|---|---|---|---|---|---|
| 10593/2 | 2 | 10593a/2 | 64 | C177stop | None |
| RIG 117 J0 | 4 | RIG 117 J56 | 32 | R16C | A40stop |
| CAS 015 J0 | 4 | CAS 015 J56 | 32 | L132stop | L39stop |
| HER 126 V1 | 2 | HER 126 V4 | 64 | None | None |
Mtr MICs for the strains were determined using Etests. The rdxA and frxA genes of each of the strains were sequenced. Mutations in the Mtr-resistant variant are given with respect to its susceptible counterpart.
Preparation of lysate fractions or cell-free protein extracts for enzyme assays.
Cells were harvested in log phase (ca. 18 h) in sterile NaCl (150 mM) and checked for purity by phase-contrast microscopy. Bacteria were washed three times by centrifugation at 14,000 × g (4 min at 4°C) and resuspended in NaCl (150 mM). The supernatants were discarded, and the pellets were resuspended in saline. Following the final wash, packed cells were resuspended in saline (109 to 1010 cells/ml) and lysed by two freeze-thaw cycles in liquid nitrogen; phase-contrast microscopy was employed to verify that greater than 99% cell lysis had occurred. To separate supernatant and particulate fractions, bacterial suspensions were centrifuged at 16,000 × g (4°C for 15 min).
Preparation of cell-free protein extracts for two-dimensional electrophoresis.
Chloramphenicol was added to bacterial suspensions to a final concentration of 128 μg/ml after 18 h of incubation. Cultures were centrifuged at 2,879 × g for 25 min at 4°C, and the pellet was washed three times with 0.2 M ice-cold sucrose. After the final wash, the cell pellet was disrupted by thrice freeze-thawing and was resuspended in 1 ml TSU buffer (50 mM Tris pH 8.0, 0.1% SDS, 2.5 M urea). Cell debris were removed by centrifugation at 14,000 × g for 20 min at 4°C.
Enzyme assays.
Proton nuclear magnetic resonance and nitrogen-14 nuclear magnetic resonance free induction decays were collected using a Bruker DMX-600 or a Bruker DMX-500 spectrometer, respectively, operating in the pulsed Fourier transform mode with quadrature detection as previously described (12). Disulfide reduction activities were measured in H. pylori cell extracts with GSSG and NADH as substrates. Metronidazole reduction activities were measured in H. pylori lysates with Mtr and NADH as substrates.
The kinetics of NADH:Fdx oxidoreductase or NADH:FAD reduction for several strains were determined in cell extracts by spectrophotometrically measuring, at 340 nm and 25°C, the rates of decrease of NADH levels in the presence of Fdx or FAD, respectively. The assay mixture consisted of Tris-HCl (20 mM, pH 7.4), 0.15 mM NADH, and Fdx at concentrations between 2.2 and 130 μg ml−1 or FAD at concentrations between 0.5 and 100 μM. The method was validated by establishing that no oxidation of NADH took place in the absence of cell extracts. NADH oxidation was observed in suspensions of extracts in the absence of Fdx or FAD. Thus, for each sample, reductase rates were calculated by subtracting the rate of NADH oxidation in the absence of substrate from the value measured in the presence of substrate.
The kinetics of nitrofurazone reduction for several strains were determined in cell extracts by measuring spectrophotometrically, at 400 nm and 25°C, the rates of the decrease of nitrofurazone levels in the presence of NADH. The assay mixture consisted of Tris-HCl (20 mM, pH 7.4), 0.15 mM NADH, and nitrofurazone at concentrations between 0.5 and 100 μM. The method was validated by establishing that no oxidation of nitrofurazone took place in the absence of cell extracts.
Michaelis constants (Km) and maximal velocities (Vmax) were calculated by nonlinear regression using the Enzyme Kinetics program (Trinity Software, Compton, NH). The errors in these calculations are determined as ±standard deviations.
Redox assays.
Tetrazolium salts are used widely for detecting the redox potential of cells in viability, proliferation, and cytotoxicity assays. The bacterial redox potential was determined using the method of Bensaid et al. (3). XTT reduction was measured at five different bacterial densities for each of the eight susceptible and resistant strains grown microaerobically or aerobically. XTT reduction was plotted as a function of bacterial density. The method was validated by measuring the change in the absorbance of XTT buffer solutions with no bacteria and including these results in the fits of the data to a straight line.
Two-dimensional gel electrophoresis and image analysis.
Two hundred micrograms of protein was suspended in 490 μl using a rehydration buffer consisting of 8 M urea, 100 mM DTT, 65 mM 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 40 mM Tris-HCl (pH 8.0), 10 μl IPG buffer (pH 4 to 7) (Amersham Biosciences, Melbourne, Victoria, Australia). Nuclease buffer (10 μl) was added, and the mixture was incubated at 4°C for 20 min. The sample was centrifuged at 14,000 × g and 4°C for 20 min, and the supernatant was loaded onto an 18-cm Immobiline DryStrip (pH 4 to 7) (Amersham Biosciences), which was sealed at room temperature and left to incubate for 20 h. Isoelectric focusing was performed using a flatbed Multiphor II unit (Amersham Biosciences) programmed for 2 h at 100 V; followed by 0.5 h at 500, 1,500, and 2,500 V; and a final 18-h step of 3,500 V. Focused Imobiline DryStrips were equilibrated sequentially in two buffers of 6 M urea, 20% (wt/wt) glycerol, 2% (wt/vol) SDS, 375 mM Tris-HCl; the first buffer also contained 130 mM DTT, and the second contained 135 mM IA (Sigma-Aldrich). SDS-polyacrylamide gel electrophoresis was performed on 11.5% acrylamide gels using the Protean II system (Bio-Rad, Sydney, New South Wales, Australia) at 50 V for 1 h, followed by 64 mA until the dye reached the bottom of the gel. Gels were fixed individually in 0.2 liters of fixing solution (50% [vol/vol] methanol, 10% [vol/vol] acetic acid) for a minimum of 1 h and subsequently were stained using a sensitive ammoniacal silver method. For comparative image analysis, statistical data were acquired and analyzed using Z3 software (Compugen, Sunnyvale, CA).
MS identification of proteins.
Excised gel slices were washed twice with 0.2 ml of 100 mM NH4HCO3 for 10 min, reduced with 50 μl of 10 mM DTT at 37°C for 1 h, alkylated in 50 μl of 10 mM IA at 37°C for 1 h, washed three times for 10 min with 0.2 ml milli-Q water, washed with 0.2 ml of 10 mM NH4HCO3 for 10 min, dehydrated in acetonitrile, and rehydrated in a buffer containing 10.5 ng μl−1 trypsin. After digestion for 14 h, peptides were extracted by washing the gel slice with 25 μl 1% formic acid for 15 min, followed by dehydration with acetonitrile. Digests were separated by nano-liquid chromatography using an Ultimate/Famos/Switchos system. Samples (5 μl) were loaded on to a Micron C18 precolumn (500 μm by 2 mm) with buffer A (H2O:CH3CN [98:2, vol/vol] and 0.1% formic acid) at 25 μl min−1. After a 4-min wash, the flow was switched into line with a PEPMAP C18 RP analytical column (75 μm by 15 cm) and eluted using buffer A (H2O:CH3CN [40:60, vol/vol] and 0.1% formic acid) at 200 nl min−1 for 30 min. The nano-electrospray needle was positioned ≈1 cm from the orifice of an API QStar Pulsar I tandem MS instrument. The QStar was operated in information-dependent acquisition mode. A time-of-flight MS survey scan was acquired (m/z 350 to 1,700, 0.5 s), and the two largest precursors (counts of >10) were selected sequentially by Q1 for tandem MS analysis (m/z 50 to 2,000, 2.5 s). A processing script generated data suitable for submission to the database search programs. Collision-induced dissociation spectra were analyzed using the Mascot MS/MS ion search tool (Matrix Science; http://www.matrixscience.com/) with the following parameters: trypsin digestion allowing up to one missed cleavage, oxidation of methionine, peptide tolerance of 1.0 Da, and MS/MS tolerance of 0.8 Da. Protein searches were performed on the NCBI nr database.
Bioinformatics.
BLASTp searches were performed using the complete protein sequences available at the NCBI database (http://www.ncbi.nlm.nih.gov/). The Kyoto Encyclopedia of Genes and Genomes (KEGG), available at www.genome.jp/kegg, was employed to determine the biochemical pathways to which the proteins were assigned. The MicrobesOnline website, available at www.microbesonline.org, was used to determine the predicted operons and regulons to which genes and proteins belong. The Search Tool for the Retrieval of Interacting Proteins (STRING) is a database of known and predicted protein-protein interactions (http://string.embl.de/). STRING was employed to examine interactions between differentially expressed proteins.
Protein determination.
The estimation of the protein content of samples was made by the bicinchoninic acid method employing a microtiter protocol (Pierce, Rockford, IL). Absorbances were measured using a Beckman Du 7500 spectrophotometer.
RESULTS AND DISCUSSION
Helicobacter pylori matched pairs of susceptible and resistant strains.
Several factors besides RdxA and FrxA affect H. pylori resistance to Mtr, but none have been considered causes of resistance. An example is the oxygen content of the growth atmosphere, which made resistant bacteria grown under oxygen-depleted conditions become susceptible to Mtr (31, 32). The majority of studies of Mtr resistance have focused on two genes involved in the resistance phenotype of the bacterium, rdxA and frxA. The emergence of resistant strains that have no mutations in either gene implies that other factors not only affect Mtr resistance but also caused the resistance phenotype.
The rdxA and frxA genes of matched pairs of strains isolated from patients after a failed treatment with Mtr were sequenced. H. pylori matched pairs were chosen to eliminate any bias arising from differences in the genetic backgrounds of the strains studied. The Mtr susceptibilities of the matched pairs were tested using Etests repeated in triplicate. The resistance threshold of 8 μg ml−1 was employed to classify strains as susceptible or resistant by following standard clinical practice (22). The Mtr MICs for susceptible strains were less than 0.38 μg ml−1 and for resistant strains ranged between 128 and 256 μg ml−1.
Four matched pairs were chosen, because they contained the four different possible combinations of changes in the two genes of interest. In the 10593/2 matched pair, the 10593a/2 drug-resistant counterpart contains a mutation that deactivated the rdxA gene; in the RIG 117 matched pair, the resistant variant contains a mutation that deactivated the frxA gene; in the CAS 015 matched pair, the resistant variant contains a mutation that deactivated both genes; and in the HER 126 matched pair, the resistant variant contains no mutations in either gene (Table 1). Thus, the matched pairs include four susceptible parent strains and four resistant mutants.
Metronidazole reduction in matched pairs of strains.
Metronidazole reduction was measured in lysate suspensions of each of the eight strains grown microaerobically and aerobically (Table 2). Mtr reduction also was measured in the same lysates that had been subjected to argon treatment, which was employed to displace oxygen from the samples (Table 2). The measurements under aerobic conditions served to establish the effect of oxygen in the growth atmosphere on the enzymes involved in the Mtr-resistant phenotype. The measurements of the samples subjected to argon treatment served to determine if oxygen in the samples modulated the activities of the enzymes involved in the reduction of the nitro group of Mtr.
TABLE 2.
Mtr reduction activities of H. pylori lysates from matched pairs of susceptible and resistant variants prepared from cells grown under microaerobic and aerobic conditionsa
| H. pylori strain | Mtr reduction velocities (nmol min−1 mg−1)
|
|||
|---|---|---|---|---|
| Microaerobic
|
Aerobic
|
|||
| Oxygenated | Argonized | Oxygenated | Argonized | |
| 10593/2 | 14 ± 2 | 32 ± 4 | 16 ± 2 | 36 ± 4 |
| 10593a/2 | 9 ± 1 | 13 ± 2 | 10 ± 1 | 15 ± 2 |
| RIG 117 J0 | 19 ± 2 | 32 ± 3 | 27 ± 3 | 33 ± 4 |
| RIG 117 J56 | 13 ± 1 | 16 ± 2 | 19 ± 2 | 20 ± 2 |
| CAS 015 J0 | 15 ± 2 | 29 ± 3 | 18 ± 2 | 31 ± 3 |
| CAS 015 J56 | 9 ± 1 | 14 ± 1 | 13 ± 1 | 15 ± 1 |
| HER 126 V1 | 18 ± 2 | 20 ± 2 | 22 ± 2 | 25 ± 3 |
| HER 126 V4 | 10 ± 1 | 11 ± 1 | 14 ± 1 | 18 ± 2 |
Lysates were suspended in phosphate buffer and, where stated, subjected to argon treatment for 30 min. Initial substrate concentrations were 12 mM Mtr and 30 mM NADH. Errors were calculated from the straight-line fitting of the values used to determine the Mtr reduction rates.
The results indicated that all resistant strains reduced Mtr less than their susceptible parent strains, whether grown microaerobically or aerobically (Table 2). The general hypothesis is that H. pylori strains become resistant when the activation of the drug, i.e., reduction, is inhibited. The finding that resistant strains reduce Mtr less than their susceptible counterparts supported this observation.
The effect of the oxygen tension in the growth atmosphere on reduction rates was variable. Mtr reduction increased in some H. pylori strains grown aerobically, but in other cases the increase was not significant (Table 2). In the 10593/2 matched pair, no significant difference was observed between cells grown microaerobically or aerobically, while a difference was measured for the RIG 117 matched pair. Only the resistant strains of the CAS 015 and HER 126 matched pairs showed significantly different rates when grown aerobically. These findings indicated that H. pylori cells grown aerobically become slightly more susceptible to Mtr than cells grown microaerobically. A study of Trichomonas vaginalis found that susceptibilities of strains also were modulated by the oxygen content of the growth atmosphere (24). Interestingly, two of the T. vaginalis strains showed characteristics similar to those of some H. pylori strains, where their resistance followed a bell-shaped curve when plotted against the oxygen content of the growth atmosphere. Other T. vaginalis strains showed no change between microaerobically and aerobically grown cells, similarly to the other H. pylori strains.
Metronidazole reduction increased dramatically in most samples subjected to argon treatment (Table 2); hence, the availability of oxygen in the sample correlated inversely with drug reduction. Subjecting a sample to argon treatment created a more reducing environment and would explain the higher reduction rates of Mtr. These findings supported a role for oxygen and the redox potential in metronidazole activation. In addition, they provided further evidence that resistant strains grown anaerobically become susceptible. Interestingly, subjecting the sample to argon treatment had little or no effect on Mtr reduction in the HER 126 matched pair of strains. This may reflect differences in the intracellular redox status of the HER 126 strains relative to those of the other strains.
The ratios between the rates of the reduction of susceptible and resistant strains grown microaerobically or aerobically and not subjected to argon were approximately 1.5. This result indicated that irrespective of the oxygen tension of the growth atmosphere, susceptible strains not subjected to argon treatment reduced Mtr about 1.5 times more than their resistant counterparts for all matched pairs. Also, the data indicated that Mtr reduction increased under aerobic conditions in both susceptible and resistant strains, reflecting an increase in the activity of Mtr-reducing enzyme(s) under aerobic conditions. Note that these enzymes are not directly involved in the formation of the Mtr-resistant phenotype.
The ratios of the rates of reduction between susceptible and resistant strains grown microaerobically or aerobically and subjected to argon treatment were higher than those for untreated strains, except for the HER 126 matched pair, for which the ratio did not change. Excluding these strains, the results demonstrated that argon treatment affected the activities of susceptible strains more than those of the resistant strains. This revealed a direct association between the displacement of oxygen from the samples and the activities of enzymes involved in the resistant phenotype. In addition, the findings provided evidence of differences between the HER 126 Mtr-resistant phenotype and those of the other matched pairs. An explanation is that the enzymes of the HER 126 Mtr-susceptible strain, which reduce Mtr, have unique characteristics, and the modulation of their activity was sufficient to develop resistance to Mtr. Thus, the resistant phenotype does not need the inactivation of the rdxA or frxA gene. Moreover, it is possible that in the other matched pairs, the combined effects of the regulation of these enzyme activities and mutations of rdxA and/or frxA were the cause of resistance.
Redox status of matched pairs of strains.
Previous work implicated the intracellular redox status of the cells in H. pylori Mtr resistance (12). Tetrazolium salts were used for determining the redox potential of cells; thus, the redox status of the bacterial cells was assessed using XTT.
XTT reduction was measured for each strain at several bacterial densities. Graphs of XTT reduction versus bacterial density were plotted for cells grown microaerobically or aerobically. The experimental points fitted linear equations with squared regression coefficient values equal to or greater than 0.95. The slopes of the lines for each strain and condition represented the rate of XTT reduction with respect to bacterial density and are given in Table 3. The slope values increased from susceptible to resistant strains for each matched pair, although for the HER matched pair of strains grown microaerobically it did not increase significantly. The slope values also increased from cells grown microaerobically to aerobically, except for that of the CAS 015 Mtr-susceptible strain, which remained the same.
TABLE 3.
XTT reduction as a function of bacterial density for each of the four matched pairs of susceptible and resistant variants grown microaerobically and aerobicallya
| H. pylori strain | XTT reduction as a function of bacterial density (R2)
|
|
|---|---|---|
| Microaerobic | Aerobic | |
| 10593/2 | 0.08 (0.998) | 0.11 (0.997) |
| 10593a/2 | 0.55 (0.997) | 0.64 (0.962) |
| RIG 117 J0 | 0.18 (0.954) | 0.21 (0.997) |
| RIG 117 J56 | 0.24 (0.996) | 0.69 (0.999) |
| CAS 015 J0 | 0.18 (0.996) | 0.18 (0.998) |
| CAS 015 J56 | 0.35 (0.985) | 0.55 (0.950) |
| HER 126 V1 | 0.09 (0.980) | 0.13 (0.986) |
| HER 126 V4 | 0.09 (0.990) | 0.18 (0.989) |
XTT reduction was measured at five different bacterial densities as described by Bensaid et al. (3). The method was validated by measuring the change in absorbance with no bacteria.
XTT reduction is linked to the electron transport chain of the bacterium. An increase in XTT reduction reflects activated aerobic respiration. This in turn is a sign of an elevated intracellular redox potential (as in aerobes), which results in the decrease in activation of Mtr (−415 mV).
The experiments identified variations in the reduction of a redox potential indicator between Mtr-susceptible and -resistant strains of different genetic backgrounds.
Metabolic changes in the matched pairs of strains.
Several enzyme activities were measured to determine if there are differences in the metabolism of the matched pairs. The four substrates chosen were GSSG, FAD, Fdx, and nitrofurazone. Glutathione is a redox metabolite and was shown to inhibit Mtr reduction (12); FAD is a substrate of FrxA; Fdx is a low-potential protein and was shown to inhibit Mtr reduction (12); and nitrofurazone is a high-redox-potential nitrogen-based drug and a substrate of RdxA.
The Km value for GSSG reduction did not change between susceptible and resistant strains for the RIG 117 and CAS 015 matched pairs (Table 4). A slight increase in the Km of the resistant strain was observed for the 10593/2 matched pair (Table 4), and the resistant strain in the HER 126 matched pair had a significantly higher Km than its susceptible counterpart. An increase in Km meant that the enzymes reducing GSSG had less affinity for the substrate, suggesting a difference in the modulation of the redox environment of the resistant strain.
TABLE 4.
Michaelis constants for GSSG, FAD, Fdx, and nitrofurazone reduction activities in H. pylori Mtr-susceptible and -resistant matched pairs of strainsa
| H. pylori strain | Michaelis constants for reduction activities
|
|||
|---|---|---|---|---|
| GSSG (mM) | FAD (μM) | Fdx (μg ml−1) | Nitrofurazone (μM) | |
| 10593/2 | 2.3 ± 0.2 | 2.1 ± 0.3 | 3.0 ± 0.4 | 2.8 ± 0.3 |
| 10593a/2 | 2.8 ± 0.2 | 95 ± 10 | No activity | 8.1 ± 1.4 |
| RIG 117 J0 | 4.9 ± 0.4 | 2.4 ± 0.3 | 44 ± 5 | 8.8 ± 1.8 |
| RIG 117 J56 | 4.5 ± 0.5 | 6 ± 1 | 24 ± 4 | 13.1 ± 2.1 |
| CAS 015 J0 | 5.0 ± 0.5 | 5 ± 1 | 23 ± 4 | 6.0 ± 0.9 |
| CAS 015 J56 | 4.7 ± 0.5 | 11 ± 2 | 40 ± 5 | 8.5 ± 2.1 |
| HER 126 V1 | 3.0 ± 0.2 | 24 ± 2 | 44 ± 7 | 3.3 ± 0.6 |
| HER 126 V4 | 5.2 ± 0.6 | 17 ± 2 | 11 ± 2 | 6.8 ± 1.1 |
GSSG concentrations ranged from 0.5 to 70 mM, FAD and nitrofurazone concentrations ranged from 0.5 μM to 100 μM, and Fdx concentrations ranged from 0 to 80 μg ml−1. NADH concentrations were 0.15 mM for FAD, Fdx, and nitrofurazone assays and 50 mM for GSSG assays. Kinetic fits were performed using 10 to 13 rates. Errors were calculated using the Enzyme Kinetics program from nonlinear regression fits to the data.
The Km value for FAD reduction was higher in the resistant strains than in the susceptible strains, except for the HER 126 matched pair (Table 4). The change in Km did not correlate with the inactivation of FrxA, suggesting that several other enzymes were involved in the reduction of FAD. The difference in Km between susceptible and resistant strains suggested that at least some of those other enzymes also participated in Mtr reduction. For example, in the 10593/2 matched pair, which contains an active FrxA, the Km value was approximately 50 times higher in the resistant strain than in the susceptible strain.
The Km values of Fdx reduction did not follow any pattern. In the 10593/2 matched pair, no activity was detected in the resistant strain (Table 4). In the RIG 117 and HER 126 matched pairs, the Km values decreased in the resistant strain, and in the CAS 015 pair the value increased in the resistant strain (Table 4). These results suggest that H. pylori has a system of enzymes involved in Fdx reduction, some of which also participated in Mtr reduction.
The Km value for nitrofurazone reduction was higher in the resistant strains; however, the increase in the CAS 015 matched pair was not significant (Table 4). Nitrofurazone is a medium-redox-potential (−257 mV) nitrogen-based compound with a nitro group similar to that of Mtr. Thus, the enzymes capable of reducing Mtr would be capable of reducing nitrofurazone. The higher Km values in the resistant strains meant that the enzymes had less affinity for the substrate, which would result in a lower level of drug reduction. The data support our previous findings in which resistant strains decreased Mtr activation.
These data demonstrated that the reduction of substrates that compete with Mtr involved a more complex metabolism than the activities of RdxA and FrxA. Hence, a more global approach was undertaken to investigate the resistant phenotype in H. pylori.
Proteomic analyses of Helicobacter pylori strains HER 126 V1 and HER 126 V4.
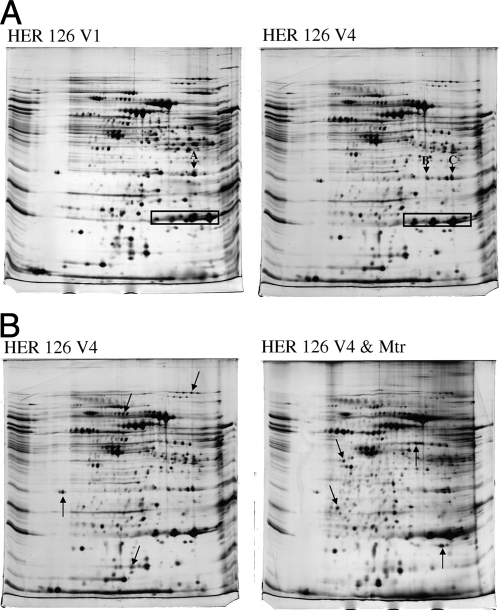
Two-dimensional gel electrophoresis and tandem MS were employed to identify the differences in protein expression between H. pylori strains HER 126 V1 and HER 126 V4 (Fig. 1A and Table 5). Eight proteins were downregulated and 10 were upregulated in the resistant strain relative to the same proteins in the susceptible one, and three proteins showed a change in pI (Table 5).
FIG. 1.
Two-dimensional proteomes (pI 4 to 7) of (A) Helicobacter pylori HER 126 V1 (left) and HER 126 V4 (right) and (B) Helicobacter pylori HER 126 V4 cells grown without Mtr (left) or in the presence of 8 μg ml−1 Mtr (right). Proteins differentially expressed between the two growth conditions are listed on Table 5 (A) and Table 6 (B). The protein spots labeled in both gels represent (A) thioredoxin reductase and fructose-1,6-bisaldolase; (B) thioredoxin reductase; and (C) fructose-1,6-bisaldolase. Examples of regulated proteins in panel B are shown with arrows.
TABLE 5.
Differences in the proteomes of the H. pylori strains HER 126 V1 and its Mtr-resistant variant HER 126 V4 mapped using two-dimensional gel electrophoresisa
| Gene open reading frame and regulation status | Protein name |
|---|---|
| Downregulated | |
| hp0500 | DNA polymerase III subunit beta |
| hp0512 | Glutamine synthetase |
| hp0591 | Ferredoxin oxidoreductase γ subunit |
| hp0691 | 3-Oxoadipate coenzyme A transferase subunit A |
| hp0695 | Hydantoin utilization protein A |
| hp0697 | Hypothetical protein |
| hp1110 | Pyruvate ferredoxin oxidoreductase |
| hp1293 | DNA-directed RNA polymerase, alpha subunit |
| Upregulated | |
| hp0002 | Riboflavin synthase subunit beta |
| hp0049 | Hypothetical protein |
| hp0105 | S-ribosyl homocysteinase |
| hp0107 | Cysteine synthase |
| hp0115 | Flagellin B |
| hp0601 | Flagellin A |
| hp0783 | Hypothetical protein |
| hp0870 | Flagellar hook protein |
| hp1046 | Hypothetical protein |
| hp1134 | ATP synthase subunit A |
| pI change | |
| hp0389 | Superoxide dismutase |
| hp0825 | Thioredoxin reductase |
| hp1563 | Alkyl hydroperoxide reductase |
Protein spots with changes in their intensity (≤0.5-fold or ≥2-fold) or in their position along the horizontal axes (pI) were identified by MS analyses.
Glutamine synthetase, which synthesizes glutamine from glutamate and ammonia, was downregulated in the HER 126 V4 Mtr-resistant variant. The enzyme's downregulation would lead to increased levels of glutamate and decreased levels of glutamine in the cell. Glutamine is a precursor of ornithine, which can be converted to citrulline in a reaction that requires ATP (17); lower concentrations of glutamine could lead to a decline in ornithine levels. Bacterial cells may compensate for an ornithine deficit by converting arginine to ornithine through citrulline (27). The upregulation of HP0049, a hypothetical protein (predicted to be peptidyl arginine deaminase) responsible for the conversion of arginine to citrulline, suggested that this was the mechanism employed by H. pylori to restore ornithine levels. HP0691, a subunit of 3-oxoadipate-coenzyme A transferase and a component of benzoate hydroxylation, was downregulated in the HER 126 V4 variant. Benzoate hydroxylation also has been used to determine hydroxyl radical production in cells (5, 8). Thus, the downregulation of this transferase in H. pylori may be related to an increase in oxidative stress. The beta subunit of riboflavin synthase, encoded by hp0002, was upregulated in the Mtr-resistant variant. This enzyme is involved in the generation of riboflavin, a central component of FAD and flavin mononucleotide. FAD is a substrate of FrxA, and increased levels of the dinucleotide would compete with Mtr as a substrate of this enzyme, resulting in the inhibition of Mtr reduction.
Cysteine synthase, which is capable of generating cysteine (40), was upregulated in the resistant HER 126 V4 variant. An increased requirement for cysteine arising from its role as a reductant in redox metabolism may explain the upregulation of the synthase. S-ribosyl homocysteinase (LuxS) is found in the same predicted operon as cysteine synthase and was upregulated too. LuxS has a critical role in the global regulation of flagellar gene transcription in H. pylori (28). This finding, together with the upregulation of the flagellin proteins encoded by hp0115 and hp0601 and of the flagellar hook protein, suggested an increased motility in the resistant phenotype. The hydantoin-metabolizing proteins encoded by hp0695 and hp0697 were downregulated in the resistant variant. Hydantoin has a chemical structure similar to that of imidazolidine, the hydrogen-saturated analogue of imidazole (38). These proteins may be able to metabolize compounds with structures similar to that of imidazole, such as Mtr. Thus, the resistant variant could downregulate these hydantoin-utilizing proteins to decrease the activation of the drug.
Fdx oxidoreductase γ subunit and pyruvate Fdx oxidoreductase were downregulated in the resistant variant. Both proteins can reduce the low-potential Fdx. Reduced Fdx is capable of activating Mtr (30); hence, the downregulation of enzymes capable of converting Fdx to its reduced form would result in a decrease in the concentration of reduced Fdx in the system, leading to a decrease in the reduction of Mtr. Moreover, if both enzymes were able to reduce Mtr directly, a decrease in their levels also would result in a decrease in the concentration of activated Mtr.
Thioredoxin reductase, superoxide dismutase, and alkyl hydroperoxide reductase were identified in different positions on both gels (Fig. 1A). The redox potential of a protein is associated with its pI (26, 37). Thus, the change of the pI of three redox proteins could reflect a modification to suit the intracellular milieu of the Mtr-resistant strain. The changes in XTT reduction rates between susceptible and resistant strains measured in three matched pairs but absent in the HER 126 strains could be a result of alterations in the pI of individual proteins in the resistant HER 126 V4 strain and not necessarily an absence of change in the overall intracellular redox status of the cell.
Analyses of the proteome of H. pylori strain HER 126 V4 grown with Mtr.
Eleven proteins were identified as downregulated and eight as upregulated in the resistant variant exposed to 8 μg ml−1 Mtr (Fig. 1B and Table 6). This concentration is taken as the threshold of Mtr resistance in H. pylori (7, 22), and it produced a significant change in the proteome without subjecting the cells to excessive stress.
TABLE 6.
H. pylori HER 126 V4 proteins (pI 4 to 7) whose expression is modulated in the presence of 8 μg ml−1 Mtra
| Gene open reading frame and regulation status | Protein name |
|---|---|
| Downregulated | |
| hp0106 | Cystathionine gamma-synthase |
| hp0152 | Hypothetical protein |
| hp0171 | Peptide chain release factor 2 |
| hp0197 | S-adenosylmethionine synthetase |
| hp0220 | Cysteine desulfurase |
| hp0264 | Heat shock protein (ClpB) |
| hp0589 | Fdx oxidoreductase α subunit |
| hp0795 | Trigger factor |
| hp0900 | Hydrogenase expression/formation protein |
| hp1164 | NADPH reductase |
| hp1495 | Transaldolase |
| Upregulated | |
| hp0072 | Urease beta subunit |
| hp0115 | Flagellin B |
| hp0570 | Leucyl aminopeptidase |
| hp0601 | Flagellin A |
| hp0653 | Ferritin |
| hp0837 | Conserved hypothetical protein |
| hp1134 | ATP synthase F1, subunit alpha |
| hp1563 | Alkyl hydroperoxide reductase AhpC |
Proteins with changes in their intensity (≤0.5-fold or ≥2-fold) were identified by MS analyses.
Cystathionine gamma-synthase (MetS) was downregulated under Mtr stress. MetS converts O-phosphohomoserine and cysteine to cystathionine in the first reaction of a pathway that converts cysteine to methionine (13). The downregulation of this enzyme will contribute to maintain the levels of cysteine, which serves to combat the oxidative stress caused by Mtr. A decrease in methionine concentrations owing to the downregulation of MetS could lead to the observed downregulation of S-adenosylmethionine synthetase. Cysteine desulfurase is involved in the conversion of l-cysteine into l-alanine (25). The downregulation of this enzyme also could be intended to conserve cysteine, which can be used as an antioxidant. The upregulation of alkyl hydroperoxide reductase supported the hypothesis of a general cell response to counteract oxidative and/or nitrosive stress, since this enzyme is important for defending bacteria against both stresses (2, 20). Exposure to Mtr also induced an upregulation of ferritin, which is involved in iron storage and performs a function in controlling the production of hydroxyl radicals arising from Fenton reactions. Leucyl aminopeptidase, which has a role in glutathione turnover in the cell owing to its ability to hydrolyze Cys-Gly (4), was upregulated under Mtr stress. The increased requirement of cysteine, the possible increase in disulfide turnover, and the upregulation of proteins involved in oxireduction suggested that an important component of the response of H. pylori was to defend itself against the oxidative and nitrosive stresses that arose from Mtr activation.
Flagellin A and B and the flagellin hook protein were upregulated in the resistant strain but not in the susceptible one (Table 5). Exposure to Mtr upregulated the expression of both flagellins in the resistant strain (Table 6). These proteins appear to be induced by stress to help the bacterium escape the harmful effects of the drug. Protein HP0837, annotated as a hypothetical protein, was upregulated in the presence of Mtr. The gene encoding this protein is predicted to be in the same operon as hp0840, which encodes FlaA1, and in the same regulon as several other genes encoding flagellum-associated proteins, such as flagellar hook assembly proteins FliQ and FliI. This supported the view that Mtr stress induced a response in the flagellar system of H. pylori. The hydrogenase expression/formation protein was downregulated under Mtr stress; hydrogenases have been implicated in the activation of Mtr in other organisms (10) and may have a similar role in H. pylori.
The expression of the NADPH reductase FqrB and the α subunit of Fdx oxidoreductase, which is involved in the Fdx reduction cycle, was found to be downregulated in the presence of Mtr. It has been hypothesized that FqrB functions in providing electrons for pyruvate ferredoxin oxidoreductase (PFOR) activity (34), and PFOR directly reduces Fdx in the presence of pyruvate (11), supplying reduced Fdx for Mtr activation. Mtr inhibits Fdx-associated processes (35), and these resistance mechanisms in various organisms are dependent on the availability of reduced Fdx. The downregulation of the two proteins involved in Fdx reduction suggested a decreased capacity to reduce Fdx and a decrease in the activation of Mtr. The more than twofold downregulation of PFOR in a rdxA knockout mutant upon the addition of Mtr (20) indicated that the phenotype is associated not only with the HER 126 V4 strain but also with other resistant strains with mutations in these genes.
Finally, the results of enzymatic investigations showed that the addition of Fdx inhibited the reduction of Mtr in the H. pylori 10593/2 matched pair of susceptible and resistant variants (12) as well as in strains RIG 117 J0, J99, RSB6, and 26695 (data not shown). Interestingly, after the addition of Fdx, Mtr reduction levels in susceptible strains were equal to levels found in the resistant counterpart (12). The enzymatic and proteomic data provided evidence for a novel Mtr resistance mechanism in H. pylori and revealed that Fdx-reducing enzymes could be potential therapeutic targets for both Mtr-susceptible and -resistant strains, since they are essential to H. pylori (11).
This study provided a conclusive association between Mtr reduction and resistance in H. pylori. It showed that the resistance phenotype is more complex than the inactivation of the rdxA and frxA genes and supported a role for oxygen and the intracellular redox status in the resistant phenotype. Evidence for this conclusion was found in the observation of changes in the pI of proteins involved in oxireduction reactions. Novel aspects of the resistance mechanisms of H. pylori to Mtr, namely, the downregulation of hydantoin-utilizing proteins and Fdx reduction, were identified as well. Finally, given the established relationships between the pathogenicity of H. pylori and flagellar expression (21), the upregulation of flagellum-associated proteins and urease in a resistant variant opened the possibility that Mtr-resistant strains are more pathogenic than their susceptible counterparts, but this hypothesis requires further studies.
Acknowledgments
This work was made possible by the support of the Australian Research Council.
Footnotes
Published ahead of print on 17 February 2009.
REFERENCES
- 1.Alarcón, T., D. Domingo, and M. Lopez-Brea. 1999. Antibiotic resistance problems with Helicobacter pylori. Int. J. Antimicrob. Agents 12:19-26. [DOI] [PubMed] [Google Scholar]
- 2.Baker, L. M., A. Raudonikiene, P. S. Hoffman, and L. B. Poole. 2001. Essential thioredoxin-dependent peroxiredoxin system from Helicobacter pylori: genetic and kinetic characterization. J. Bacteriol. 183:1961-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bensaid, A., J. Thierie, and M. Penninckx. 2000. The use of the tetrazolium salt XTT for the estimation of biological activity of activated sludge cultivated under steady-state and transient regimes. J. Microbiol. Methods 40:255-263. [DOI] [PubMed] [Google Scholar]
- 4.Cappiello, M., A. Lazzarotti, F. Buono, A. Scaloni, C. D'Ambrosio, P. Amodeo, B. L. Mendez, P. Pelosi, A. Del Corso, and U. Mura. 2004. New role for leucyl aminopeptidase in glutathione turnover. Biochem. J. 378:35-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, S.-X., and P. Schopfer. 1999. Hydroxyl-radical production in physiological reactions: a novel function of peroxidase. Eur. J. Biochem. 260:726-735. [DOI] [PubMed] [Google Scholar]
- 6.Debets-Ossenkopp, Y. J., R. G. J. Pot, D. J. Van Westerloo, A. Goodwin, C. M. J. E. Vandenbroucke-Grauls, D. Berg, P. S. Hoffman, and J. G. Kusters. 1999. Insertion of mini-IS605 and deletion of adjacent sequences in the nitroreductase (rdxA) gene cause metronidazole resistance in Helicobacter pylori NCTC11637. Antimicrob. Agents Chemother. 43:2657-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodwin, A., D. Kersulyte, G. Sisson, S. J. O. Veldhuyzen van Zanten, D. E. Berg, and P. S. Hoffman. 1998. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxA) that encodes an oxygen-insenstive NADPH nitroreductase. Mol. Microbiol. 28:383-393. [DOI] [PubMed] [Google Scholar]
- 8.Gronow, G., W. Kähler, A. Koch, and N. Klause. 2005. Benzoate hydroxylation: a measure of oxidative stress in divers. Adv. Exp. Med. Biol. 566:223-229. [DOI] [PubMed] [Google Scholar]
- 9.Hoffman, P. S., A. Goodwin, J. Johnson, K. Magee, and S. J. O. Veldhuyzen van Zanten. 1996. Metabolic activities of metronidazole-sensitive and -resistant strains of Helicobacter pylori: repression of pyruvate oxidoreductase and expression of isocitrate lyase activity correlate with resistance. J. Bacteriol. 178:4822-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hrdý, I., R. Cammack, P. Stopka, J. Kulda, and J. Tachezy. 2005. Alternative pathway of metronidazole activation in Trichomonas vaginalis hydrogenosomes. Antimicrob. Agents Chemother. 49:5033-5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hughes, N. J., C. L. Clayton, P. A. Chalk, and D. J. Kelly. 1998. Helicobacter pylori porCDAB and oorDABC genes encode distinct pyruvate:flavodoxin and 2-oxoglutarate:acceptor oxidoreductases which mediate electron transport to NADP. J. Bacteriol. 180:1119-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaakoush, N. O., and G. L. Mendz. 2005. Helicobacter pylori disulphide reductases: role in metronidazole reduction. FEMS Immunol. Med. Microbiol. 44:137-142. [DOI] [PubMed] [Google Scholar]
- 13.Kim, J., M. Lee, R. Chalam, M. N. Martin, T. Leustek, and W. Boerjan. 2002. Constitutive overexpression of cystathionine γ-synthase in Arabidopsis leads to accumulation of soluble methionine and S-methylmethionine. Plant Physiol. 128:95-107. [PMC free article] [PubMed] [Google Scholar]
- 14.Kwon, D. H., F. A. K. El-Zaatari, M. Kato, M. S. Osato, R. Reddy, Y. Yamaoka, and D. Y. Graham. 2000. Analysis of rdxA and involvement of additional genes encoding NAD(P)H flavin oxidoreductase (FrxA) and ferrodoxin-like protein (FdxB) in metronidazole resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 44:2133-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwon, D. H., M. Kato, F. A. K. El-Zaatari, M. S. Osato, and D. Y. Graham. 2000. Frame-shift mutations in NAD(P)H flavin oxidoreductase encoding gene (frxA) from metronidazole resistant Helicobacter pylori ATCC43505 and its involvement in metronidazole resistance. FEMS Microbiol. Lett. 118:197-202. [DOI] [PubMed] [Google Scholar]
- 16.Land, K. M., and P. J. Johnson. 1999. Molecular basis of metronidazole resistance in pathogenic bacteria and protozoa. Drug Resist. Updat. 2:289-294. [DOI] [PubMed] [Google Scholar]
- 17.Leigh, J. A., and J. A. Dodsworth. 2007. Nitrogen regulation in Bacteria and Archaea. Annu. Rev. Microbiol. 61:349-377. [DOI] [PubMed] [Google Scholar]
- 18.Marais, A., C. Bilardi, F. Cantet, G. L. Mendz, and F. Mégraud. 2003. Characterization of the genes rdxA and frxA involved in metronidazole resistance in Helicobacter pylori. Res. Microbiol. 154:137-144. [DOI] [PubMed] [Google Scholar]
- 19.Marshall, B. J., and J. R. Warren. 1983. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet i:1311-1315. [DOI] [PubMed] [Google Scholar]
- 20.McAtee, C. P., P. S. Hoffman, and D. E. Berg. 2001. Identification of differentially regulated proteins in metronidazole resistant Helicobacter pylori by proteome techniques. Proteomics 1:516-521. [DOI] [PubMed] [Google Scholar]
- 21.McGee, D. J., and H. L. Mobley. 2000. Pathogenesis of Helicobacter pylori infection. Curr. Opin. Gastroenterol. 16:24-31. [DOI] [PubMed] [Google Scholar]
- 22.Mendz, G. L., and F. Mégraud. 2002. Is the molecular basis of metronidazole resistance in microaerophilic organisms understood? TRENDS Microbiol. 10:370-375. [DOI] [PubMed] [Google Scholar]
- 23.Mendz, G. L., and M. A. Trend. 2001. Intracellular redox status and antibiotic resistance in enterogastric microaerophilic bacteria: evidence for the scavenging of oxygen hypothesis. Redox Rep. 6:179-181. [DOI] [PubMed] [Google Scholar]
- 24.Meri, T., T. S. Jokiranta, L. Suhonen, and S. Meri. 2000. Resistance of Trichomonas vaginalis to metronidazole: report of the first three cases from Finland and optimization of in vitro susceptibility testing under various oxygen concentrations. J. Clin. Microbiol. 38:763-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mihara, H., and N. Esaki. 2002. Bacterial cysteine desulfurases: their function and mechanisms. Appl. Microbiol. Biotechnol. 60:12-23. [DOI] [PubMed] [Google Scholar]
- 26.Moore, G. R., G. W. Pettigrew, and N. K. Rogers. 1986. Factors influencing redox potentials of electron transfer proteins. Proc. Natl. Acad. Sci. USA 83:4998-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poolman, B., A. J. Driessen, and W. N. Konings. 1987. Regulation of arginine-ornithine exchange and the arginine deiminase pathway in Streptococcus lactis. J. Bacteriol. 169:5597-5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rader, B. A., S. R. Campagna, M. F. Semmelhack, B. L. Bassler, and K. Guillemin. 2007. The quorum-sensing molecule Autoinducer 2 regulates motility and flagellar morphogenesis in Helicobacter pylori. J. Bacteriol. 189:6109-6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scarpignato, C. 2004. Towards the ideal regimen for Helicobacter pylori eradication: the search continues. Dig. Liver Dis. 36:243-247. [DOI] [PubMed] [Google Scholar]
- 30.Sisson, G., J.-Y. Jeong, A. Goodwin, L. Bryden, N. Rossler, S. Lim-Morrison, A. Raudonikiene, D. E. Berg, and P. S. Hoffman. 2000. Metronidazole activation is mutagenic and causes DNA fragmentation in Helicobacter pylori and in Escherichia coli containing a cloned H. pylori rdxA+ (nitroreductase) gene. J. Bacteriol. 182:5091-5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith, M. A., and D. I. Edwards. 1995. Redox potential and oxygen concentration as factors in the susceptibility of Helicobacter pylori to nitroheterocyclic drugs. J. Antimicrob. Chemother. 35:751-764. [DOI] [PubMed] [Google Scholar]
- 32.Smith, M. A., and D. I. Edwards. 1997. Oxygen scavenging, NADH-oxidase and metronidazole resistance in Helicobacter pylori. J. Antimicrob. Chemother. 39:347-353. [DOI] [PubMed] [Google Scholar]
- 33.Smith, M. A., M. A. Jorgensen, G. L. Mendz, and S. L. Hazell. 1998. Metronidazole resistance and microaerophily in Campylobacter species. Arch. Microbiol. 170:279-284. [DOI] [PubMed] [Google Scholar]
- 34.St. Maurice, M., N. Cremades, M. A. Croxen, G. Sisson, J. Sancho, and P. S. Hoffman. 2007. Flavodoxin:quinone reductase (FqrB): a redox partner of pyruvate:ferredoxin oxidoreductase that reversibly couples pyruvate oxidation to NADPH production in Helicobacter pylori and Campylobacter jejuni. J. Bacteriol. 189:4764-4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tetley, R. M., and N. I. Bishop. 1979. The differential action of metronidazole on nitrogen fixation, hydrogen metabolism, photosynthesis and respiration in Anabaena and Scenedesmus. Biochim. Biophys. Acta 546:43-53. [DOI] [PubMed] [Google Scholar]
- 36.Trend, M. A., M. A. Jorgensen, S. L. Hazell, and G. L. Mendz. 2001. Oxidases and reductases are involved in metronidazole sensitivity in Helicobacter pylori. Int. J. Biochem. Cell Biol. 33:143-153. [DOI] [PubMed] [Google Scholar]
- 37.Wait, R., S. Begum, D. Brambilla, A. M. Carabelli, F. Conserva, A. Rocco Guerini, I. Eberini, R. Ballerio, M. Gemeiner, I. Miller, and E. Gianazza. 2005. Redox options in two-dimensional electrophoresis. Amino Acids 28:239-272. [DOI] [PubMed] [Google Scholar]
- 38.Ware, E. 1950. The chemistry of the hydantoins. Chem. Rev. 46:403-470. [DOI] [PubMed] [Google Scholar]
- 39.Warren, J. R. 1983. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet i:1273-1275. [PubMed] [Google Scholar]
- 40.Warrilow, A. G. S., and M. J. Hawkesford. 2000. Cysteine synthase (O-acetylserine (thiol) lyase) substrate specificities classify the mitochondrial isoform as a cyanoalanine synthase. J. Exp. Bot. 51:985-993. [DOI] [PubMed] [Google Scholar]