Abstract
We describe the characterization of a novel CTX-M β-lactamase from Salmonella enterica. Four S. enterica isolates (three of serotype Westhampton and one of serotype Senftenberg) resistant to extended-spectrum cephalosporins (cefotaxime and ceftazidime) were recovered in 2004 from living cockles in three supermarkets located in distant geographic areas in France, which got their supplies from the same fishery. The isolates were found to produce a novel extended-spectrum β-lactamase (ESBL) belonging to the CTX-M-1 phylogenetic group and named CTX-M-53. The CTX-M-53 β-lactamase harbored the substitution Asp240Gly, like the CTX-M-15 enzyme, which is specifically implicated in a higher catalytic efficiency against ceftazidime. The blaCTX-M-53 gene was located on a mobilizable 11-kb plasmid, pWES-1. The complete sequence of pWES-1 revealed the presence of a novel insertion sequence, ISSen2, and an IS26 element upstream and downstream of the blaCTX-M-53 gene, respectively; however, transposition assays of the blaCTX-M-53 gene were unsuccessful. IS26 elements may have contributed to the acquisition of the blaCTX-M-53 gene. Interestingly, the mobilization module of the pWES-1 plasmid was similar to that of quinolone resistance plasmids (carrying the qnrS2 gene) from aquatic sources. Although belonging to two serotypes differentiated on the basis of the O-antigen structure (E1 or E4 groups), the isolates were found to be genetically indistinguishable by pulsed-field gel electrophoresis. Multilocus sequence typing showed that the isolates of serotype Westhampton had a sequence type, ST14, common among isolates of serotype Senftenberg. This is the first characterization of the CTX-M-53 ESBL, which represents an additional ceftazidime-hydrolyzing CTX-M enzyme.
Salmonella enterica is a frequent pathogen of animals and humans. Food-borne diseases caused by this species represent an important public health problem worldwide. Extended-spectrum cephalosporins (ESC) and fluoroquinolones are often used in the treatment of invasive cases of salmonellosis. However, the emergence of extended-spectrum β-lactamases (ESBLs) in Enterobacteriaceae is an increasing problem worldwide, compromising the utilization of these drugs in the treatment of complicated Salmonella infections. Moreover, there is an increasing number of reports of ESBL-producing S. enterica strains throughout the world (3, 36). These strains, isolated mostly in hospitalized patients, produced plasmid-mediated class A ESBLs belonging to the TEM, SHV, cefotaximase (CTX-M), or PER families (3).
CTX-M ESBLs are class A ESBLs that in general possess a higher level of hydrolytic activity against cefotaxime (and ceftriaxone) than against ceftazidime but that are inhibited by clavulanic acid, sulbactam, and tazobactam (5, 9, 26). On the basis of their amino acid sequences, the CTX-M enzymes have been classified into five major phylogenetic branches, namely the CTX-M-1, -2, -8, -9, and -25 groups (5, 9, 26; http://www.lahey.org/Studies/other.asp). CTX-M ESBLs are a rapidly growing group, which contains ESBLs encoded by more than 80 identified CTX-M genes (http://www.lahey.org/Studies/other.asp). In the genus Salmonella, 14 different CTX-M β-lactamases have been reported in several serotypes and over wide geographic areas (3, 5, 20, 21, 36-38). We report here the characterization of the novel CTX-M-53 ESBL in S. enterica serotypes Westhampton and Senftenberg in France. The S. enterica strains showing an ESBL phenotype were recovered from cockles from the Etel River (Morbihan, France) in August 2004. A molecular characterization of the β-lactamase gene was done by PCR, cloning, and sequencing. The genomic diversity of the isolates was determined by pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST). The novel CTX-M enzyme was characterized by MIC determination of the β-lactams, isoelectric focusing (IEF), and kinetic parameters. The CTX-M-carrying plasmid was fully sequenced, and blaCTX-M mobilization experiments were performed.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The four S. enterica isolates were collected from cockles fished from the Etel River (Morbihan, France) and distributed in three supermarkets across the country from August to September 2004. The detection of the Salmonella sp. was performed by using an enzyme-linked immunosorbent assay-based test, Transia Plate Salmonella (Raisio Diagnostics SAS, Lyon, France), and by the reference cultural method EN ISO 6579:2002. The Salmonella strains were identified using API 20E strips (bioMérieux, Marcy l'Etoile, France) and serotyped at the AFSSA laboratory (French Agency for Food Safety) on the basis of somatic O and phase 1 and phase 2 flagellar antigens by agglutination tests with antisera (Bio-Rad, Marnes la Coquette, France, and the WHO Collaborative Center for Reference and Research on Salmonella, Institut Pasteur, Paris, France) as specified by the White-Kauffmann-Le Minor scheme (19). Table 1 shows details of all the bacterial strains and plasmids used in this study.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Phenotype, genotype, and/or characteristicsa | Reference or source (year of isolation) |
|---|---|---|
| Strains | ||
| S. enterica serotype Westhampton | ||
| 04CEB8118SAL | ESBL, blaCTX-M-53 | Living cockles, fishery, Morbihan, Franceb (2004) |
| 04CEB8273SAL | ESBL, blaCTX-M-53 | Living cockles, supermarket, Puy-de-Dôme, Franceb (2004) |
| 04CEB8276SAL | ESBL, blaCTX-M-53 | Living cockles, supermarket, Vienne, Franceb (2004) |
| S. enterica serotype Senftenberg | ||
| 04CEB8275SAL | ESBL, blaCTX-M-53 | Living cockles, supermarket, Vienne, Franceb (2004) |
| E. coli | ||
| ATCC 25922 | Control in disk diffusion method and in MIC determinations | CDC, Atlanta, GA |
| DH1 | F−endA1 gyrA96 thi-1 hsdR17(rK− mK+) supE44 relA1 | |
| DH5α | DH1 genotype and F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 | |
| DH10B | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 endA1 araΔ139 Δ(ara, leu)7697 galU galK-rpsL nupG tonA | Invitrogen |
| HB101 | hsd20(rB− mB−) recA13 rpsL20(Strr) leu proA2 | |
| Plasmids | ||
| pBK-CMV phagemid | Neor Kanr | Boehringer-Mannheim |
| pOX38-Neor | F derivative Tra+ Neor, 58.2 kb | 18 |
| pGBG1 | pBBR1 derivative Chlr, 7.6 kb | 35 |
| pWES-1 | 11-kb natural plasmid from S. enterica 04CEB8273SAL electrotransformed in E. coli DH10B | This study |
| pCTX-M-53 | 1.5-kb PCR fragment from S. enterica 04CEB8273SAL that contained blaCTX-M-53 in the XbaI-PstI sites of pBK-CMV | This study |
Strr, streptomycin resistant; Neor, neomycin resistant; Kanr, kanamycin resistant; Chlr, chloramphenicol resistant.
Département, French administrative subdivision.
Antimicrobial susceptibility testing.
Antibiotic susceptibility was determined by the disk diffusion method with 32 antimicrobial drugs (Bio-Rad), as previously described (38). The MICs of ceftriaxone and ceftazidime were determined by Etest (AB Biodisk, Solna, Sweden). The ESBL phenotype was detected by using the ESBL detection Etest strips (AB Biodisk) and the double-disk synergy method (16). The isolates were categorized as susceptible, intermediate, or resistant according to Antibiogram Committee of the French Society for Microbiology (CA-SFM) clinical breakpoints (http://www.sfm.asso.fr/nouv/general.php?pa=2). The clinical breakpoints used for ceftriaxone and ceftazidime are slightly different from those determined by the Clinical and Laboratory Standards Institute (CLSI); susceptible strains were thus defined by a MIC of ≤4 μg/ml (CLSI, ≤8 μg/ml) and resistant strains by a MIC of >32 μg/ml (CLSI, ≥64 μg/ml for ceftriaxone and ≥32 μg/ml for ceftazidime).
Escherichia coli ATCC 25922 was used as a control for the disk diffusion method and for the MIC determinations.
PCR amplification of β-lactamase genes and sequence analysis.
The total DNA of S. enterica isolates was extracted using the InstaGene matrix kit (Bio-Rad) in accordance with the manufacturer's recommendations. PCR amplifications of the blaTEM, blaSHV, blaOXA-1 group, and blaCTX-M group genes were performed using TEM-F and TEM-R, SHV-F and SHV-R, OXA-1-F and OXA-1-R, CTX-M-F and CTX-M-R, or CTX-M-1-F and CTX-M3/M15-R primers, respectively, as described previously (37, 38). Sequencing was performed at Genome Express (Meylan, France). The nucleotide sequences and the deduced protein sequences were analyzed with EditSeq and Megalign software (Dnastar, Madison, WI). The BLASTN program of NCBI was used for database searches (http://www.ncbi.nlm.nih.gov/BLAST/).
PFGE.
The genetic diversity of the Salmonella isolates was assessed by the PFGE of genomic DNA digested with XbaI (Roche, Mannheim, Germany), as described previously (22). The running conditions and the molecular size marker were as described in the standardized PulseNet protocol (34). BioNumerics 4.0 (Applied Maths, Sint-Martens-Latem, Belgium) was used for image normalization and the construction of similarity matrices. Bands were assigned manually. Clustering was carried out by the unweighted-pair group method with arithmetic averages based on the Dice similarity index, using a 1% optimization parameter and 1% band position tolerance.
MLST.
The genomic DNA was prepared from a bacterial culture plate using the Wizard kit (Promega, Madison, WI). The seven genes for the MLST analysis, aroC, dnaN, hemD, hisD, purE, sucA, and thrA, were amplified using the primers and PCR conditions described elsewhere (http://mlst.ucc.ie/mlst/dbs/Senterica/documents/primersEnterica_html). Both strands of purified amplicons were sequenced by the Genotyping of Pathogens and Public Health platform (PF8; Institut Pasteur) and nucleotide sequences obtained with BigDye version 3.1 chemistry (Applied Biosystems, Foster City, CA) on an ABI 3700 apparatus (Applied Biosystems). Alleles were assigned by comparing the sequences to those in the Salmonella MLST database hosted by University College Cork, Cork, Ireland.
ESBL resistance transfer and plasmid analysis.
A resistance transfer experiment was carried out on liquid or solid media as described previously (37). E. coli DH1 resistant to nalidixic acid (Nal) was used as the recipient strain. Transconjugants were selected on Drigalski agar (Bio-Rad) supplemented with cefotaxime (2 μg/ml) and Nal (50 μg/ml). The electroporation of plasmid DNA, extracted by the QIAfilter Plasmid Midi kit (Qiagen), from S. enterica isolates to E. coli DH10B was performed using a GenePulser apparatus (Bio-Rad). The transformants were selected on Mueller-Hinton agar containing ceftazidime (4 μg/ml). Plasmid DNA was extracted from E. coli transformants by an alkaline lysis procedure (37) and subjected to 0.8% agarose gel electrophoresis. The molecular sizes of the plasmids were determined by reference to plasmids of known sizes, RP4 (54 kb) and pIP173 (126 kb), mixed with a supercoiled DNA ladder (Invitrogen, Groningen, The Netherlands). Plasmid DNAs from the E. coli transformants were digested using the AvaI restriction enzyme (Promega) and compared by agarose gel electrophoresis, as previously described (37). The pWES-1 plasmid from S. enterica isolate 04CEB8273SAL was fully sequenced on both strands by directional genome walking using internal primers (at Genome Express). The nucleotide sequences and the deduced protein sequences were analyzed with EditSeq and Megalign software (Dnastar). The BLASTN program of NCBI was used for database searches.
Determination and cloning of the novel blaCTX-M-53 gene.
The sequences of the entire blaCTX-M-53 gene and the surrounding DNA were obtained by directional genome walking of the pWES-1 plasmid. The blaCTX-M-53 entire gene was PCR amplified by using primers CLON05-686F/PstI (5′-GGGCTGCAGGCTGGAGCCGCACCAGAGCCAAA-3′) and CLON05-686R/XbaI (5′-CCCTCTAGAACGGAATGAGTTTCCCCATTCCG-3′) located outside the open reading frame (ORF) and comprising external endonuclease restriction sites (underlined). The amplification was performed on a 50-μl sample containing plasmidic DNA from pWES-1 (1 μl), primers (50 pmol each), deoxynucleoside triphosphates (100 μM each), DNA polymerase (1 U Isis proofreading DNA polymerase; Qbiogene, Illkirch, France) and its buffer, and dimethyl sulfoxide (10%). The cycling conditions included 10 min of denaturation at 94°C (1 cycle) and 1 min of denaturation at 94°C, 1 min of annealing at 57°C, and 1 min 30 s of polymerization at 72°C (35 cycles), followed by 10 min of extension at 72°C.
The amplified product (1,064 bp) was digested with PstI and XbaI (Roche) and cloned into PstI- and XbaI-digested phagemid pBK-CMV using T4 DNA ligase (Roche). The transformation of the recombinant plasmid was performed using heat shock transformation with DH5α-competent E. coli (Invitrogen). The transformants were selected on Mueller-Hinton agar containing cefotaxime (5 μg/ml). One E. coli transformant containing recombinant plasmid pCTX-M-53 with an insert having 100% nucleotide identity compared to the corresponding sequence of pWES-1 was selected for further studies.
IEF.
IEF was performed with polyacrylamide gels containing ampholines with a pH range of 3.5 to 10 as previously described (6). The following β-lactamases of known isoelectric points (pIs; in parentheses) were used as standards: CTX-M-14 (7.9), CTX-M-1 (8.4), and CTX-M-15 (8.6).
β-Lactamase preparation.
The CTX-M-producing E. coli DH5α(pCTX-M-53) was grown in 6 liters of brain heart infusion broth containing cefotaxime at 2 μg/ml for 18 h at 37°C. The bacteria collected by centrifugation were suspended with 20 mM MES (morpholineethanesulfonic acid)-NaOH (pH 6.0) and disrupted by ultrasonic treatment (four times for 30 s, each time at 20 W). After centrifugation (10,000 × g for 10 min at 4°C), nucleic acids were precipitated by the addition of 0.2 M (7% [vol/vol]) spermine and centrifugation at 48,000 × g for 60 min at 4°C. The clarified supernatant was dialyzed overnight against 20 mM MES-NaOH (pH 6.0). The CTX-M purification was carried out as previously described (7) by ion-exchange chromatography with an SP Sepharose column (Amersham Pharmacia Biotech, Uppsala, Sweden) and gel filtration chromatography with a Superose 12 column (Amersham Pharmacia Biotech). The total protein concentration was estimated by the Bio-Rad protein assay, with bovine serum albumin (Sigma Chemical Co., St. Louis, MO) used as a standard.
The purity of the CTX-M extracts was estimated as previously described (7) by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and staining with Coomassie blue R-250 (Sigma Chemical Co.).
Determination of β-lactamase kinetic constants.
The kinetic constants Km and kcat of the β-lactamases were obtained by a computerized microacidimetric method as previously described (25). The concentrations of the inhibitors (clavulanate and tazobactam) required to inhibit enzyme activity by 50% (IC50s) were determined as described previously with penicillin G (7). The IC50s were monitored with penicillin G (200 mM) as the reporter substrate. The kinetic constants were determined three times. The variation coefficients had a maximum of 10%, except with the CTX-M-9 enzyme for aztreonam and ceftazidime for which it had a maximum of 20%.
Transposition assay.
The mobility of a putative transposable element comprising the blaCTX-M-53 gene was studied by two different methods. (i) A blaCTX-M-53 transposition assay was done in a standard mating assay as described previously (18). The E. coli donor strain HB101 (recA, streptomycin resistant) harboring pOX38-Neor was transformed with plasmid pWES-1. The resulting strain, HB101(pOX38-Neor, pWES-1), was mated with E. coli DH1 (Nalr) either in liquid or on solid medium at different temperatures (25, 30, and 37°C). The transconjugants were selected on Drigalski medium containing Nal (50 μg/ml), kanamycin (40 μg/ml), and cefotaxime (4 μg/ml). (ii) Plasmid pGBG1 was introduced by electroporation into E. coli DH10B containing the natural plasmid pWES-1 and also into S. enterica isolate 04CEB8273SAL. Plasmid pGBG1 contains an insertion cartridge allowing positive selection for tetracycline resistance. This plasmid is dedicated to the isolation of mobile genetic elements in a wide variety of gram-negative bacteria (35). Tetracycline-resistant mutants were selected on LB plates containing the appropriate tetracycline concentration that prevents spontaneous mutants in pGBG1-free isolates. An analysis of the pGBG1 insertion cartridge was performed by PCR in tetracycline-resistant mutants, as previously described (35).
Nucleotide sequence accession number.
The nucleotide sequence of plasmid pWES-1 (11 kb) containing the blaCTX-M-53 gene has been deposited in the GenBank database under accession no. DQ268764.
RESULTS
Antimicrobial susceptibility.
S. enterica serotype Westhampton (antigenic formula 3,10:g,s,t:-) and Senftenberg (antigenic formula 1,3,19:g,s,t:-) isolates were resistant to amoxicillin, ticarcillin, piperacillin, cephalothin, cefamandole, ceftazidime, and ceftriaxone and remained in vitro susceptible to piperacillin-tazobactam, cefoxitin, and imipenem by the disk diffusion method. The isolates were susceptible to all other antibiotic classes tested. An ESBL phenotype was detected by using the double disk diffusion test (data not shown) and ESBL detection Etest strips (Table 2). S. enterica serotype Westhampton isolate 04CEB8273SAL was highly resistant both to ceftriaxone and ceftazidime (MICs of >256 mg/liter). MICs of other β-lactams are shown in Table 2.
TABLE 2.
MICs of β-lactams (Etest) for the S. enterica serotype Westhampton isolate 04CEB8273SAL, E. coli DH10B harboring natural plasmid pWES-1, E. coli DH5α harboring recombinant plasmid pCTX-M-53, E. coli DH5α harboring phagemid pBK-CMV, E. coli DH10B reference strain, and E. coli ATCC 25922 quality control strain
| β-Lactam | MIC (μg/ml) for:
|
|||||
|---|---|---|---|---|---|---|
| Serotype Westhampton 04CEB8273SAL | E. coli DH10B(pWES-1) | E. coli DH5α(pCTX-M-53) | E. coli DH5α(pBK-CMV) | E. coli DH10B | E. coli ATCC 25922 | |
| Ampicillin | >256 | >256 | >256 | 2 | 4 | 4 |
| Amoxicillin-clavulanic acida | 8 | 8 | 8 | 4 | 8 | 8 |
| Ticarcillin | >256 | >256 | >256 | 2 | 8 | 8 |
| Ticarcillin-clavulanic acidb | 128 | 32 | 32 | 2 | 2 | 4 |
| Piperacillin | >256 | >256 | >256 | 1 | 2 | 2 |
| Piperacillin-tazobactamc | 16 | 4 | 2 | 1 | 4 | 2 |
| Cefoxitin | 8 | 4 | 4 | 2 | 4 | 2 |
| Ceftazidime | >256 | 64 | 64 | 0.125 | 0.5 | 0.25 |
| Ceftazidime-clavulanic acidd | 2 | 0.5 | 0.125 | 0.125 | 0.5 | 0.125 |
| Ceftriaxone | >256 | >256 | >256 | <0.06 | 0.125 | 0.06 |
| Cefotaxime-clavulanic acidd | 1 | 0.125 | 0.06 | <0.06 | 0.125 | 0.06 |
| Cefepime | >32 | 32 | >32 | <0.06 | 0.125 | 0.06 |
| Aztreonam | >256 | 128 | 128 | <0.06 | 0.25 | 0.06 |
| Imipenem | 0.5 | 0.5 | 0.5 | 0.25 | 0.5 | 0.25 |
2:1 amoxicillin-clavulanic acid.
2 μg/ml clavulanic acid.
4 μg/ml tazobactam.
4 μg/ml clavulanic acid.
Characterization of the β-lactamase gene(s).
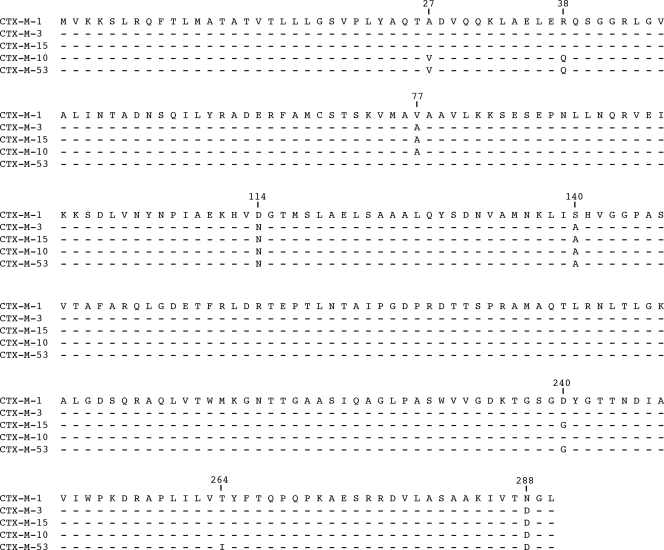
CTX-M consensus PCR performed on all the isolates gave the expected PCR product of 540 bp, whereas PCRs for blaTEM, blaSHV, and the blaOXA-1 group were negative. A CTX-M-1 group-specific PCR assay was then carried out, and the isolates gave the expected PCR product of 874 bp. A sequence analysis of the PCR product amplified from the pWES-1 plasmid revealed 99% homology with the corresponding blaCTX-M-10 gene sequence (GenBank accession number AF255298). To ensure that there were no sequence errors due to primer-directed mismatches at the extremities of the ORF, we determined the entire sequence of the blaCTX-M gene by genome walking on plasmid pWES-1. This novel blaCTX-M gene exhibited 99.5% nucleotide identity with the blaCTX-M-10 gene (AF255298). Its deduced amino acid sequence showed ≥97% identity with the members of the CTX-M-1 phylogenetic group (Fig. 1). This novel CTX-M β-lactamase represented a new member in this group, closely related to CTX-M-10, and thus was named CTX-M-53. The CTX-M-53 protein exhibited different substitutions previously described for CTX-M-1, CTX-M-10, and CTX-M-15 (Fig. 1).
FIG. 1.
Alignment of the deduced amino acid sequences encoded by the novel blaCTX-M-53 gene with those of members of the phylogenetic CTX-M-1 group. Amino acid sequences are from CTX-M-1 (X92506), CTX-M-3 (Y10278), CTX-M-15 (AY044436), CTX-M-10 (AF255298), and CTX-M-53 (DQ268764). The positions of the substitutions are indicated according to the standard numbering scheme for the class A β-lactamases (2). Dashes indicate amino acids identical to those of CTX-M-1.
Molecular typing.
All four S. enterica isolates were tested by XbaI-PFGE. A unique profile, SSFTXB0023, was obtained. Two serotype Westhampton isolates (04CEB8118SAL and 04CEB8273SAL) were tested by MLST and had a sequence type, ST14, frequently found among serotype Senftenberg isolates (http://mlst.ucc.ie/mlst/dbs/Senterica/GetTableInfo_html).
Transfer of β-lactam resistance and plasmid analysis.
β-Lactam resistance could not be transferred by conjugation from any S. enterica isolate to E. coli DH1 (Nalr), either in liquid or on solid medium. However, β-lactam resistance was transferred to E. coli DH10B by electroporation with plasmid DNAs extracted from all S. enterica isolates.
A single plasmid of approximately 11 kb was found in all selected E. coli transformants (data not shown). The β-lactam-resistant E. coli DH10B(pWES-1) exhibited a lower resistance to β-lactams than the parental strain as determined from the MIC (Table 2). These plasmids extracted from the different transformants were further characterized by AvaI restriction analysis showing that they were all identical and around 11 kb in size (data not shown).
IEF and kinetic parameters.
S. enterica isolate 04CEB8273SAL produced a single β-lactamase with a pI of approximately 8.4 (data not shown). The E. coli DH10B transformant (pWES-1) expressed the same β-lactamase (pI, 8.4) (data not shown).
The purified CTX-M proteins appeared on sodium dodecyl sulfate-polyacrylamide gels as a single band (≥97% pure) of 28.6 kDa (data not shown). The substrate profile of CTX-M-53 is shown in Table 3. Kinetic constants exhibited usual values for CTX-M-type ESBLs. Km values were lower for penicillins (10 to 40 μM) than for cephalosporins (80 to 174 μM). Cephalothin was the best substrate (kcat for cephalothin was 15- to 200-fold higher than those for penicillins). A 40- to 145-fold higher kcat value was observed for cefotaxime than for carboxy propyl oxyimino β-lactams (585 versus 4 to 14 s−1). However, the kinetic parameters were unusual, with a significant kcat value for ceftazidime (14 s−1) and a low Km value for aztreonam (14 μM). The enzyme was susceptible to tazobactam (IC50, 2.2 nM) and clavulanate (IC50, 10.0 nM).
TABLE 3.
Substrate profiles of the CTX-M-53 β-lactamases
| β-Lactamase | kcat (s−1) | Km (μM) | kcat/Km (s−1·μM−1) |
|---|---|---|---|
| Penicillin G | 135 | 10 | 13.5 |
| Amoxicillin | 85 | 40 | 2.1 |
| Ticarcillin | 10 | 18 | 0.5 |
| Piperacillin | 90 | 12 | 7.5 |
| Cephalothin | 2,000 | 122 | 16.4 |
| Cefuroxime | 292 | 80 | 3.6 |
| Cefotaxime | 585 | 180 | 3.2 |
| Cefpirome | 710 | 125 | 5.7 |
| Ceftazidime | 14 | 174 | 0.1 |
| Aztreonam | 4 | 14 | 0.3 |
Genetic environment of the blaCTX-M-53 gene.
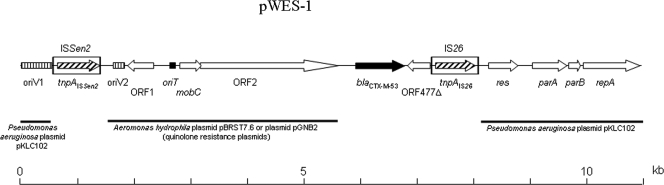
To study the genetic environment of the blaCTX-M-53 gene, we have sequenced the entire pWES-1 (11 kb) on both strands (accession number DQ268764). The plasmid was shown to be 10,908 bp in size and to contain 12 ORFs (Table 4 and Fig. 2). The backbone of the pWES-1 plasmid shares significant nucleotide identity with two different elements (Fig. 2). A 3.3-kb region of pWES-1 shared ≥80% nucleotide identity with a part of the TnCP23 transposon found in the chromosomal integrated plasmid pKLC102 of Pseudomonas aeruginosa (Table 4 and Fig. 2) (24). A second 4-kb region of pWES-1 shared ≥90% nucleotide identity with two similar plasmids carrying the quinolone resistance gene qnrS2 from Aeromonas hydrophila (GenBank accession number EU925817) and from the bacterial community of a wastewater treatment plant (GenBank accession number DQ460733), respectively (4). These two regions were shown to contain different ORFs and structures implicated in plasmid replication and mobilization (Table 4 and Fig. 2). Between these regions of the plasmid backbone are located the blaCTX-M-53 gene, ORF477Δ, and IS26 element on one side and a novel insertion sequence (IS) on the other side (Table 4, Fig. 2). This novel IS of the IS5 family has been deposited in the IS Finder database (http://www-is.biotoul.fr/is.html) and named ISSen2.
TABLE 4.
ORFs and other features of blaCTX-M-53 -carrying plasmid pWES-1
| ORF name/feature | Location (start-stop)a | Size in:
|
G+C content (%) | % Identity/ rangeb | Exhibits significant homologyc to (accession no.): | |
|---|---|---|---|---|---|---|
| bp | Amino acids | |||||
| oriV1 | 1-539 | 539 | NAd | 63.4 | 77/545 | oriV region; transposon TnCP23 of Pseudomonas aeruginosa plasmid pKLC102 (AY257539) |
| ISSen2 | 557-1402 | 846 | NA | 66.2 | 73/763 | Transposase-containing region; Methylobacterium radiotolerans plasmid (CP001006) |
| tnpAISSen2 | 629-1392 | 764 | 254 | 66.7 | 47/255 | IS869 putative transposase; Agrobacterium tumefaciens Ti plasmid (X53945) |
| oriV2 | 1593-1808 | 216 | NA | 66.2 | 95/220 | oriV region; mobilizable IncQ-related plasmid pGNB2 (DQ460733) |
| ORF1 | 2326-1859 | 468 | 155 | 59.6 | 92/155 | Hypothetical protein; plasmid pGNB2 (DQ460733) |
| nic | 2659-2670 | 12 | NA | NA | 100/12 | Putative nic site of oriT; plasmid pGNB2 (DQ460733) |
| mobC | 2783-3169 | 387 | 128 | 68.3 | 74/143 | Auxiliary mobilization protein C; mobilizable IncQ-related plasmid pGNB2 (DQ460733) |
| ORF2 | 3153-5594 | 2,442 | 813 | 66.8 | 95/813 | Relaxase/mobilization nuclease topoisomerase/primase fusion protein; IncQ-related plasmid pGNB2 |
| blaCTX-M-53 | 5892-6767 | 876 | 291 | 54.4 | 98/291 | β-Lactamase CTX-M-57 in Salmonella enterica serotype Typhimurium (DQ810789) |
| ORF477Δ | 7227-6813 | 415 | NA | 57.1 | 100/413 | ORF477 from Klebsiella pneumoniae plasmid pRYCE21 (AY598759) |
| IS26 | 7228-8047 | 820 | NA | 52.3 | 99/820 | IS26 insertion sequence (X00011) |
| tnpAIS26 | 7291-7995 | 705 | 234 | 53.7 | 99/234 | Transposase of IS26 (X00011) |
| res | 8216-8737 | 522 | 173 | 70.1 | 78/173 | Putative resolvase; transposon TnCP23 of Pseudomonas aeruginosa plasmid pKLC102 (AY257539) |
| parA | 8995-9624 | 630 | 209 | 65.6 | 97/210 | Partitioning protein; transposon TnCP23 of Pseudomonas aeruginosa plasmid pKLC102 (AY257539) |
| parB | 9645-9854 | 210 | 69 | 57.6 | 78/73 | Putative plasmid stabilization protein; transposon TnCP23 of Pseudomonas aeruginosa plasmid pKLC102 (AY257539) |
| repA | 9907-10908 | 1,002 | 333 | 64.3 | 91/337 | Replication protein; transposon TnCP23 of Pseudomonas aeruginosa plasmid pKLC102 (AY257539) |
Nucleotide position in the sequence deposited under accession no. DQ268764.
Percentage of identity as returned by BLAST search and range (number of amino acids or nucleotides and gaps) over which the identity value was calculated. Percentage of identity as returned by BLASTN search and range for oriV1, oriV2, oriT, and IS elements.
Significant homology is defined as >20% identity over at least 60% of the length of the protein.
NA, not applicable.
FIG. 2.
Linear representation of pWES-1 (10.9 kb) harboring the blaCTX-M-53 gene. IS elements are indicated by hatched arrows within boxes. The blaCTX-M-53 gene is indicated by a black arrow. Vertically striped and black boxes indicate oriV and oriT regions, respectively. Regions exhibiting significant homology to extant sequences on plasmids and a distance scale are given below the map.
Transposition of the blaCTX-M-53 gene.
The transposition of blaCTX-M-53 was assayed by using plasmid conjugation and a capture vector system. No transposition event of the blaCTX-M-53 gene was obtained despite multiple attempts either in conjugation experiments or with the pGBG1 capture system. Thus, it suggests that the plasmid fragment containing the blaCTX-M-53 gene and flanked by two different ISs (ISSen2 upstream and IS26 downstream) cannot be mobilized by transposition. However, in conjugation experiments, when transconjugants were selected on plates containing only cefotaxime and Nal, numerous transconjugants carrying the pWES-1 plasmid were obtained. This result indicates that the blaCTX-M-53-carrying plasmid pWES-1 was mobilizable in the presence of a helper plasmid (pOX38-Neor) in the donor strain.
DISCUSSION
In our study, we report the characterization of a novel CTX-M-53 ESBL identified in S. enterica serotype Westhampton and Senftenberg strains isolated from cockles in France. According to our knowledge, there were only two previous descriptions of CTX-M ESBLs (CTX-M-3) in serotype Senftenberg in Algeria and Japan (1, 29). In the CTX-M-1 phylogenetic group, the amino acid sequence of CTX-M-53 is closely related to those of CTX-M-10 (31) and CTX-M-34 (GenBank accession number AY515297), which formed a cluster of CTX-M enzymes characterized by the association of residues Val27 and Q38. The sequence of CTX-M-53 differs from those of CTX-M-10 and CTX-M-34 by substitutions Ala77Val and Asp240Gly and from those of all previously reported CTX-M enzymes by the substitution Tyr264Ile.
The substitution Asp240Gly in CTX-M-53 was previously observed in enzymes CTX-M-15 (33), CTX-M-16 (7), CTX-M-25 (28), CTX-M-27 (8), CTX-M-28 (20), CTX-M-29 (39), CTX-M-33 (17), CTX-M-41 (30), CTX-M-43 (10, 11), CTX-M-55 (23), CTX-M-57 (21), CTX-M-64 (AB284167), CTX-M-69 (EU402393), CTX-M-79 (EF426798), and CTX-M-82 (DQ256091). The kinetic study of enzymes CTX-M-15, CTX-M-16, and CTX-M-27 revealed the implication of this mutation in the improvement of the catalytic efficiency against ceftazidime (kcat/Km, 0.001, 0.04, and 0.01 s−1·μM−1, respectively) and of Km values against aztreonam (Km, 11, 17, and 17 μM, respectively) (7, 8, 33). The atomic resolution structure of CTX-M-27 showed that the Asp240Gly substitution allowed broad and coordinated vibrations of the β3 strand (14). Molecular modeling experiments also suggest coordinated motions of the β3 strand with the reactive Ser70 and residues 167 to 170 of the Ω loop which are critical for cephalosporin accommodation (14, 15). This behavior of the binding site and the absence of a negative-charged residue at position 240 probably allowed a deep insertion of ceftazidime in the catalytic pocket, as observed for a ceftazidime-like compound in the crystal structure of CTX-M-16 (14, 15). These different modifications explained the increased activity against ceftazidime observed in the Gly240-harboring CTX-M enzymes.
The residue Tyr264 is conserved in CTX-M enzymes. In the crystallographic structure of CTX-M-9, its side chain is located in the core of the protein and in contact with the CTX-M conserved residues Thr71, Val262, and Met186 (13-15). The atomic resolution of this structure revealed double conformations of the three residues Met186, Val262, and Tyr264, suggesting the high mobility of their side chains (14). In CTX-M-53, the replacement of this residue by Ile264, which is a smaller residue than Tyr264, may increase the mobility of the enzyme in this zone. This zone is located at the vicinity of the 70SXX73K conserved sequence harboring the active serine. Mobility in this zone may therefore influence the catalytic activity of CTX-M-53.
The sequence analysis of plasmid pWES-1 revealed that the plasmid backbone is composed of two different modules for replication initiation and mobilization, respectively. The replication module consists of the parA, parB, and repA genes, and a large part of the oriV region found in an IS6100 composite transposon integrated in the genomic island pKLC102 from Pseudomonas aeruginosa (24). The mobilization module harbors a mobC gene, an ORF2 coding for a putative relaxase protein, a putative oriT region, and an IncQ-like oriV region. This module has been previously described for the IncQ-related plasmid pGNB2 carrying the quinolone resistance gene qnrS2 (4). These two modules are separated by the novel ISSen2 element. The 846-bp length ISSen2 encodes two partially overlapping ORFs with a potential site for −1 frameshifting between both ORFs (5′-AAAAGGGGGGA-3′). This novel IS showed several similarities with IS elements of the IS427 subgroup in the IS5 family (12, 27): (i) 16-bp inverted repeats, (ii) TA target site duplication, and (iii) the putative transposase of ISSen2, which shared ≥40% identity with several members of this subgroup. A preferred target sequence, YTAR (often CTAG), is observed for two subgroups, IS5 and IS427. Thus, the TA target duplication found outside the inverted repeats suggested that this ISSen2 element was inserted alone in the plasmid backbone.
Immediately upstream of the blaCTX-M-53 gene, any genetic structure could explain its acquisition. Interestingly, the region immediately upstream of blaCTX-M-53 showed 99% identity with the 74-bp and 118-bp regions located just upstream of blaCTX-M-3 and blaCTX-M-10, respectively (32). The acquisition of the blaCTX-M-53 gene by the pWES-1 plasmid could be the result of a transposition event related to the downstream IS26 element. A possible explanation is that an IS26 element was originally present in the pWES-1 plasmid and a recombination event may have occurred between the IS26 elements resulting in the integration of the blaCTX-M-53 gene and ORF477Δ. Another possibility is that an IS26 composite transposon carrying the blaCTX-M-53 gene and ORF477Δ formed a cointegrate with the pWES-1 plasmid. Then, after a resolution step which is required to separate the donor and target replicons, the upstream copy of IS26 could have been lost (12, 27).
Considering the origin of CTX-M-53-producing serotype Westhampton and Senftenberg isolates (i.e., living cockles from France), it is relevant to note that this blaCTX-M-53-carrying plasmid harbored similarities with qnrS2-carrying plasmids from aquatic sources (4). Thus, the natural aquatic environment may contribute to genetic exchanges between different bacterial pathogens.
CTX-M-53-producing S. enterica isolates were indistinguishable by PFGE, whereas they were of different serotypes. Serotype Westhampton differs from serotype Senftenberg by the type of O antigens, E1 group (O:3,10) or E4 group (O:1,3,19), respectively. It has been demonstrated that the group E1 and E4 strains have the same chromosomal rfb gene cluster (encoding the enzymes for O-antigen biosynthesis), and the difference between E1 and E4 was proposed to be due to the presence of a gene(s) on a converting phage in E4, although the phage has not been observed (40). The presence of MLST type ST14, which is common in Senftenberg isolates, in the serotype Westhampton isolates under study also speaks in favor of the close genetic relationship between these two serotypes.
In 2004, no human infections due to S. enterica serotypes Westhampton or Senftenberg isolates resistant to ESC were reported by the French National Reference Center for Salmonella. It might be due to the fact that cockles are generally cooked before consumption.
In conclusion, this study reported for the first time the identification of the novel blaCTX-M-53 gene in Salmonella, an important food-borne pathogen. It was located on an 11-kb mobilizable plasmid present in serotype Westhampton and Senftenberg isolates from living cockles in France. The CTX-M-53 enzyme harbors substitutions probably implicated in a greater efficiency against ceftazidime and aztreonam than other CTX-M enzymes. The spread of plasmid-mediated CTX-M-producing strains of Salmonella is of concern, and an enhanced surveillance of ESBL-producing strains should be performed in animals as well as in humans.
Acknowledgments
We thank all the corresponding laboratories of the French Food Safety Agency Salmonella Network for their collaboration. We also thank K. Praud for expert technical assistance and L. Poirel for helpful comments regarding plasmid analysis.
Footnotes
Published ahead of print on 9 March 2009.
REFERENCES
- 1.Ahmed, A. M., H. Nakano, and T. Shimamoto. 2004. The first characterization of extended-spectrum β-lactamase-producing Salmonella in Japan. J. Antimicrob. Chemother. 54:283-284. [DOI] [PubMed] [Google Scholar]
- 2.Ambler, R. P., A. F. W. Coulson, J.-M. Frère, J.-M. Ghuysen, B. Joris, M. Forsman, R. C. Lévesque, G. Tiraby, and S. G. Waley. 1991. A standard numbering scheme for the class A β-lactamases. Biochem. J. 276:269-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arlet, G., T. J. Barrett, P. Butaye, A. Cloeckaert, M. R. Mulvey, and D. G. White. 2006. Salmonella resistant to extended-spectrum cephalosporins: prevalence and epidemiology. Microbes Infect. 8:1945-1954. [DOI] [PubMed] [Google Scholar]
- 4.Bönemann, G., M. Stiens, A. Pühler, and A. Schlüter. 2006. Mobilizable IncQ-related plasmid carrying a new quinolone resistance gene, qnrS2, isolated from the bacterial community of a wastewater treatment plant. Antimicrob. Agents Chemother. 50:3075-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonnet, R. 2004. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonnet, R., C. De Champs, D. Sirot, C. Chanal, R. Labia, and J. Sirot. 1999. Diversity of TEM mutants in Proteus mirabilis. Antimicrob. Agents Chemother. 43:2671-2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonnet, R., C. Dutour, J. L. Sampaio, C. Chanal, D. Sirot, R. Labia, C. De Champs, and J. Sirot. 2001. Novel cefotaximase (CTX-M-16) with increased catalytic efficiency due to substitution Asp-240/Gly. Antimicrob. Agents Chemother. 45:2269-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonnet, R., C. Recule, R. Baraduc, C. Chanal, D. Sirot, C. De Champs, and J. Sirot. 2003. Effect of D240G substitution in a novel ESBL CTX-M-27. J. Antimicrob. Chemother. 52:29-35. [DOI] [PubMed] [Google Scholar]
- 9.Canton, R., and T. M. Coque. 2006. The CTX-M β-lactamase pandemic. Curr. Opin. Microbiol. 9:466-475. [DOI] [PubMed] [Google Scholar]
- 10.Celenza, G., C. Luzi, M. Aschi, B. Segatore, D. Setacci, C. Pellegrini, C. Forcella, G. Amicosante, and M. Perilli. 2008. Natural D240G Toho-1 mutant conferring resistance to ceftazidime: biochemical characterization of CTX-M-43. J. Antimicrob. Chemother. 62:991-997. [DOI] [PubMed] [Google Scholar]
- 11.Celenza, G., C. Pellegrini, M. Caccamo, B. Segatore, G. Amicosante, and M. Perilli. 2006. Spread of bla(CTX-M-type) and bla(PER-2) β-lactamase genes in clinical isolates from Bolivian hospitals. J. Antimicrob. Chemother. 57:975-978. [DOI] [PubMed] [Google Scholar]
- 12.Chandler, M., and J. Mahillon. 2002. Insertion sequences revisited, p. 305-366. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, DC.
- 13.Chen, Y., B. Shoichet, and R. Bonnet. 2005. Structure, function, and inhibition along the reaction coordinate of CTX-M β-lactamases. J. Am. Chem. Soc. 127:5423-5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, Y., J. Delmas, J. Sirot, B. Shoichet, and R. Bonnet. 2005. Atomic resolution structures of CTX-M β-lactamases: extended spectrum activities from increased mobility and decreased stability. J. Mol. Biol. 348:349-362. [DOI] [PubMed] [Google Scholar]
- 15.Delmas, J., Y. Chen, F. Prati, F. Robin, B. K. Shoichet, and R. Bonnet. 2008. Structure and dynamics of CTX-M enzymes reveal insights into substrate accommodation by extended-spectrum β-lactamases. J. Mol. Biol. 375:192-201. [DOI] [PubMed] [Google Scholar]
- 16.Drieux, L., F. Brossier, W. Sougakoff, and V. Jarlier. 2008. Phenotypic detection of extended-spectrum β-lactamase production in Enterobacteriaceae: review and bench guide. Clin. Microbiol. Infect. 14:90-103. [DOI] [PubMed] [Google Scholar]
- 17.Galani, I., M. Souli, Z. Chryssouli, and H. Giamarellou. 2007. Detection of CTX-M-15 and CTX-M-33, a novel variant of CTX-M-15, in clinical Escherichia coli isolates in Greece. Int. J. Antimicrob. Agents 29:598-600. [DOI] [PubMed] [Google Scholar]
- 18.Galimand, M., S. Sabtcheva, P. Courvalin, and T. Lambert. 2005. Worldwide disseminated armA aminoglycoside resistance methylase gene is borne by composite transposon Tn1548. Antimicrob. Agents Chemother. 49:2949-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grimont, P. A. G., and F.-X. Weill. 2007. Antigenic formulas of the Salmonella serovars, 9th ed. W. H. O. Collaborating Center for Reference and Research on Salmonella, Institut Pasteur, Paris, France.
- 20.Hasman, H., D. Mevius, K. Veldman, I. Olesen, and F. M. Aarestrup. 2005. β-Lactamases among extended-spectrum β-lactamase (ESBL)-resistant Salmonella from poultry, poultry products and human patients in The Netherlands. J. Antimicrob. Chemother. 56:115-121. [DOI] [PubMed] [Google Scholar]
- 21.Hopkins, K. L., E. Karisik, and J. K. Wardle. 2008. Identification of novel plasmid-mediated extended-spectrum β-lactamase CTX-M-57 in Salmonella enterica serovar Typhimurium. Int. J. Antimicrob. Agents 31:85-86. [DOI] [PubMed] [Google Scholar]
- 22.Kérouanton, A., M. Marault, R. Lailler, F. X. Weill, C. Feurer, E. Espie, and A. Brisabois. 2007. Pulsed-field gel electrophoresis subtyping database for foodborne Salmonella enterica serotype discrimination. Foodborne Pathog. Dis. 4:293-303. [DOI] [PubMed] [Google Scholar]
- 23.Kiratisin, P., A. Apisarnthanarak, P. Saifon, C. Laesripa, R. Kitphati, and L. M. Mundy. 2007. The emergence of a novel ceftazidime-resistant CTX-M extended-spectrum β-lactamase, CTX-M-55, in both community-onset and hospital-acquired infections in Thailand. Diagn. Microbiol. Infect. Dis. 58:349-355. [DOI] [PubMed] [Google Scholar]
- 24.Klockgether, J., O. Reva, K. Larbig, and B. Tümmler. 2004. Sequence analysis of the mobile genome island pKLC102 of Pseudomonas aeruginosa C. J. Bacteriol. 186:518-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Labia, R., J. Andrillon, and F. Le Goffic. 1973. Computerized microacidimetric determination of β-lactamase Michaelis-Menten constants. FEBS Lett. 33:42-44. [DOI] [PubMed] [Google Scholar]
- 26.Livermore, D. M., R. Canton, M. Gniadkowski, P. Nordmann, G. Maria Rossolini, G. Arlet, T. M. Coque, I. Kern-Zdanowicz, F. Luzzaro, L. Poirel, and N. Woodford. 2007. CTX-M: changing the face of ESBLs in Europe. J. Antimicrob. Chemother. 59:165-174. [DOI] [PubMed] [Google Scholar]
- 27.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munday, C. J., D. A. Boyd, N. Brenwald, M. Miller, J. M. Andrews, R. Wise, M. R. Mulvey, and P. M. Hawkey. 2004. Molecular and kinetic comparison of the novel extended-spectrum β-lactamases CTX-M-25 and CTX-M-26. Antimicrob. Agents Chemother. 48:4829-4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naas, T., A. Lezzar, C. Bentchouala, F. Smati, J.-M. Scheftel, H. Monteil, and P. Nordmann. 2005. Multidrug-resistant Salmonella enterica serotype Senftenberg isolates producing CTX-M β-lactamases from Constantine, Algeria. J. Antimicrob. Chemother. 56:439-440. [DOI] [PubMed] [Google Scholar]
- 30.Navon-Venezia, S., I. Chmelnitsky, A. Leavitt, and Y. Carmeli. 2008. Dissemination of the CTX-M-25 family β-lactamases among Klebsiella pneumoniae, Escherichia coli and Enterobacter cloacae and identification of the novel enzyme CTX-M-41 in Proteus mirabilis in Israel. J. Antimicrob. Chemother. 62:289-295. [DOI] [PubMed] [Google Scholar]
- 31.Oliver, A., J. C. Perez-Diaz, T. M. Coque, F. Baquero, and R. Canton. 2001. Nucleotide sequence and characterization of a novel cefotaxime-hydrolyzing β-lactamase (CTX-M-10) isolated in Spain. Antimicrob. Agents Chemother. 45:616-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oliver, A., T. M. Coque, D. Alonso, A. Valverde, F. Baquero, and R. Canton. 2005. CTX-M-10 linked to a phage-related element is widely disseminated among Enterobacteriaceae in a Spanish hospital. Antimicrob. Agents Chemother. 49:1567-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poirel, L., M. Gniadkowski, and P. Nordmann. 2002. Biochemical analysis of the ceftazidime-hydrolysing extended-spectrum β-lactamase CTX-M-15 and of its structurally related β-lactamase CTX-M-3. J. Antimicrob. Chemother. 50:1031-1034. [DOI] [PubMed] [Google Scholar]
- 34.Ribot, E. M., M. A. Fair, R. Gautom, D. N. Cameron, S. B. Hunter, B. Swaminathan, and T. J. Barrett. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59-67. [DOI] [PubMed] [Google Scholar]
- 35.Schneider, D., D. Faure, M. Noirclerc-Savoye, A. C. Barriere, E. Coursange, and M. Blot. 2000. A broad-host-range plasmid for isolating mobile genetic elements in Gram-negative bacteria. Plasmid 44:201-207. [DOI] [PubMed] [Google Scholar]
- 36.Su, L. H., C. Chu, A. Cloeckaert, and C. H. Chiu. 2008. An epidemic of plasmids? Dissemination of extended-spectrum cephalosporinases among Salmonella and other Enterobacteriaceae. FEMS Immunol. Med. Microbiol. 52:155-168. [DOI] [PubMed] [Google Scholar]
- 37.Weill, F.-X., J.-D. Perrier-Gros-Claude, M. Demartin, S. Coignard, and P. A. D. Grimont. 2004. Characterization of extended-spectrum β-lactamase (CTX-M-15)-producing strains of Salmonella enterica isolates in France and Senegal. FEMS Microbiol. Lett. 238:353-358. [DOI] [PubMed] [Google Scholar]
- 38.Weill, F.-X., R. Lailler, K. Praud, A. Kérouanton, L. Fabre, A. Brisabois, P. A. D. Grimont, and A. Cloeckaert. 2004. Emergence of extended-spectrum-β-lactamase (CTX-M-9)-producing multiresistant strains of Salmonella enterica serotype Virchow in poultry and humans in France. J. Clin. Microbiol. 42:5767-5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu, Y., S. Ji, Y. Chen, W. Zhou, Z. Wei, L. Li, and Y. Ma. 2007. Resistance of strains producing extended-spectrum β-lactamases and genotype distribution in China. J. Infect. 54:53-57. [DOI] [PubMed] [Google Scholar]
- 40.Xiang, S. H., A. M. Haase, and P. R. Reeves. 1993. Variation of the rfb gene clusters in Salmonella enterica. J. Bacteriol. 175:4877-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]