Abstract
Retrospective analysis of 189 nonredundant strains of Pseudomonas aeruginosa sequentially recovered from the sputum samples of 46 cystic fibrosis (CF) patients over a 10-year period (1998 to 2007) revealed that 53 out of 189 (28%) samples were hypersusceptible to the β-lactam antibiotic ticarcillin (MIC ≤ 4 μg/ml) (phenotype dubbed Tichs). As evidenced by trans-complementation and gene inactivation experiments, the mutational upregulation of the efflux system MexXY was responsible for various degrees of resistance to aminoglycosides in a selection of 11 genotypically distinct strains (gentamicin MICs from 2 to 64 μg/ml). By demonstrating for the first time that the MexXY pump may evolve in CF strains, we found that a mutation leading to an F1018L change in the resistance-nodulation-cell division (RND) transporter MexY was able to increase pump-promoted resistance to aminoglycosides, cefepime, and fluoroquinolones twofold. The inactivation of the mexB gene (which codes for the RND transporter MexB) in the 11 selected strains showed that the Tichs phenotype was due to a mutational or functional loss of function of MexAB-OprM, the multidrug efflux system known to contribute to the natural resistance of P. aeruginosa to β-lactams (e.g., ticarcillin and aztreonam), fluoroquinolones, tetracycline, and novobiocin. Two of the selected strains synthesized abnormally low amounts of the MexB protein, and 3 of 11 strains expressed truncated MexB (n = 2) or MexA (n = 1) polypeptide as a result of mutations in the corresponding genes, while 7 of 11 strains produced wild-type though nonfunctional MexAB-OprM pumps at levels similar to or even higher than that of reference strain PAO1. Overall, our data indicate that while MexXY is necessary for P. aeruginosa to adapt to the hostile environment of the CF lung, the MexAB-OprM pump is dispensable and tends to be lost or inactivated in subpopulations of P. aeruginosa.
The chronic colonization of the airways by Pseudomonas aeruginosa is often associated with a decline in respiratory function and higher rates of morbidity in cystic fibrosis (CF) patients (44). As antibiotic chemotherapy remains the cornerstone of the management of CF lung infection, many studies have attempted to correlate the results of in vitro methods for susceptibility testing to patients' outcomes in order to optimize individual treatments. However, clinical practice brings evidence that the administration of antibiotics predicted to be poorly efficient by in vitro susceptibility tests may actually improve the condition of some CF patients (16, 67, 73). On the other hand, strains that are susceptible to many antibiotics in vitro may turn out to be impossible to eradicate in vivo by “appropriate” antibiotic regimens. The reasons why conventional parameters (MIC and MBC) fail to reliably predict clinical success in the treatment of pulmonary exacerbations are complex and related to both host and bacterial factors (21). For instance, the mode of life of P. aeruginosa in CF airways is believed to contribute to the higher resistance of the pathogen in vivo (recently discussed in reference 55). Alternatively, the great phenotypic diversity of bacterial populations at the stage of chronic infection may be underestimated when routine susceptibility tests are performed on a colony morphotype basis (14, 30, 61, 69).
More than three decades ago, May and Ingold (51) reported the existence of an intriguing subpopulation of P. aeruginosa in the sputum samples of CF patients that is hypersusceptible to carbenicillin in vitro (MIC ≤ 6 μg/ml). The strains exhibiting this particular phenotype, dubbed Tichs in the present paper (for hypersusceptibility to ticarcillin), accounted for 33% of the selected isolates. A subsequent study confirmed the high prevalence of these strains (45%) and their even distributions among the mucoid and nonmucoid populations of P. aeruginosa (30). The Tichs phenotype, which extends to other penicillins (e.g., azlocillin and piperacillin), tetracycline, and trimethoprim but not to aminoglycosides, was attributed to qualitative variations in outer membrane proteins (30) and later on was associated with mutations in a genetic locus closely linked to nalB (15). Interestingly, studies in the 1990s demonstrated that the nalB gene encodes a negative regulator of MexAB-OprM (63), a polyspecific efflux system which contributes to the natural resistance of P. aeruginosa toward a wide range of antibiotics including β-lactams, tetracyclines, trimethoprim, fluoroquinolones, and novobiocin (36, 40). In parallel, another efflux pump, MexXY, which is encoded by a distinct operon (mexXY) on the bacterial chromosome, was found to provide CF isolates with moderate resistance to aminoglycosides, fluoroquinolones, and the zwitterionic cephalosporin cefepime when stably overproduced upon various mutations (31, 50, 83, 84).
The present study revisits the prevalence of Tichs subpopulations of P. aeruginosa in a cohort of 46 CF patients. Analysis of 11 representative Tichs strains shows the divergent roles played by the efflux systems MexAB-OprM and MexXY in the adaptation of P. aeruginosa to the specific environment of CF lungs.
MATERIALS AND METHODS
Bacteria, growth conditions, and drug susceptibility tests.
The laboratory strains and plasmids used in this study are listed in Table 1. Strains 72.1 and 100.1 were isolated during a French national survey of P. aeruginosa-associated bloodstream infections and were found to be genotypically different (25). An environmental strain, P. aeruginosa E1, was isolated from surface waters in the east of France. The 189 CF strains of P. aeruginosa cited in the text were obtained from 19 children and 27 adult CF patients monitored at the Besançon teaching hospital in France between 1998 and 2007. These nonredundant isolates were selected from standard sputum cultures on the basis of both patient and resistance profiles. We considered all the strains from the same individual patient that differed by at least one major difference (from the category “susceptible” to the category “resistant”) in their profiles of susceptibility to a panel of 16 antibiotics according to the breakpoints defined by the Comité de l'Antibiogramme de la Société Française de Microbiologie (http://www.sfm.asso.fr/) to be nonredundant. Routine susceptibility testings with the disk diffusion method were performed on Mueller-Hinton agar (MHA) plates (Bio-Rad) as recommended by the Clinical and Laboratory Standards Institute (CLSI) (8). Strains 615R, 3020R, 2715, 2716, 2721, 2729, 2804, 2858, 2933, 2998, and 3066 were selected for further analysis because of their hypersusceptibility to ticarcillin (MIC ≤ 2 μg/ml). All these isolates exhibited very different random amplified polymorphic DNA banding patterns (data not shown) (46). Random amplified polymorphic DNA banding pattern analysis showed that 615R was clonally related to an aminoglycoside-susceptible isolate, 615S, occurring in a same sputum sample (83). Similarly, 3020R was found in mixed populations with a genotypically identical counterpart, 3020S, exhibiting wild-type susceptibility to antibiotics. Lipopolysaccharide O serotyping was performed by slide agglutination with fresh colonies and specific antisera supplied by Bio-Rad. The strains were routinely cultured at 37°C in Mueller-Hinton broth (MHB; Bio-Rad) or on MHA plates. Where necessary and unless otherwise stated, the media were rendered selective by the addition of 50 μg/ml ampicillin for Escherichia coli and 150 μg/ml ticarcillin or 200 μg/ml gentamicin for P. aeruginosa. Electrotransformation of competent cells with plasmid DNA was performed as reported elsewhere previously (74). The MICs of selected antibiotics were determined by the conventional serial twofold macrodilution method in MHA with adjusted concentrations of Mg2+ and Ca2+ (BBL, Cockeysville, MD), by using a Steers replicator and inocula of ca. 104 CFU per spot (7). Inoculated plates were incubated for 18 h at 37°C ± 1°C before bacterial growth was assessed visually.
TABLE 1.
Bacterial strains and plasmidsa
| Strain or plasmid | Phenotype or genotype | Reference or source |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| PAO1 | Wild-type reference strain | 82 |
| MutGR1 | mexXY overexpressing mutant of PAO1 | 83 |
| FE60 | ΔmexXY mutant of PAO1 | 12 |
| PT629 | mexAB-oprM overexpressing mutant of PAO1 | 11 |
| FB1 | mexB::FRT mutant of PAO1 | This study |
| K1119 | ΔmexAB-oprM mutant of PAO1 | 39 |
| P. putida KT2440 | Plasmid-free derivative of strain mt-2; hsdR1 (r− m+) | 2 |
| E. coli | ||
| S17.1 | recA thi pro hsdR−M+ RP4-2-Tc::Mu Km::Tn7 Tpr Smr | 72 |
| DH5α | F−supE44 endA1 hsdR17 (rK− mK+) thi-1 recA1 Δ(argF-lacZYA) U169 φ80dlacZΔM15 phoA gyrA96 relA1 deoR λ− | Invitrogen |
| Plasmids | ||
| pAK1900 | Broad-host-range expression vector; Ampr Ticr | 62 |
| pRK2013 | Broad-host-range helper plasmid; Tcr | 35 |
| pRSP17 | mexAB-oprM operon cloned into broad-host-range vector pRK415; Tcr | 79 |
| pAZ17 | mexZ gene cloned into pAK1900; Ampr Ticr | 83 |
| pAGH97 | mexXY operon cloned into pAK1900; Ampr Ticr | 65 |
| pAGH29 | pAGH97 encoding an F29S substitution in MexY; Ampr Ticr | This study |
| pAGH1018 | pAGH97 encoding an F1018L substitution in MexY; Ampr Ticr | This study |
| pEX100Tlink | Gene replacement vector with multiple-cloning site from pUC18; oriT+sacB+ Ampr | 64 |
| pPS858 | Source of FRT gene sequences, green fluorescent protein gene, and Genr cassette; Ampr | 24 |
| pFLP2 | Source of Flp recombinase; Ampr | 24 |
| pEXB | 1-kb PCR fragment of the mexB gene cloned into pEX100Tlink; Ampr | This study |
| pEXBR | FRT cassette from pPS858 inserted into pEXB; Ampr | This study |
| pUC18 | Multipurpose cloning vector; Ampr | 85 |
| pUCΔY | 1.1-kb internal fragment of mexY cloned into pUC18; Ampr | This study |
Abbreviations: Tpr, trimethoprim resistance; Smr, streptomycin resistance; Ampr, ampicillin resistance; Ticr, ticarcillin resistance; Tcr, tetracycline; Genr, gentamicin resistance.
Complementation experiments.
The complementation of MexAB-OprM deficient strains 2804, 2933, and K1119 with broad-host-range plasmid pRSP17(Tcr), which carries the wild-type mexAB-oprM operon from PAO1 (79), was carried out by triparental mating essentially as indicated previously by Srikumar et al. (80). In short, cultures of donor strain E. coli S17-1(pRSP17), of helper strain E. coli HB101(pRK2013), and of a recipient P. aeruginosa strain grown overnight were mixed together (50 μl:50 μl:100 μl, respectively); pelleted in a microcentrifuge for 20 s; and resuspended in 25 μl of MHB. The bacterial mixture was spotted onto the surface of an MHA plate and left during 4 h at 37°C before dispersion in 1 ml MHB. MHA plates containing 200 μg/ml cetrimide (to counterselect the E. coli strains) and tetracycline at twofold the MIC (to select for the P. aeruginosa transconjugants) were inoculated with 100-μl fractions of the suspension and incubated for 48 to 72 h at 37°C. The presence of plasmid pRSP17 in selected colonies was checked by agarose gel electrophoresis after small-scale extraction. The susceptibility of transconjugants to ticarcillin and aztreonam, two specific substrates of the MexAB-OprM pump (49), was subsequently assayed in MHB without IPTG (isopropyl-β-d-thiogalactopyranoside) since mexAB-oprM is constitutively expressed from the Plac promoter in pRSP17 (79).
Molecular biology methods.
Standard protocols were used for DNA restriction, fragment ligation, plasmid transformation, and agarose gel electrophoresis (1). Plasmids were extracted and purified with the Qiagen (Hilden, Germany) Midi kit. Chromosomal DNA was prepared with the Wizard Genomic DNA purification kit (Promega, Madison, WI). PCR amplifications were carried out in a 50-μl final volume with 0.5 U of BioTaq Red (Bioline, Paris, France). The reactions were performed using a DNA thermal cycler (Biometra, Göttingen, Germany) for 35 cycles, each consisting of 30 s at 94°C, 30 s at 60°C, and 1 min at 72°C. DNA amplicons were sequenced on both strands in a 3130 genetic analyzer (Applied Biosystems, Courtaboeuf, France) with the BigDye Terminator v3 cycle sequencing kit (Applied Biosystems). Data were subsequently edited with SeqScape software v2.5 (Applied Biosystems).
QRDR sequencing.
The search for mutations in the quinolone resistance-determining regions (QRDRs) encoded by the genes gyrA, gyrB, parC, and parE was carried out in strains 2716, 2804, and 3066, as described previously (26). Isolates 2716 and 3066 exhibited wild-type QRDRs, while 2804 showed the canonical T83I substitution in GyrA that is known to confer fluoroquinolone resistance (56).
Quantitative real-time PCR.
The expression levels of the operons mexAB-oprM, mexCD-oprJ, mexEF-oprN, mexGHI-opmD, mexJK, mexVW, and mexXY were assessed by reverse transcription real-time PCR (RT-PCR) with the fluorescent dye Sybr green (Qiagen Sciences, MD) in a RotorGene RG3000 apparatus (Corbett Research, Sydney, Australia), as described previously by Dumas et al. (11). The primers used for the amplification of the mexB (primers mexB1 and mexB2), mexC (mexC3 and mexC4), mexE (mexE4 and mexE5), mexG (mexG1 and mexG2), mexJ (mexJ1 and mexJ2), mexV (mexV1 and mexV2), and mexY (mexY1a and mexY1b) genes are listed in Table 2. The gene transcription levels were normalized in each strain to that of the housekeeping gene uvrD (34) and expressed as ratios to the values of strain PAO1 (by definition set at 1). The RT-PCR data presented here are means of four determinations from two independent experiments. Well-characterized mutants overexpressing MexAB-OprM (PT629) (38), MexCD-OprJ (EryR) (52), MexEF-OprN (PAO7H) (37), MexJK (PAO318) (6), and MexXY (MutGR1) (83) were used as positive controls. None of the CF isolates exhibited mRNA levels of the mexC and mexE genes greater than 5% of those of EryR and PAO7H, respectively. The transcript levels of the mexG and mexV genes in the CF strains were found to be identical or rather close to those of wild-type strain PAO1 (from 1- to 3.8-fold and from 0.4- to 1.7-fold, respectively).
TABLE 2.
Primers used in the study
| Function and primer | Nucleotide sequence (5′-3′) | Reference or source |
|---|---|---|
| Gene expression | ||
| mexB1 | ATC CGC CAG ACC ATC GCC A | 27 |
| mexB2 | CAT CAC CAG GAA CAC GAG GAG G | 27 |
| mexC3 | GTA CCG GCG TCA TGC AGG GTT C | 11 |
| mexC4 | TTA CTG TTG CGG CGC AGG TGA CT | 11 |
| mexE4 | CCA GGA CCA GCA CGA ACT TCT TGC | 11 |
| mexE5 | CGA CAA CGC CAA GGG CGA GTT CAC C | 11 |
| mexG1 | GCA ACT GGC TCT GGC TGA CC | 27 |
| mexG2 | ACG GCG GTG GCG ATG TTG AA | 27 |
| mexJ1 | GCC CTG TCC CTG TTT TCC TCC C | 27 |
| mexJ2 | CCT TCT TTA CCC GCT CGC CG | 27 |
| mexV1 | CGT CAG CAG ATC GCC CTT TTC AGC | 42 |
| mexV2 | CGC TTT TCG AGA TGG CCT TGC TGC | 42 |
| mexY1a | TTA CCT CCT CCA GCG GC | 33 |
| mexY1b | GTG AGG CGG GCG TTG TG | 33 |
| uvrD1 | CAC GCC TCG CCC TAC AGC A | 34 |
| uvrD2 | GGA TCT GGA AGT TCT GCT CAG C | 34 |
| Gene inactivationa | ||
| mexBrec1 | CTC GGA TCC GTC GGT GAC TTC CAG GTG TT (BamHI) | This study |
| mexBrec2 | CTC AAG CTT GAA AGG AAC ATC CGG TTG AA (HindIII) | This study |
| mexYb1 | CTC GGA TCC GGT CTA CAC CCT GGT CAT CG (BamHI) | This study |
| mexYb2 | CTC AAG CTT GGC CGA CCT TGA AGT AGA TG (HindIII) | This study |
| Mutagenesis experimentsb | ||
| F29S sup | GCG ATC CGC TCC CTG CCG GTC | This study |
| F29S down | GAC CGG CAG GGA GCG GAT CGC | This study |
| F1018 up | CTG GTA CCG CTG CTC TTC CTG GTG GTC | This study |
| F1018 down | GAC CAC CAG GAA GAG CAG CGG TAC CAG | This study |
The restriction sites introduced into primers are underlined in the sequences, with the corresponding endonucleases indicated in parentheses.
The nucleotide substitutions introduced into primers are indicated in boldface type in the sequences.
Immunodetection of MexB, MexY, and OprM.
Bacterial membranes (whole-membrane fractions for MexB and MexY and outer membrane fractions for OprM) were isolated, subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and analyzed by Western blotting with MexB-, MexY-, and OprM-specific polyclonal antisera (diluted 1:1,000, 1:20,000, and 1:5,000, respectively), as reported previously (28).
Gene inactivation experiments.
The sacB-based strategy described previously by Hoang et al. (24) was used here to inactivate the mexB gene. Briefly, a ca. 1-kb BamHI-HindIII PCR fragment carrying mexB from strain PAO1 (primers mexBrec1 and mexBrec2) (Table 2) was cloned into BamHI-HindIII-restricted vector pEX100Tlink(sacB+), yielding pEXB. This plasmid was cleaved inside the insert with endonuclease SplI, and the resultant fragment was blunt ended with Klenow enzyme. The 1.7-kb SmaI fragment, which contains the gentamicin cassette, the green fluorescent protein gene, and the Flp recognition target (FRT) gene sequences from plasmid pPS858 (24), was then ligated into linearized pEXB. This recombinant plasmid, named pEXBR, was conjugally transferred from E. coli S17.1 to the P. aeruginosa strains. Recombinant clones were selected on M9 minimal medium (1) supplemented with gentamicin, and merodiploids were subsequently resolved by culture on MHA medium containing 5% (wt/vol) sucrose and gentamicin. Flippase-promoted excision of the chromosomally integrated FRT cassette (gentamicin resistance and green fluorescent protein markers) was finally achieved by the transfer of plasmid pLFP2, as described previously (12). The disruption of mexB by the FRT sequences was verified by PCR and DNA sequencing experiments.
For unknown reasons, the above-described sacB-based strategy with plasmid pEXΔXYR (12) failed to inactivate the mexXY operon in the CF strains. A suicide plasmid derived from multicopy vector pUC18(Ticr) was thus constructed in E. coli DH5α cells by cloning a ca. 1.1-kb BamHI-HindIII PCR fragment internal to the mexY gene (primers mexYb1and mexYb2) (Table 2). Transformants of CF isolates 3020S, 3020R, and 2804 with crossover recombination of the resultant plasmid pUCΔY with the chromosomally located mexY gene were obtained on MHA medium supplemented with ticarcillin. PCR experiments confirmed the disruption of mexY by pUCΔY in these bacteria.
Mutagenesis experiments.
Site-directed mutagenesis of the mexY gene was performed with the QuikChange II site-directed mutagenesis kit (Stratagene). Plasmid pAGH97, which carries the mexXY operon from strain PAO1 (65), was used as the target DNA. The oligonucleotide primers, each complementary to opposite strands of pAGH97 and harboring the desired nucleotide substitution (Table 2), were extended during temperature cycling by Pfu Turbo polymerase (Stratagene). Two pairs of primers, designated F29S-up/F29S-down and F1018L-up/F1018L-down (Table 2), were used to introduce the amino acid substitutions F29S and F1018L, respectively, in plasmid-encoded MexY in vitro. DNA sequence analysis confirmed that the proper nucleotide changes had been successfully engineered in the resultant plasmids pAGH29 and pAGH1018, respectively. Transformants of a ΔmexXY mutant from PAO1, named FE60, and of Pseudomonas putida reference strain KT2440 were obtained by electrotransformation and subsequent selection on MHA with ticarcillin. We used the same strategy to generate additional mutations in pAGH1018, leading to K329Q and W358R substitutions in the MexX protein and T543A substitution in the MexY protein (data not shown).
β-Lactamase activities.
Enzymatic activities were measured on crude French press lysates by a spectrophotometric assay using nitrocefin as a chromogenic substrate (26). Briefly, CF strains were cultured to mid-log phase both in 200 ml MHB (uninduced culture) and in 200 ml MHB (induced culture) supplemented with 50 μg/ml cefoxitin, a β-lactam antibiotic that is able to strongly induce the expression of chromosomally encoded AmpC β-lactamase in P. aeruginosa cells. Spectrophotometric measurements were performed on each bacterial lysate in triplicates.
RESULTS
Prevalence of ticarcillin-hypersusceptible strains among CF patients.
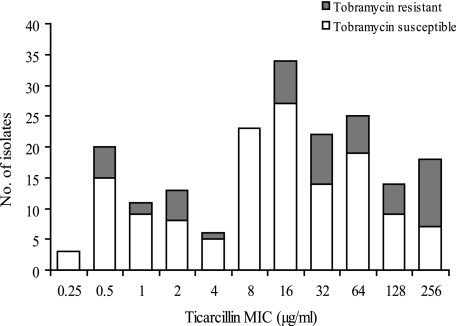
Forty-six CF patients with P. aeruginosa-positive sputum samples (19 children and 27 adults) were monitored on a regular basis between 1998 and 2007 at the teaching hospital of Besançon, France. Analysis of the drug resistance patterns of 189 nonredundant (as defined in Materials and Methods) isolates sequentially collected from these patients during the survey showed that 25 of 46 patients (54.3%) were colonized with P. aeruginosa strains that were hypersusceptible to ticarcillin (at least fourfold more susceptible than wild-type strains such as PAO1) (MIC ≤ 4 μg/ml) (Fig. 1). Interestingly, many of the strains displaying this particular phenotype, named Tichs, appeared to exhibit various degrees of resistance to aminoglycosides (gentamicin, amikacin, tobramycin, and netilmicin) (data not shown). For instance, 32 of 53 (60.4%) of the Tichs isolates were at least fourfold more resistant to tobramycin than was PAO1 (i.e., MIC ≥ 2 μg/ml). However, these rates were not very different from those of the isolates with ticarcillin MICs of ≥8 μg/ml (98/136 isolates; 72%), supporting the notion that the Tichs phenotype and aminoglycoside resistance result from independent mechanisms. In order to further characterize the Tichs subpopulation, we selected 11 genotypically distinct Tichs strains showing various levels of resistance to aminoglycosides from different patients. In four patients, the Tichs strains constituted the only P. aeruginosa population detected in the sputum samples over the course of the survey. In the other seven patients, the Tichs isolates were found in mixed populations with one (n = 3), two (n = 1), or more (n = 3) strains for which ticarcillin MICs were ≥8 μg/ml. In one case, a Tichs isolate with moderate resistance to aminoglycosides (615R) was present in a mixed culture with a genotypically identical counterpart (615S) showing wild-type susceptibility to these antibiotics. Finally, in another patient, the Tichs isolate (3020R) was cocultured with a clonally related parent exhibiting wild-type susceptibility to both ticarcillin and aminoglycosides (3020S). As expected from long-term colonizing strains (23), only 2 of 11 of the selected isolates were serotypeable (O:3 and O:11 for 2716 and 2729, respectively), while 3 of 11 isolates produced mucoid colonies (2715, 2858, and 2933). These data confirmed that the Tichs phenotype is not necessarily associated with a loss of O-type lipopolysaccharides or mucoidy.
FIG. 1.
Susceptibilities of selected CF isolates to ticarcillin and tobramycin. One hundred eighty-nine isolates recovered from 46 patients were tested for drug susceptibility with the standard agar dilution method. White and gray bars represent isolates that are susceptible (MIC ≤ 4 μg/ml) and resistant (MIC ≥ 8 μg/ml) to tobramycin according to CLSI breakpoints, respectively.
Role of the MexXY-OprM pump in aminoglycoside resistance.
As indicated in Table 3, the selected Tichs strains exhibited various levels of resistance to antipseudomonal aminoglycosides such as gentamicin (2- to 64-fold), amikacin (4- to 64-fold), and tobramycin (2- to 128-fold) as well as to enzyme-recalcitrant test compounds like fortimicin (2- to >16-fold) (data not shown) and apramycin (2- to 64-fold) (data not shown) (71). These results were fully consistent with previously published data showing the absence of horizontally acquired aminoglycoside-modifying enzymes in most CF isolates of P. aeruginosa (29, 45, 70, 83). As the efflux system MexXY-OprM is known to play a major role in emergence of aminoglycoside resistance in CF strains (31, 83, 84), we assessed its expression at the gene (mexY) and the protein (MexY) levels by reverse transcription RT-PCR and Western blotting, respectively. As expected, all the Tichs strains were found to overexpress both the mexY gene (11.4- to 58.8-fold) (data not shown) and the MexY protein compared with aminoglycoside-susceptible strains PAO1, 615S, and 3020S (Fig. 2).
TABLE 3.
Drug susceptibilities of the P. aeruginosa strains
| Strain | MIC (μg/ml)a
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| GENb | AMK | TOB | TIC | ATMc | CAZ | FEP | CIP | NOVc | |
| PAO1 and derivatives | |||||||||
| PAO1 | 1 (0.125) | 2 | 0.5 | 32 | 4 (0.12) | 1 | 2 | 0.25 | 512 (32) |
| MutGR1 | 2 | 8 | 1 | 32 | 4 | 1 | 8 | 0.5 | 512 |
| FE60(pAK1900) | 0.125 | 0.5 | 0.06 | ND | ND | ND | 2 | 0.25 | ND |
| FE60(pAGH97) | 1 | 4 | 0.5 | ND | ND | ND | 8 | 0.5 | ND |
| FE60(pAGH29) | 1 | 4 | 0.5 | ND | ND | ND | 8 | 0.5 | ND |
| FE60(pAGH1018) | 2 | 8 | 1 | ND | ND | ND | 16 | 1 | ND |
| Clinical strains | |||||||||
| 615S | 1 (0.125) | 2 | 0.25 | 1 | 0.12 (—)d | 1 | 2 | 4 | 64 (—) |
| 615R | 8 (0.125) | 16 | 2 | 2 | 0.25 (0.25) | 1 | 4 | 1 | 4 (4) |
| 3020S | 2 (0.125) | 2 | 0.5 | 16 | 2 (0.25) | 1 | 2 | 0.25 | 512 (8) |
| 3020R | 16 (0.25) | 32 | 4 | 2 | 0.25 (0.25) | 1 | 4 | 0.5 | 16 (16) |
| 2715 | 4 (0.125) | 8 | 2 | 2 | 0.5 (0.25) | 1 | 16 | 1 | 16 (8) |
| 2716 | 2 (0.125) | 8 | 1 | 2 | 0.25 (0.25) | 1 | 8 | 2 | 32 (32) |
| 2721 | 16 (0.125) | 32 | 4 | 2 | 0.25 (0.12) | 0.5 | 8 | 1 | 32 (32) |
| 2729 | 8 (0.25) | 16 | 2 | 0.5 | 0.25 (0.25) | 1 | 8 | 1 | 32 (32) |
| 2804 | 64 (0.5) | 128 | 64 | 0.5 | 0.25 (—) | 2 | 32 | 16 | 8 (—) |
| 2858 | 4 (0.125) | 8 | 1 | 2 | 0.25 (—) | 4 | 4 | 0.5 | 64 (—) |
| 2933 | 16 (—) | 32 | 4 | 0.5 | 0.25 (0.12) | 0.5 | 16 | 0.5 | 8 (4) |
| 2998 | 8 (0.125) | 16 | 4 | 0.5 | 0.12 (0.12) | 0.5 | 8 | 1 | 32 (32) |
| 3066 | 64 (0.125) | 128 | 16 | 2 | 0.25 (0.25) | >8 | 32 | 16 | 32 (32) |
Values in boldface type (or underlined) are at least fourfold higher (or fourfold lower) than those for wild-type strain PAO1. Abbreviations: GEN, gentamicin; AMK, amikacin; TOB, tobramycin; TIC, ticarcillin; ATM, aztreonam; CAZ, ceftazidime; FEP, cefepime; CIP, ciprofloxacin; NOV, novobiocin; ND, not determined.
Values in parentheses indicate MICs after the mexXY operon has been repressed in trans by plasmid pAZ17 (in isolates 3020S and 3020R, the inactivation of mexXY was achieved by the chromosomal integration of plasmid pUCΔY). Preliminary experiments showed that the addition of 50 μg/ml ticarcillin to selectively maintain pAZ17 in cultures did not influence MICs.
Values in parentheses indicate MICs after the mexB gene has been inactivated by the FRT cassette from plasmid pEXBR.
—, the inactivation of mexXY or mexB was unsuccessful in these strains.
FIG. 2.
Expression of efflux pumps in CF isolates. The production of the MexY, MexB, and OprM proteins was assessed by Western blotting with specific antibodies after extraction of cell membranes and SDS-PAGE. Mutants MutGR1 and PT629 were used as positive controls (C+) for the overexpression of MexXY and MexAB-OprM efflux systems, respectively. Twenty micrograms of whole (outer and inner) membranes was subjected to SDS-PAGE for detection of MexY and MexB. Ten micrograms of outer membrane protein was used per lane for immunodetection of OprM.
An upregulation of the mexXY operon may result from mutations occurring in the regulatory gene mexZ, which codes for a TetR-like repressor, or in as-yet-undetermined loci (31, 43, 77, 83, 84). Modulating previous conclusions that CF strains overexpress mexXY mostly as a result of mutations in the mexZ gene (31, 83), only 5 of 11 strains exhibited alterations (frameshifts) in the coding sequence of mexZ (Table 4). The nucleotide sequences of mexZ and of the mexZ-mexXY intergenic region were identical to that of PAO1 in the other six strains.
TABLE 4.
Mutations and amino acid changes in CF strains
| Strain | Alteration(s)a
|
||||
|---|---|---|---|---|---|
| mexZ | MexX | MexY | mexA | mexB | |
| Control strains | |||||
| E1 | —b | —c,d,e,f | —g,h,i,j | ND | ND |
| 72.1 | +6 nt at position 166 | —b | —h | ND | ND |
| 100.1 | —b | —d,e,f | —g,h,i | ND | ND |
| CF strains | |||||
| 615S | —b | —d,e,f | —h | —b | —b |
| 615R | Δ386 nt (from A248 to A633) | —d,e,f | A254G, Q282Rh | —b | —b |
| 3020S | —b | —d,e,f | —h | —b | —b |
| 3020R | —b | —d,e,f | —h | —b | —b |
| 2715 | —b | —d,e,f | —h | —b | —b |
| 2716 | —b | R351Sd,f | E152Dh | —b | —b |
| 2721 | —b | L22M, D135Yd,e,f | S46G, Q282R, A596V, K692Mk | —b | —b |
| 2729 | Δ15 nt (from C595 to C609) | —d,e,f | I536Ph | —b | —b |
| 2804 | Δ81 nt (from C217 to C297) IS Pa1635 | —d,f | F1018Lh | —b | Δ1 nt at position 2147 |
| 2858 | —b | —c,d,e,f | G1002Ah,j | —b | —b |
| 2933 | —b | —c,d,e,f | —h | Δ1 nt at position 870 | —b |
| 2998 | +1 nt at position 27 | —d,e,f | —h | —b | —b |
| 3066 | Δ25 nt (from C217 to G241) | —d,e,f | F29S | —b | G2364A (nonsense) |
Nucleotide or amino acid positions refer to strain PAO1. Abbreviations: nt, nucleotide; ND, not determined.
Sequence identical to that of PAO1.
Contains amino acid substitution A30T of PA14.
Contains amino acid substitution K329Q of PA14.
Contains amino acid substitution L331V of PA14.
Contains amino acid substitution W358R of PA14.
Contains amino acid substitution I536V of PA14.
Contains amino acid substitution T543A of PA14.
Contains amino acid substitution G589A of PA14.
Contains amino acid substitution Q840E of PA14.
Contains amino acid substitution N1036T of PA14.
Variations in the amino acid sequence of the MexXY pump.
While stable MexXY overproduction is usually associated with a modest two- to fourfold increase in aminoglycoside MICs in in vitro mutants such as MutGR1 (as seen here for CF strains 2715, 2716, and 2858) (Table 3), most of the selected Tichs isolates displayed much stronger resistance to these antibiotics (for example, 2804 and 3066 fall into the “resistant” category according to CLSI breakpoints). To evaluate the contribution of the upregulated MexXY proteins to aminoglycoside resistance, we turned off the expression of the mexXY operon in the Tichs strains by trans-complementation with a plasmid-encoded repressor, MexZ (construct pAZ17). The transformation of the strains with pAZ17 was successful in all the strains but 3020R and 2933. Subsequent RT-PCR experiments provided evidence that the mexY gene was strongly repressed following pAZ17 transfer (data not shown). As expected, the MexZ-dependent repression of mexXY resulted in a decrease in aminoglycoside MICs in the pAZ17-transformed strains (Table 3). The residual resistance that was supposed to result from MexXY-independent mechanisms was actually very low and comparable between the CF isolates and PAO1(pAZ17) (gentamicin MICs from 0.125 to 0.5 μg/ml versus 0.125 μg/ml, respectively), thus suggesting a major role of the efflux process in the high level of aminoglycoside resistance exhibited by some strains (2721, 2804, and 3066). Of note, pAZ17-dependent repression of mexXY also strongly reduced the MIC of ciprofloxacin (from 16 to 0.5 μg/ml) in 3066, a strain showing wild-type QRDRs in the DNA gyrase and topoisomerase IV enzymes. The transfer of pAZ17 had similar effects on ciprofloxacin resistance (from 16 to 1 μg/ml) in 2804, which contains a T83I substitution in the QRDR of GyrA. However, one could argue that pAZ17-encoded MexZ may well sensitize P. aeruginosa to antibiotics by mechanisms other than repressing mexXY. To address this issue, we carried out the inactivation of the mexY gene in several isolates (PAO1, 3020S, 3020R, and 2804) by homologous recombination with suicide plasmid pUCΔY. As with the pAZ17 strategy, the disruption of mexY rendered 3020R (MIC equal to 0.25 μg/ml) and 2804 (0.5 μg/ml) almost as susceptible to gentamicin as 3020S::pUCΔY (0.125 μg/ml) and PAO1::pUCΔY (0.125 μg/ml), thereby confirming the absence of mechanisms other than drug efflux providing significant resistance to aminoglycosides (more than fourfold) in the Tichs strains. Residual resistance to ciprofloxacin in 2804 following the inactivation of mexY (1 μg/ml) was identical to that provided by pAZ17.
Since our RT-PCR and immunoblotting experiments did not show evident differences in levels of MexXY expression among the clinical strains, we wondered whether specific amino acid substitutions in these proteins would account for the variations in aminoglycoside MICs. We thus sequenced the mexXY operon in all the 11 CF strains as well as in the two susceptible strains 615S and 3020S (Table 4). We next aligned these sequences with those of reference strains PAO1 and PA14 (available at http://v2.pseudomonas.com/), those of two bacteremic, non-CF isolates (72.1 and 100.1) (25), and that of one environmental strain, named E1. Interestingly, all the CF strains appeared to contain the same amino acid substitutions in the predicted proteins MexX (A30T, K329Q, L331V, and/or W358R) and MexY (T543A, Q840E, and/or N1036T) compared with PAO1 (Table 4). However, since these variations were present in susceptible strains PA14, 100.1, and E1, they were considered to be nonsignificant with respect to aminoglycoside resistance. On the other hand, a number of strain-specific changes in MexXY could be identified in bacteria exhibiting low to moderate resistance to gentamicin such as 615R (MIC of 8 μg/ml), 2716 (2 μg/ml), 2721 (16 μg/ml), 2729 (8 μg/ml), and 2858 (4 μg/ml). While it remains unclear whether these amino acid changes in the MexXY translocase actually improve the efflux of aminoglycosides and resistance, this finding demonstrates that the MexXY proteins may be subject to evolution in CF strains (compare 615S and 615R in Table 4).
To gain an insight into the adaptation of the pump to the CF lung environment, we focused our attention on strains 2804 and 3066, which combine a strong resistance to aminoglycosides (gentamicin MIC of 64 μg/ml) with a single-amino-acid substitution in MexY (F1018L and F29S, respectively). These strain-specific mutations were engineered by directed mutagenesis into the mexXY operon from PAO1 previously cloned in a proper orientation downstream of the lac promoter on broad-host-range vector pAK1900 (yielding construct pAGH97) (65). The resultant constructs, named pAGH1018 (encoding an F1018L change) and pAGH29 (encoding an F29S change), and their parent plasmid, pAGH97, were transferred by electroporation into a ΔmexXY mutant, FE60, derived from PAO1. Control RT-PCR experiments confirmed that the three transformants of FE60 expressed similar mRNA levels of the mexY gene (56.6 ± 6 times that of PAO1). As indicated in Table 3, FE60(pAGH1018) turned out to be consistently more resistant (twofold) than FE60(pAGH97) or FE60(pAGH29) to all of the MexXY substrates including aminoglycosides, cefepime, and ciprofloxacin. To confirm these results, we introduced pAGH97 and pAGH1018 into P. putida reference strain KT2440 (2) and measured the levels of resistance of the resultant transformants to gentamicin and cefepime. Again, pAGH1018 provided levels of resistance to both agents that were twofold greater than that provided by pAGH97 (4 versus 2 μg/ml and 4 versus 2 μg/ml, respectively). These data provide clear evidence that specific mutations may improve the drug transport activity of the MexXY translocase. However, since the resistance levels of FE60(pAGH1018) were much lower than those of 2804, we wondered whether the additional substitutions detected in the MexX (K329Q and W358R) and MexY (T543A) proteins from 2804 might cooperatively improve the efflux activity provided by the F1018L mutation (as pAGH1018 carries the mexXY operon from strain PAO1). The K329Q, W358R (MexX), and T543A (MexY) changes were thus engineered into pAGH1018 in addition to F1018L. The resultant plasmid was found to confer the same levels of resistance to mutant FE60 as pAGH1018, ruling out a cooperative effect of the four amino acid residues in pump functioning (data not shown).
Role of the MexAB-OprM pump in the Tichs phenotype.
As indicated in Table 3, the hypersusceptibility of the 11 selected strains to ticarcillin (16- to 64-fold more than reference strain PAO1) also extended to other antipseudomonal β-lactams such as aztreonam (8- to 32-fold) and piperacillin (four- to eightfold) (data not shown) but was not correlated with lower resistance to ceftazidime or cefepime. Since the MexAB-OprM efflux system strongly contributes to the natural resistance of P. aeruginosa to ticarcillin, carbenicillin, aztreonam, and piperacillin but has a poor impact on intrinsic resistance to ceftazidime (40, 58), we hypothesized that the selected Tichs strains might have impaired MexAB-OprM pumps. Supporting this notion, all the strains proved to be highly susceptible to novobiocin, a hydrophobic antibiotic known to be specifically extruded by the pump (41, 47) (Table 3). Furthermore, a disruption of the mexB gene in these bacteria (except 615S, 2804, and 2858, for which the inactivation experiments with plasmid pEXBR were unsuccessful) did not result in a more-than-twofold reduction in MICs of ticarcillin (data not shown), aztreonam, and novobiocin (Table 3). In comparison, mexB null mutant FB1 was 64-, 32-, and 16-fold more susceptible than its parent, PAO1, to these agents, respectively (data not shown). It should mentioned here that cefepime MICs may be influenced by the expression of other efflux systems such as MexCD-OprJ and MexXY-OprM independently of MexAB-OprM (49).
Because the activity of MexAB-OprM is thought to be impaired when the MexCD-OprJ (20, 32) or MexEF-OprN (47) pump is upregulated, we measured the transcript levels of the mexC and mexE genes (as representatives of the mexCD-oprJ and mexEF-oprN operons, respectively) by reverse transcription RT-PCR. However, none of the 11 CF strains significantly overexpressed these operons compared to wild-type strain PAO1 (data not shown). Similar negative results were obtained when the transcript levels of the mexGHI-opmD, mexJK, and mexVW operons, which code for other efflux systems operating with resistance-nodulation-cell division (RND) transporters (data not shown), were assessed.
More interestingly, immunoblotting analysis of bacterial membranes revealed the presence of smaller amounts of the MexB and OprM proteins in strain 3020R compared with its wild-type counterpart, 3020S, and the lack of visible MexB bands in strains 2804, 2858, and 3066 (Fig. 2). Surprisingly, the latter bacteria were able to express the OprM protein, the exit duct which, together with MexAB, enables the extrusion of substrates to the external milieu. The other 7 of 11 Tichs strains (namely, 615R, 2715, 2716, 2721, 2729, 2933, and 2998) were found to produce significant amounts of both MexB and OprM. Assessment of gene transcription by RT-PCR confirmed that mexB was underexpressed in 3020R (0.2-fold) and 2858 (0.3-fold) compared with PAO1 or 3020S (onefold) and was expressed at wild-type levels or higher (0.9- to 2.3-fold) in the other strains (data not shown). However, RT-PCR experiments also showed significant levels of mexB transcripts in MexB-deficient isolates 2804 and 3066 (0.9- and 3.1-fold that of PAO1, respectively), suggesting the presence of mutations disrupting mexB in these bacteria.
Nucleotide sequencing of (i) the repressor gene mexR, whose product downregulates the mexAB-oprM operon (63); (ii) the intergenic region between mexR and mexA, which carries the two promoters of mexAB-oprM (13, 68); (iii) the PA3721 gene (5), which negatively controls the expression of a protein (coded by PA3719) that is able to bind and inactivate MexR (9); and (iv) the PA3574 gene, which codes for a second repressor of mexAB-oprM (76), did not show significant mutations in strains 2858 and 3020R, compared with PAO1 and 3020S, that would explain their reduced levels of expression of MexB. In addition, no differences were observed between strains 2858, 3020S, 3020R, and PAO1 with respect to mRNA levels of the mexR, PA3719, and PA3574 genes (data not shown).
Alterations in the MexAB-OprM pump.
Several studies have shown that amino acid substitutions in the transporter MexB at positions essential for proton translocation (22), proper compaction of transmembrane stretches (TMSs) (86), trimerization (53), or interactions with the periplasmic adaptor MexA (53, 60) may impair the transport activity of MexAB-OprM and thus increase the susceptibility of resultant mutants to the pump substrates. Similarly, mutations in the mexA gene may compromise the oligomerization of MexA or its binding to MexB and thus alter the functioning of the efflux system (59). To determine if such alterations could be responsible for the Tichs phenotype, we sequenced the mexAB-oprM operon in the 11 CF strains as well as in 615S and 3020S. This operon appeared strictly conserved and identical to that of reference strain PAO1 in all the isolates except in three strains (Table 4). Strains 2804 and 3066 exhibited mutations in mexB resulting in premature stop codons and truncated polypeptides of 719 and 787 amino acids, respectively, instead of 1,046 residues for the wild-type MexB protein. These polypeptides, which lacked 5 of 12 transmembrane segments (from TMS-8 to TMS-12) were not detected in whole-membrane extracts by Western blotting (Fig. 2), likely because of their inability to insert into the cytoplasmic membrane. As mentioned above, mRNAs of the corresponding mexB genes were amplified by RT-PCR. Strain 2933 displayed a C870 deletion in the mexA gene, generating a truncated polypeptide of 311 amino acids lacking 72 residues at the C-terminal end of the MexA protein. Finally, nucleotide sequencing of the oprM genes demonstrated that all of the Tichs isolates produced a strictly conserved OprM protein that was 100% identical to that of PAO1.
To confirm the impact of mutations on pump activity in strains 2933, 2804, and 3066, we attempted to complement the bacteria with plasmid pRSP17, which carries the entire mexAB-oprM operon from PAO1 in the proper orientation downstream of the Plac promoter. The transfer of pRSP17 was successful with 2933, 2804, and a ΔmexAB-oprM derivative of PAO1 named K1119 but not with 3066. The overexpression of mexAB-oprM from pRSP17 dramatically increased the resistance to ticarcillin (from 0.5 to 64 μg/ml) and aztreonam (from 0.25 to 16 to 32 μg/ml) in 2933, 2804, and K1119, thus clearly indicating that in these strains, the MexAB-OprM function was lost mutationally.
Because the chromosomally encoded, large-spectrum AmpC β-lactamase contributes to the natural resistance of P. aeruginosa to many β-lactam antibiotics together with MexAB-OprM (48), we measured the β-lactamase activities expressed by the CF strains. Both their basal (from 9 to 51 nmol nitrocefin hydrolyzed min−1 mg−1 protein) and cefoxitin-induced (from 1,241- to 6,604 nmol min−1 mg−1 protein) enzymatic levels were comparable to those of reference strain PAO1 (33 and 3,900 nmol min−1 mg−1 protein, respectively). These results provide evidence that the Tichs phenotype was not associated with deficient production in AmpC β-lactamase, especially in those strains producing intact MexA, MexB, and OprM proteins.
DISCUSSION
This study shows that many CF patients are colonized and/or infected by populations of P. aeruginosa that are strongly deficient in MexAB-OprM-dependent efflux activity. Bacteria expressing the typical phenotype (Tichs) due to inactive MexAB-OprM are mostly recovered during chronic colonization but may also emerge rapidly at the stage of early colonization (e.g., strain 3020R). For instance, we observed that 17 of 27 adults (63%) versus 8 of 19 children (42%) harbored Tichs isolates. This bacterial adaptation to the CF lung is intriguing, as the loss of MexAB-OprM function results in in vitro hypersusceptibility to a number of antibiotics that are widely prescribed for the treatment of exacerbations of pulmonary infection, such as ticarcillin, aztreonam, piperacillin, and ciprofloxacin (66). It is interesting that the increasing use of “newer” β-lactam molecules (e.g., ceftazidime, cefepime, and meropenem) over the years has not reduced the prevalence of Tichs strains compared with data reported in the initial article by May and Ingold in the early 1970s (51). There is little doubt that the Tichs phenotype expressed in vitro by so many persistent P. aeruginosa isolates does not reflect the real susceptibility of bacteria in CF hosts. It has been well documented that some lung populations of P. aeruginosa adapt to the strong selective pressure exerted by repeated cures of β-lactams through the stable or transient upregulation of intrinsic AmpC β-lactamase (3, 17), decreased outer membrane permeability (4), or alterations in penicillin binding proteins (19). Preexisting subpopulations with stable, partially derepressed AmpC may thus rapidly expand under treatment with agents such as ceftazidime, piperacillin, or imipenem (17). Partial release of their β-lactamase content in sputum samples could contribute to antibiotic inactivation in situ (18). Whether these partially derepressed mutants would provide more susceptible bacterial populations with efficient protection against β-lactams is unclear. AmpC-overproducing mutants were not detected in the sputum samples of 10 of 25 of our patients, suggesting that at least in these patients, the persistence of Tichs populations involves nonhydrolytic mechanisms. It is conceivable that hypersusceptible bacteria may survive in the CF lung if physically protected from antibiotics by mucus and/or biofilm-like materials (10). However, our observation that most of the Tichs isolates were resistant to aminoglycosides, a class of antibiotics known to diffuse poorly in exopolymer matrices (10), does not support this hypothesis (Table 3). In addition, strain 3066 turned out to be highly resistant to ceftazidime as a consequence of repeated courses of chemotherapy with this product. Because of the high prevalence of the Tichs populations, the loss of MexAB-OprM is likely to confer a decisive advantage to P. aeruginosa for its survival in the hostile environment of CF airways. Time-kill studies with ticarcillin in our laboratory failed to demonstrate a tolerance of the selected isolates to β-lactams under standard laboratory conditions (i.e., exponentially growing bacteria in rich medium) (data not shown). However, other conditions that more closely resemble those of the CF lung (microaerobiosis, biofilm mode of growth, and nutrient limitation) should be tested to determine which factors specifically contribute to the resistance of Tichs strains in vivo (78, 81).
Confirming the results of previous studies on CF strains (31, 83, 84), all the Tichs strains exhibiting some degree of resistance to aminoglycosides (at least twofold that of reference strain PAO1) (Table 3) proved to overproduce the MexXY proteins, which interact with OprM to form a functional tripartite efflux system (65). However, strain 615S provides evidence that the Tichs phenotype is not linked to MexXY upregulation (Tables 3 and 4). RT-PCR analysis of another strain, named 1710, exhibiting wild-type susceptibility to tobramycin (MIC of 0.25 μg/ml) and hypersusceptibility to ticarcillin (MIC of 0.25 μg/ml) (Fig. 1) confirmed this result (data not shown). More importantly, complementation experiments with plasmid pAZ17 (the mexZ gene) demonstrated for the first time that MexXY can be responsible for strong aminoglycoside resistance in CF strains (2804 and 3066). Although the factors that modulate MexXY-OprM functioning remain poorly understood (77, 83), we could establish that specific mutations in the transporter MexY are able to increase the efflux of aminoglycosides, cefepime, and fluoroquinolones [compare FE60(pAGH97) and FE60(pAGH1018) in Table 3]. The F1018L substitution of strain 2804 is located in TMS-12 of MexY, at the groove delimited by TMS-7, TMS-8, and TMS-9. Based on the crystal structure of the homolog transporter AcrB, this groove is supposed to be an efflux pathway for substrates from the cytosol or inner membrane (57). Additional site-directed mutagenesis studies have been carried out to elucidate how the F1018L mutation may facilitate the export of antibiotics predicted to be captured from the periplasm (87).
To our knowledge, this is the first example of the in vivo emergence of resistant mutants overproducing a “modified” efflux pump. Interestingly, in this study, the two Tichs strains displaying the highest levels of resistance to aminoglycosides (2804 and 3066) both appeared to lack the MexB protein. It is tempting to assume that these strains form chimeric MexAY-OprM pumps that contribute to the resistance in addition to MexXY-OprM. Against this hypothesis, pull-down assays reported previously by Mokhonov et al. (54) did not evidence an interaction between MexA, MexY, and OprM. Alternatively, the loss of MexB might allow more recruitment of OprM by the tandem MexXY.
The suppression of MexAB-OprM drug transport activity was associated with mutations disrupting the mexA (2933) and mexB (2804 and 3066) genes in 3 of 11 of our strains. Consistent with our conclusions that pulmonary populations of P. aeruginosa tend to abolish MexAB-OprM efflux during long-term colonization, another study showed that isolates from 11 of 29 (38%) CF patients harbored nonsynonymous mutations in the mexA gene (75). Whereas 2 of 11 of our Tichs strains (3020R and 2858) were partially deficient in MexB production, 6 of 11 were unexpectedly found to express the wild-type pump at levels similar to those of PAO1. Reminiscent of this, recent data from our laboratory strongly suggest that, while normally produced, the MexAB-OprM system is functionally impaired in MexCD-OprJ-overproducing nfxB mutants (32). In the present study, none of the Tichs strains appeared to overexpress the mexC gene; nevertheless, it is clear that still unknown factors may strongly influence the drug transport activity of MexAB-OprM. Ongoing experiments are investigating the role of TonB1 in the emergence of the Tichs phenotype, since mutations in defined regions of this energy-coupling periplasmic protein may compromise the operation of the MexAB-OprM efflux pump without affecting iron acquisition (88).
In conclusion, our data demonstrate the existence in CF strains of an unbalance between the efflux system MexAB-OprM, which seems to be dispensable in the context of the CF lung environment, and MexXY-OprM, whose upregulation is necessary for P. aeruginosa to stand the strong selective pressure exerted by aminoglycosides. We believe that the MexXY-OprM pump should be the primary target for the development of efflux inhibitors in adjunctive therapy of CF pulmonary infection.
Acknowledgments
This work was supported by the French Cystic Fibrosis Association Vaincre la Mucoviscidose and the Conseil Régional de Franche Comté.
We are grateful to Christiane Bailly for collecting the P. aeruginosa CF isolates, Gérard Couetdic for recovery of clinical data, Thilo Köhler for providing strain PT629, Keith Poole for sharing strain K1119 and plasmid pRSP17, Fabrice Poncet for DNA sequencing, and Katy Jeannot for helpful assistance.
Footnotes
Published ahead of print on 2 March 2009.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2000. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, NY.
- 2.Bagdasarian, M., R. Lurz, B. Rückert, F. C. H. Franklin, M. M. Bagdasarian, J. Frey, and K. N. Timmis. 1981. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors and a host-vector system for gene cloning in Pseudomonas. Gene 16:237-247. [DOI] [PubMed] [Google Scholar]
- 3.Bagge, N., M. Hentzer, J. B. Andersen, O. Ciofu, M. Givskov, and N. Høiby. 2004. Dynamic and spatial distribution of β-lactamase expression in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 48:1168-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ballestero, S., A. Fernández-Rodríguez, R. Villaverde, H. Escobar, J. C. Pérez-Díaz, and F. Baquero. 1996. Carbapenem resistance in Pseudomonas aeruginosa from cystic fibrosis patients. J. Antimicrob. Chemother. 38:39-45. [DOI] [PubMed] [Google Scholar]
- 5.Cao, L., R. Srikumar, and K. Poole. 2004. MexAB-OprM hyperexpression in NalC-type multidrug-resistant Pseudomonas aeruginosa: identification and characterization of the nalC gene encoding a repressor of PA3720-PA3719. Mol. Microbiol. 53:1423-1436. [DOI] [PubMed] [Google Scholar]
- 6.Chuanchuen, R., C. T. Narasaki, and H. P. Schweizer. 2002. The MexJK efflux pump of Pseudomonas aeruginosa requires OprM for antibiotic efflux but not for efflux of triclosan. J. Bacteriol. 184:5036-5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard, 7th ed. M7-A7. Clinical and Laboratory Standards Institute, Wayne, PA.
- 8.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial disk susceptibility tests. Approved standard, 9th ed., M2-A9. Clinical and Laboratory Standards Institute, Wayne, PA.
- 9.Daigle, D. M., L. Cao, S. Fraud, M. S. Wilke, A. Pacey, R. Klinoski, N. C. Strynadka, C. R. Dean, and K. Poole. 2007. Protein modulator of multidrug efflux gene expression in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 189:5441-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drenkard, E. 2003. Antimicrobial resistance of Pseudomonas aeruginosa biofilms. Microbes Infect. 5:1213-1219. [DOI] [PubMed] [Google Scholar]
- 11.Dumas, J.-L., C. vanDelden, K. Perron, and T. Köhler. 2006. Analysis of antibiotic resistance gene expression in Pseudomonas aeruginosa by quantitative real-time-PCR. FEMS Microbiol. Lett. 254:217-225. [DOI] [PubMed] [Google Scholar]
- 12.El'Garch, F., K. Jeannot, D. Hocquet, C. Llanes-Barakat, and P. Plésiat. 2007. Cumulative effects of several nonenzymatic mechanisms on the resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 51:1016-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans, K., L. Adewoye, and K. Poole. 2001. MexR repressor of the mexAB-oprM multidrug efflux operon of Pseudomonas aeruginosa: identification of MexR binding sites in the mexA-mexR intergenic region. J. Bacteriol. 183:807-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foweraker, J. E., C. R. Laughton, D. F. J. Brown, and D. Bilton. 2005. Phenotypic variability of Pseudomonas aeruginosa in sputa from patients with acute infective exacerbation of cystic fibrosis and its impact on the validity of antimicrobial susceptibility testing. J. Antimicrob. Chemother. 55:921-927. [DOI] [PubMed] [Google Scholar]
- 15.Fyfe, J. A., and J. R. Govan. 1984. Chromosomal loci associated with antibiotic hypersensitivity in pulmonary isolates of Pseudomonas aeruginosa. J. Gen. Microbiol. 130:825-834. [DOI] [PubMed] [Google Scholar]
- 16.Gaillard, J. L., P. Cahen, C. Delacourt, C. Silly, M. Le Bourgeois, C. Coustère, J. de Blic, G. Lenoir, and P. Scheinmann. 1995. Correlation between activity of beta-lactam agents in vitro and bacteriological outcome in acute pulmonary exacerbations of cystic fibrosis. Eur. J. Clin. Microbiol. Infect. Dis. 14:291-296. [DOI] [PubMed] [Google Scholar]
- 17.Giwercman, B., P. A. Lambert, V. T. Rosdahl, G. H. Shand, and N. Høiby. 1990. Rapid emergence of resistance in Pseudomonas aeruginosa in cystic fibrosis patients due to in-vivo selection of stable partially derepressed β-lactamase producing strains. J. Antimicrob. Chemother. 26:247-259. [DOI] [PubMed] [Google Scholar]
- 18.Giwercman, B., C. Meyer, P. A. Lambert, C. Reinert, and N. Høiby. 1992. High-level β-lactamase activity in sputum samples from cystic fibrosis patients during antipseudomonal treatment. Antimicrob. Agents Chemother. 36:71-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Godfrey, A. J., L. E. Bryan, and H. R. Rabin. 1981. β-Lactam-resistant Pseudomonas aeruginosa with modified penicillin-binding proteins emerging during cystic fibrosis treatment. Antimicrob. Agents Chemother. 19:705-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gotoh, N., H. Tsujimoto, M. Tsuda, K. Okamoto, A. Nomura, T. Wada, M. Nakahashi, and T. Nishino. 1998. Characterization of the MexC-MexD-OprJ multidrug efflux system in delta mexA-mexB-oprM mutants of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 42:1938-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Govan, J. W. R. 2006. Multidrug-resistant pulmonary infection in cystic fibrosis—what does “resistant” mean? J. Med. Microbiol. 55:1615-1617. [DOI] [PubMed] [Google Scholar]
- 22.Guan, L., and T. Nakae. 2001. Identification of essential charged residues in transmembrane segments of the multidrug transporter MexB of Pseudomonas aeruginosa. J. Bacteriol. 183:1734-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hancock, R. E. W., L. M. Mutharia, L. Chan, R. P. Darveau, D. P. Speert, and G. B. Pier. 1983. Pseudomonas aeruginosa isolates from patients with cystic fibrosis: a class of serum-sensitive, nontypeable strains deficient in lipopolysaccharide O side chains. Infect. Immun. 42:170-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 25.Hocquet, D., P. Berthelot, M. Roussel-Delvallez, R. Favre, K. Jeannot, O. Bajolet, N. Marty, F. Grattard, P. Mariani-Kurkdjian, E. Bingen, M.-O. Husson, G. Couedic, and P. Plésiat. 2007. Pseudomonas aeruginosa may accumulate drug resistance mechanisms without losing its ability to cause bloodstream infections. Antimicrob. Agents Chemother. 51:3531-3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hocquet, D., X. Bertrand, T. Köhler, D. Talon, and P. Plésiat. 2003. Genetic and phenotypic variations of a resistant Pseudomonas aeruginosa epidemic clone. Antimicrob. Agents Chemother. 47:1887-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hocquet, D., P. Nordmann, F. El'Garch, L. Cabanne, and P. Plésiat. 2006. Involvement of the MexXY-OprM efflux system in emergence of cefepime resistance in clinical strains of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 50:1347-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hocquet, D., C. Vogne, F. El'Garch, A. Vejux, N. Gotoh, A. Lee, O. Lomovskaya, and P. Plésiat. 2003. MexXY-OprM efflux pump is necessary for adaptive resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 47:1371-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hurley, J. C., G. H. Miller, and A. L. Smith. 1995. Mechanism of amikacin resistance in Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Diagn. Microbiol. Infect. Dis. 22:331-336. [DOI] [PubMed] [Google Scholar]
- 30.Irvin, R. T., J. W. R. Govan, J. A. M. Fyfe, and J. W. Costerton. 1981. Heterogeneity of antibiotic resistance in mucoid isolates of Pseudomonas aeruginosa obtained from cystic fibrosis patients: role of outer membrane proteins. Antimicrob. Agents Chemother. 19:1056-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Islam, S., S. Jalal, and B. Wretling. 2004. Expression of the MexXY efflux pump in amikacin-resistant isolates of Pseudomonas aeruginosa. Clin. Microbiol. Infect. 10:877-883. [DOI] [PubMed] [Google Scholar]
- 32.Jeannot, K., S. Elsen, T. Köhler, I. Attree, C. vanDelden, and P. Plésiat. 2008. Resistance and virulence of Pseudomonas aeruginosa clinical strains overproducing the MexCD-OprJ efflux pump. Antimicrob. Agents Chemother. 52:2455-2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeannot, K., M. L. Sobel, F. El Garch, K. Poole, and P. Plésiat. 2005. Induction of the MexXY efflux pump in Pseudomonas aeruginosa is dependent on drug-ribosome interaction. J. Bacteriol. 187:5341-5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jo, J. T. H., F. S. L. Brinkman, and R. E. W. Hancock. 2003. Aminoglycoside efflux in Pseudomonas aeruginosa: involvement of novel outer membrane proteins. Antimicrob. Agents Chemother. 47:1101-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 36.Köhler, T., M. Kok, M. Michéa-Hamzehpour, P. Plésiat, N. Gotoh, T. Nishino, L. K. Curty, and J. C. Pechère. 1996. Multidrug efflux in intrinsic resistance to trimethoprim and sulfamethoxazole in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 40:2288-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Köhler, T., M. Michéa-Hamzehpour, U. Henze, N. Gotoh, L. Kocjancic Curty, and J.-C. Pechère. 1997. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol. Microbiol. 23:345-354. [DOI] [PubMed] [Google Scholar]
- 38.Köhler, T., M. Michéa-Hamzehpour, P. Plésiat, A. L. Kahr, and J. C. Pechère. 1997. Differential selection of multidrug efflux systems by quinolones in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 41:2540-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li, X.-Z., L. Zhang, R. Srikumar, and K. Poole. 1998. β-Lactamase inhibitors are substrates for the multidrug efflux pumps of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 42:399-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li, X. Z., H. Nikaido, and K. Poole. 1995. Role of MexA-MexB-OprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:1948-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li, X. Z., L. Zhang, and K. Poole. 2000. Interplay between the MexA-MexB-OprM multidrug efflux system and the outer membrane barrier in the multiple antibiotic resistance of Pseudomonas aeruginosa. J. Antimicrob. Chemother. 45:433-436. [DOI] [PubMed] [Google Scholar]
- 42.Li, Y., T. Mima, Y. Komori, Y. Morita, T. Kuroda, T. Mizushima, and T. Tsuchiya. 2003. A new member of the tripartite multidrug efflux pumps, MexVW-OprM, in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 52:572-575. [DOI] [PubMed] [Google Scholar]
- 43.Llanes, C., D. Hocquet, C. Vogne, D. Benali-Baitich, C. Neuwirth, and P. Plésiat. 2004. Clinical strains of Pseudomonas aeruginosa overproducing MexAB-OprM and MexXY efflux pumps simultaneously. Antimicrob. Agents Chemother. 48:1797-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lyczack, J. B., C. L. Cannon, and G. B. Pier. 2002. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15:194-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.MacLeod, D. L., L. E. Nelson, R. M. Shawar, B. B. Lin, L. G. Lockwood, J. E. Dirk, G. H. Miller, J. L. Burns, and R. L. Garber. 2000. Aminoglycoside-resistance mechanisms for cystic fibrosis Pseudomonas aeruginosa isolates are unchanged by long-term, intermittent, inhaled tobramycin treatment. J. Infect. Dis. 181:1180-1184. [DOI] [PubMed] [Google Scholar]
- 46.Mahenthiralingam, E., M. E. Campbell, J. Foster, J. S. Lam, and D. P. Speert. 1996. Random amplified polymorphic DNA typing of Pseudomonas aeruginosa isolates recovered from patients with cystic fibrosis. J. Clin. Microbiol. 34:1129-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maseda, H., H. Yoneyama, and T. Nakae. 2000. Assignment of the substrate-selective subunits of the MexEF-OprN multidrug efflux pump of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:658-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Masuda, N., N. Gotoh, C. Ishii, E. Sakagawa, S. Ohya, and T. Nishino. 1999. Interplay between chromosomal β-lactamase and the MexAB-OprM efflux system in intrinsic resistance to β-lactams in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:400-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, H. Tsujimoto, and T. Nishino. 2000. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-OprM efflux pumps in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:3322-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matsuo, Y., S. Eda, N. Gotoh, E. Yoshihara, and T. Nakae. 2004. MexZ-mediated regulation of mexXY multidrug efflux pump expression in Pseudomonas aeruginosa by binding on the mexZ-mexX intergenic DNA. FEMS Microbiol. Lett. 238:23-28. [DOI] [PubMed] [Google Scholar]
- 51.May, J. R., and A. Ingold. 1973. Sensitivity of respiratory strains of Pseudomonas aeruginosa to carbenicillin. J. Med. Microbiol. 6:77-82. [DOI] [PubMed] [Google Scholar]
- 52.Michéa Hamzehpour, M., J. C. Pechère, P. Plésiat, and T. Köhler. 1995. OprK and OprM define two genetically distinct multidrug efflux systems in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:2392-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Middlemiss, J. K., and K. Poole. 2004. Differential impact of MexB mutations on substrate selectivity of the MexAB-OprM multidrug efflux pump of Pseudomonas aeruginosa. J. Bacteriol. 186:1258-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mokhonov, V. V., E. I. Mokhonova, H. Akama, and T. Nakae. 2004. Role of the membrane fusion protein in the assembly of resistance-nodulation-cell division multidrug efflux pump in Pseudomonas aeruginosa. Biochem. Biophys. Res. Com. 322:483-489. [DOI] [PubMed] [Google Scholar]
- 55.Moreau-Marquis, S., B. A. Stanton, and G. A. O'Toole. 2008. Pseudomonas aeruginosa biofilm formation in the cystic fibrosis airway. Pulm. Pharmacol. Ther. 21:595-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mouneimné, H., J. Robert, V. Jarlier, and E. Cambau. 1999. Type II topoisomerase mutations in ciprofloxacin-resistant strains of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:62-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murakami, S., R. Nakashima, E. Yamashita, and A. Yamaguchi. 2002. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419:587-593. [DOI] [PubMed] [Google Scholar]
- 58.Nakae, T., A. Nakajima, T. Ono, K. Saito, and H. Yoneyama. 1999. Resistance to β-lactam antibiotics in Pseudomonas aeruginosa due to interplay between the MexAB-OprM efflux pump and β-lactamase. Antimicrob. Agents Chemother. 43:1301-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nehme, D., X.-Z. Li, R. Elliot, and K. Poole. 2004. Assembly of the MexAB-OprM multidrug efflux system of Pseudomonas aeruginosa: identification and characterization of mutations in mexA compromising MexA multimerization and interaction with MexB. J. Bacteriol. 186:2973-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nehme, D., and K. Poole. 2005. Interaction of the MexA and MexB components of the MexAB-OprM multidrug efflux system of Pseudomonas aeruginosa: identification of MexA extragenic suppressors of a T578I mutation in MexB. Antimicrob. Agents Chemother. 49:4375-4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Perry, J. D., L. Laine, S. Hughes, A. Nicholson, A. Galloway, and F. K. Gould. 2008. Recovery of antimicrobial-resistant Pseudomonas aeruginosa from sputa of cystic fibrosis patients by culture on selective media. J. Antimicrob. Chemother. 61:1057-1061. [DOI] [PubMed] [Google Scholar]
- 62.Poole, K., D. E. Heinrichs, and S. Neshat. 1993. Cloning and sequence analysis of an EnvCD homologue in Pseudomonas aeruginosa: regulation by iron and possible involvement in the secretion of the siderophore pyoverdine. Mol. Microbiol. 10:529-544. [DOI] [PubMed] [Google Scholar]
- 63.Poole, K., K. Tetro, Q. X. Zhao, S. Neshat, D. E. Heinrichs, and N. Bianco. 1996. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob. Agents Chemother. 40:2021-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Quenee, L., D. Lamotte, and B. Polack. 2005. Combined sacB-based negative selection and cre-lox antibiotic marker recycling for efficient deletion in Pseudomonas aeruginosa. BioTechniques 38:63-67. [DOI] [PubMed] [Google Scholar]
- 65.Ramos Aires, J., T. Köhler, H. Nikaido, and P. Plésiat. 1999. Involvement of an efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 43:2624-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ramsey, B. W. 1996. Management of pulmonary disease in patients with cystic fibrosis. N. Engl. J. Med. 335:179-188. [DOI] [PubMed] [Google Scholar]
- 67.Ramsey, B. W., M. S. Pepe, J. M. Quan, K. L. Otto, A. B. Montgomery, J. Williams-Warren, M. Vasiljev, D. Borowitz, C. M. Bowman, B. C. Marshall, S. Marshall, and A. L. Smith. 1999. Intermittent administration of inhaled tobramycin in patients with cystic fibrosis. N. Engl. J. Med. 340:23-30. [DOI] [PubMed] [Google Scholar]
- 68.Saito, K., S. Eda, H. Maseda, and T. Nakae. 2001. Molecular mechanism of MexR-mediated regulation of MexAB-OprM efflux pump expression in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 195:23-28. [DOI] [PubMed] [Google Scholar]
- 69.Seale, T. W., H. Thirkill, M. Tarpay, M. Flux, and O. M. Rennert. 1979. Serotypes and antibiotic susceptibilities of Pseudomonas aeruginosa isolates from single sputa of cystic fibrosis patients. J. Clin. Microbiol. 9:72-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shawar, R. M., D. L. MacLeod, R. L. Garber, J. L. Burns, J. R. Stapp, C. R. Clausen, and S. K. Tanaka. 1999. Activities of tobramycin and six other antibiotics against Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Antimicrob. Agents Chemother. 43:2877-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shimizu, K., T. Kumada, W.-C. Hsieh, H.-Y. Chung, Y. Chong, R. S. Hare, G. H. Miller, F. J. Sabatelli, and J. Howard. 1985. Comparison of aminoglycoside resistance patterns in Japan, Formosa, and Korea, Chile, and the United States. Antimicrob. Agents Chemother. 28:282-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Simon, R., U. Prieffer, and A. Pühler. 1983. Vector plasmids for in vivo and in vitro manipulations of gram-negative bacteria, p. 98-106. In A. Pühler (ed.), Molecular genetics of the bacteria-plant interaction. Springer-Verlag, Berlin, Germany.
- 73.Smith, A. L., S. B. Fiel, N. Mayer-Hamblett, B. Ramsey, and J. L. Burns. 2003. Susceptibility testing of Pseudomonas aeruginosa isolates and clinical response to parenteral antibiotic administration. Chest 123:1495-1502. [DOI] [PubMed] [Google Scholar]
- 74.Smith, A. W., and B. H. Iglewski. 1989. Transformation of Pseudomonas aeruginosa by electroporation. Nucleic Acids Res. 17:10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smith, E. E., D. G. Buckley, Z. Wu, C. Saenphimmachak, L. R. Hoffman, D. A. D'Argenio, S. I. Miller, B. W. Ramsey, D. P. Speert, S. M. Moskowitz, J. L. Burns, R. Kaul, and M. V. Olson. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. USA 103:8487-8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sobel, M. L., D. Hocquet, L. Cao, P. Plésiat, and K. Poole. 2005. Mutations in PA3574 (nalD) lead to increased MexAB-OprM expression and multidrug resistance in laboratory and clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 49:1782-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sobel, M. L., G. A. McKay, and K. Poole. 2003. Contribution of the MexXY multidrug transporter to aminoglycoside resistance in Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 47:3202-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Spoering, A. L., and K. Lewis. 2001. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 183:6746-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Srikumar, R., T. Kon, N. Gotoh, and K. Poole. 1998. Expression of Pseudomonas aeruginosa multidrug efflux pumps MexA-MexB-OprM and MexC-MexD-OprJ in a multidrug-sensitive Escherichia coli strain. Antimicrob. Agents Chemother. 42:65-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Srikumar, R., X. Z. Li, and K. Poole. 1997. Inner membrane efflux components are responsible for β-lactam specificity of multidrug efflux pumps in Pseudomonas aeruginosa. J. Bacteriol. 179:7875-7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sriramulu, D. D., H. Lünsdorf, J. S. Lam, and U. Römling. 2005. Microcolony formation: a novel biofilm model of Pseudomonas aeruginosa for the cystic fibrosis lung. J. Med. Microbiol. 54:667-676. [DOI] [PubMed] [Google Scholar]
- 82.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K.-S. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 83.Vogne, C., J. Ramos Aires, C. Bailly, D. Hocquet, and P. Plésiat. 2004. Role of the multidrug efflux system MexXY in the emergence of moderate resistance to aminoglycosides among Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Antimicrob. Agents Chemother. 48:1676-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Westbrock-Wadman, S., D. R. Sherman, M. J. Hickey, S. N. Coulter, Y. Q. Zhu, P. Warrener, L. Y. Nguyen, R. M. Shawar, K. R. Folger, and C. K. Stover. 1999. Characterization of a Pseudomonas aeruginosa efflux pump contributing to aminoglycoside impermeability. Antimicrob. Agents Chemother. 43:2975-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yanisch-Perron, C., J. Vieria, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13 mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 86.Yoneyama, H., H. Maseda, T. Yamabayashi, S. Izumi, and T. Nakae. 2002. Secondary-site mutations restore the transport defect caused by the transmembrane domain mutation of the xenobiotic transporter MexB in Pseudomonas aeruginosa. Biochem. Biophys. Res. Com. 292:513-518. [DOI] [PubMed] [Google Scholar]
- 87.Yu, E. W., J. Ramos Aires, and H. Nikaido. 2003. AcrB multidrug efflux pump of Escherichia coli: composite substrate-binding cavity of exceptional flexibility generates its extremely wide substrate specificity. J. Bacteriol. 185:5657-5664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhao, Q., and K. Poole. 2002. Differential effects of mutations in tonB1 on intrinsic multidrug resistance and iron acquisition in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 184:2045-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]