Abstract
Since the discovery of qnrA in 1998, two additional qnr genes, qnrB and qnrS, have been described. These three plasmid-mediated genes contribute to quinolone resistance in gram-negative pathogens worldwide. A clinical strain of Proteus mirabilis was isolated from an outpatient with a urinary tract infection and was susceptible to most antimicrobials but resistant to ampicillin, sulfamethoxazole, and trimethoprim. Plasmid pHS10, harbored by this strain, was transferred to azide-resistant Escherichia coli J53 by conjugation. A transconjugant with pHS10 had low-level quinolone resistance but was negative by PCR for the known qnr genes, aac(6′)-Ib-cr and qepA. The ciprofloxacin MIC for the clinical strain and a J53/pHS10 transconjugant was 0.25 μg/ml, representing an increase of 32-fold relative to that for the recipient, J53. The plasmid was digested with HindIII, and a 4.4-kb DNA fragment containing the new gene was cloned into pUC18 and transformed into E. coli TOP10. Sequencing showed that the responsible 666-bp gene, designated qnrC, encoded a 221-amino-acid protein, QnrC, which shared 64%, 42%, 59%, and 43% amino acid identity with QnrA1, QnrB1, QnrS1, and QnrD, respectively. Upstream of qnrC there existed a new IS3 family insertion sequence, ISPmi1, which encoded a frameshifted transposase. qnrC could not be detected by PCR, however, in 2,020 strains of Enterobacteriaceae. A new quinolone resistance gene, qnrC, was thus characterized from plasmid pHS10 carried by a clinical isolate of P. mirabilis.
Plasmid-mediated quinolone resistance was first described for a ciprofloxacin-resistant strain of Klebsiella pneumoniae in 1998 (15). The responsible gene, qnr (later named qnrA), was located on plasmid pMG252, which encodes multidrug resistance proteins. qnrB and qnrS were discovered in 2005 and 2006, respectively, and mediated similar levels of ciprofloxacin resistance (9, 11). Qnr proteins belong to the pentapeptide repeat protein (PRP) family and protect DNA gyrase and topoisomerase IV from quinolone inhibition (26, 27, 28). qnr genes show a high level of diversity; there are at least 6 qnrA, 20 qnrB, and 3 qnrS alleles reported, with one or more amino acid alterations within each family (12; http://www.lahey.org/qnrStudies). More recently, qnrD was found in Salmonella isolates (3). qnr genes are widely distributed in clinical Enterobacteriaceae isolates around the world and are usually associated with mobile elements (21). There were also qnr-like genes found on the chromosomes of Vibrio vulnificus, Vibrio parahaemolyticus, Photobacterium profundum, Stenotrophomonas maltophilia, and gram-positive genera such as Enterococcus, Listeria, Clostridium, and Bacillus (1, 17, 22, 24). The wide distribution of qnr genes in different species of Enterobacteriaceae and their high degree of diversity raise the concern that there might be more qnr genes that have not yet been discovered. In this study, a new plasmid-mediated quinolone resistance gene, qnrC, was found on and cloned from a transferable plasmid, pHS10, in a clinical isolate of Proteus mirabilis. The qnrC gene, however, is rare and was not detected by PCR in 2,020 strains of Enterobacteriaceae isolated from Shanghai.
MATERIALS AND METHODS
Strains and plasmids.
Proteus mirabilis 06-489 was isolated from a urine specimen of an outpatient with a urinary tract infection in 2006 in Huashan Hospital, a teaching hospital of Fudan University in Shanghai. Escherichia coli J53 Azir (resistant to azide) was used as the recipient strain in conjugation experiments. Plasmids pUC18 (Ampr [resistance to ampicillin]) and pHSG398 (Chlr [resistance to chloramphenicol]) (Takara Bio, Otsu, Japan) were used as cloning vectors. E. coli TOP10 (Invitrogen) was used for cloning. Cultures were routinely grown in Luria-Bertani broth. Culture plates contained tryptic soy agar (TSA) or Mueller-Hinton agar (Oxoid, Basingstoke, England). Selective media contained sodium azide (200 μg/ml) together with either ampicillin (100 μg/ml), ciprofloxacin (0.06 μg/ml), or sulfamethoxazole (300 μg/ml).
A total of 2,020 nonduplicate clinical isolates of Enterobacteriaceae, regardless of their susceptibilities to antimicrobials, were collected from Huashan Hospital between 2005 and 2007, including 496 strains of K. pneumoniae, 63 strains of Klebsiella oxytoca, 492 strains of E. coli, 204 strains of Enterobacter spp. (186 strains of Enterobacter cloacae and 18 strains of Enterobacter aerogenes), 259 strains of P. mirabilis, 34 strains of Proteus vulgaris, 203 strains of Serratia marcescens, 137 strains of Morganella morganii, 63 strains of Citrobacter spp., 33 strains of Providencia spp., and 36 strains of other Enterobacteriaceae.
Cloning and nucleotide sequence analysis.
Plasmid DNA was isolated from an E. coli J53 derivative containing plasmid pHS10 by use of a Plasmid Midi kit (Qiagen GmbH, Hilden, Germany) and then ligated, after digestion with EcoRI or HindIII, into pUC18. The recombinants were transformed into E. coli TOP10, with selection on TSA plates containing ampicillin and ciprofloxacin. A clone carrying a plasmid with an approximately 4.4-kb HindIII insert, designated pHS11, was isolated, and the nucleotide sequence of the 4,409-bp insert was determined. A PCR fragment of 904 bp, which was internal to the HindIII fragment and encompassed the entire transcription unit of the PRP gene, was amplified with pHS10 as the template, using primers qnrCBam and qnrCSal (Table 1). The PCR product was digested with BamHI and SalI, cloned into pHSG398, and transformed into E. coli TOP10. The resultant construct, plasmid pHS12, was isolated, and its nucleotide sequence was verified. A gene encoding the PRP carried by pHS12 was designated qnrC.
TABLE 1.
Primers designed and used in this study
| Primer | Sequence |
|---|---|
| qnrCBam | GGTGGATCCGTTTAACAACCGTCGGCT |
| qnrCSal | AATGTCGACGCCTTGAAGATGATTCGCT |
| qnrC-ATGtoACG-F | GATGCTAAATTCACGGGTTGTACAT |
| qnrC-ATGtoACG-R | ATGTACAACCCGTGAATTTAGCATC |
| qnrC-TTGtoATG-F | GAGGTTATAACAATGAATTATTCCC |
| qnrC-TTGtoATG-R | GGGAATAATTCATTGTTATAACCTC |
| qnrC-TTGtoTCG-F | GAGGTTATAACATCGAATTATTCCC |
| qnrC-TTGtoTCG-R | GGGAATAATTCGATGTTATAACCTC |
| qnrC-TTGtoTTA-F | GAGGTTATAACATTAAATTATTCCCATAAAACGTACG |
| qnrC-TTGtoTTA-R | CGTACGTTTTATGGGAATAATTTAATGTTATAACCTC |
| qnrC-ATTtoATG-F | GGCTGTAGATGTTAGTCTTAATTTAAATGAATCAAGAGGTTATAACATTG |
| qnrC-ATTtoATG-R | CAATGTTATAACCTCTTGATTCATTTAAATTAAGACTAACATCTACAGCC |
| qnrC-ATTtoTTA-F | GGCTGTAGATGTTAGTCTTAATTTAATTAAATCAAGAGGTTATAACATTG |
| qnrC-ATTtoTTA-R | CAATGTTATAACCTCTTGATTTAATTAAATTAAGACTAACATCTACAGCC |
| qnrC-F | GGGTTGTACATTTATTGAATC |
| qnrC-R | TCCACTTTACGAGGTTCT |
Determination of qnrC start codon.
The putative translation initiation codons ATG, TTG, and ATT of qnrC were mutated using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) to identify the start codon (Fig. 1). Mutagenesis was carried out on the pHS12 plasmid following the experimental protocol of the manufacturer, and the transformant was selected with chloramphenicol. Complementary forward and reverse primers were designed to alter the DNA sequence from 5′ATG to 5′ACG, from 5′TTG to 5′ATG, 5′TTA, and 5′TCG, and from 5′ATT to 5′ATG and 5′TTA (Table 1). The resultant plasmid constructs were called pHS12-ATGtoACG, pHS12-TTGtoATG, pHS12-TTGtoTTA, pHS12-TTGtoTCG, pHS12-ATTtoATG, and pHS12-ATTtoTTA, respectively.
FIG. 1.
Three putative start codons of qnrC found using the alternative initiation codon finder in ORF Finder (http://www.ncbi.nlm.nih.gov/projects/gorf/). Putative start codons are underlined, and TTG is suggested as the start codon of qnrC.
Susceptibility testing.
MICs were determined by the CLSI agar dilution methodology and interpreted according to CLSI guidelines (7). The Etest (Biodisk AB, Solna, Sweden) was used to detect minimal changes in nalidixic acid, ciprofloxacin, and levofloxacin susceptibilities for P. mirabilis 06-489 and E. coli strains containing pHS10 and recombinant plasmids. E. coli ATCC 25922 was used as a quality control strain in susceptibility testing experiments.
Screening for qnrC in clinical strains.
PCR was used to investigate the prevalence of qnrC in 2,020 clinical strains, using primers qnrC-F and qnrC-R (Table 1). PCR conditions were 94°C for 30 s, 50°C for 30 s, and 72°C for 30 s for 30 cycles. Amplification products were detected by electrophoresis on a 1% agarose gel with ethidium bromide and were photographed under UV light. P. mirabilis 06-489 was used as a positive control and generated a 447-bp PCR product.
Nucleotide sequence accession number.
The nucleotide sequence in plasmid pHS10 containing qnrC has been submitted to GenBank and assigned accession number EU917444.
RESULTS
Cloning of qnrC.
A clinical strain of P. mirabilis, strain 06-489, was susceptible to quinolones and to cephalosporins, such as cefuroxime, cefotaxime, and ceftazidime, but resistant to ampicillin, gentamicin, chloramphenicol, tetracycline, sulfamethoxazole, and trimethoprim. Low-level ciprofloxacin resistance could be transferred from P. mirabilis 06-489 to E. coli J53 Azir by conjugation. The plasmid responsible was termed pHS10. The ciprofloxacin MIC for the clinical strain and a J53/pHS10 transconjugant was 0.25 μg/ml, an increase of 32-fold relative to that for J53 (Table 2). pHS10 was about 120 kb in size and was negative for known plasmid-mediated quinolone resistance determinants, i.e., qnrA, qnrB, qnrS, aac(6′)-Ib-cr, and qepA, by PCR amplification. A 4,409-bp HindIII DNA fragment and a 904-bp PCR product were cloned into pUC18 and pHSG398, respectively, to obtain pHS11 and pHS12. E. coli TOP10 cells containing pHS11 and pHS12 had the same MICs for ciprofloxacin (0.125 μg/ml) and levofloxacin (0.19 μg/ml) (Table 3). The DNA sequence of the 4,409-bp HindIII DNA insert was determined with recombinant plasmid pHS11. Four open reading frames (ORFs) were found by ORF Finder (http://www.ncbi.nlm.nih.gov/projects/gorf/). The ORFs showed that there was a conserved domain belonging to the PRP family, with relatively high identities (>40%) with qnrA, qnrB, and qnrS, so we considered this a qnr-like gene. Three putative start codons were found in the ORF: peptide synthesis of 178, 221, or 228 amino acids (aa) may start from ATG, TTG, or ATT, respectively (Fig. 1).
TABLE 2.
Susceptibilities of P. mirabilis 06-489 and E. coli transconjugant to three quinolones and other antimicrobialsa
| Strain | MIC (μg/ml)
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NAL | CIP | LEV | AMP | CXM | CTX | CAZ | FEP | FOX | GM | AMK | SMZ | TMP | CHL | TET | |
| P. mirabilis 06-489 | 16 | 0.25 | 0.50 | 64 | 4 | ≤0.06 | ≤0.06 | ≤0.06 | 2 | 16 | 8 | >1,024 | >128 | 16 | 128 |
| E. coli J53/pHS10 | 16 | 0.25 | 0.25 | 128 | 8 | ≤0.06 | 0.06 | ≤0.06 | 4 | 4 | 0.5 | >1,024 | >128 | 64 | 128 |
| E. coli J 53 | 4 | 0.008 | 0.023 | 4 | 8 | ≤0.06 | ≤0.06 | ≤0.06 | 4 | 0.125 | 0.25 | 16 | 0.25 | 8 | 1 |
AMK, amikacin; AMP, ampicillin; CAZ, ceftazidime; CHL, chloramphenicol; CIP, ciprofloxacin; CTX, cefotaxime; CXM, cefuroxime; FEP, cefepime; FOX, cefoxitin; GEN, gentamicin; LEV, levofloxacin; NAL, nalidixic acid; SMZ, sulfamethoxazole; TET, tetracycline; TMP, trimethoprim.
TABLE 3.
Susceptibilities of E. coli TOP10 cells harboring pHS12 derivatives to nalidixic acid, ciprofloxacin, and levofloxacin
| Plasmid in E. coli TOP10 | Amino acid changea | Changed to another potential start codon | MIC (μg/ml)
|
||
|---|---|---|---|---|---|
| Nalidixic acid | Ciprofloxacin | Levofloxacin | |||
| No plasmid | NA | NA | 1.0 | 0.002 | 0.006 |
| pHS12 | NA | NA | 4.0 | 0.125 | 0.19 |
| pHS12-ATGtoACG | M to T | No | 4.0 | 0.125 | 0.19 |
| pHS12-TTGtoATG | L to Mb | Yes | 4.0 | 0.19 | 0.19 |
| pHS12-TTGtoTCG | L to S | No | 1.5 | 0.006 | 0.016 |
| pHS12-TTGtoTTA | L to L | No | 1.0 | 0.003 | 0.006 |
| pHS12-ATTtoATG | I to Mb | Yes | 4.0 | 0.125 | 0.125 |
| pHS12-ATTtoTTA | I to L | No | 3.0 | 0.125 | 0.125 |
NA, not applicable.
When TTG or ATT is used as an initiation codon, it is decoded as methionine (M).
Determination of start codon of qnrC.
The 178-aa PRP protein expressed starting at ATG of qnrC was 40 or 48 amino acids shorter than QnrA1, QnrB1, and QnrS1. When ATG was changed to ACG, a noninitiation codon, the MICs of E. coli TOP10 harboring pHS12-ATGtoACG for ciprofloxacin and levofloxacin were similar to those with pHS12 (Table 3), indicating that the translation of mRNA still occurred and the function of the QnrC protein was not affected by the mutation. Thus, this ATG was not the start codon of qnrC.
A 221-aa protein translated starting at TTG of qnrC had substantial similarity to QnrA1 (64%), QnrB1 (42%), and QnrS1 (59%) (Fig. 2). When TTG was mutated to ATG, the MICs of E. coli TOP10 harboring pHS12-TTGtoATG for ciprofloxacin and levofloxacin were similar to those with pHS12. When TTG was replaced with TCG or TTA, which are not start codons, the ciprofloxacin MIC for E. coli TOP10 harboring pHS12-TTGtoTCG or pHS12-TTGtoTTA decreased from 0.125 μg/ml to 0.003 or 0.006 μg/ml (Table 3). Furthermore, a predicted Shine-Dalgarno sequence (AAGAGG) was found to be located eight nucleotides upstream of the TTG initiation codon, an optimum distance for initiation of translation.
FIG. 2.
Sequence alignment of QnrC and its structural homologs, QnrA1, QnrB1, QnrS1, and QnrD. Residues conserved in at least two proteins are shown with a gray background, and the pentapeptide repeat sequences in QnrC are underlined. The residue (G56) that appears to link the two distinct domains of Qnr proteins is boxed for all proteins.
Another possible start codon, ATT, 21 bp upstream of TTG in qnrC, was mutated to ATG or the non-start-codon TTA. The MICs of E. coli TOP10 harboring pHS12-ATTtoATG or pHS12-ATTtoTTA for ciprofloxacin and levofloxacin were similar to those with pHS12 (Table 3), indicating that ATT was also not a start codon in qnrC.
Therefore, TTG appeared to be the start codon of qnrC, which was 666 bp in length and encoded a 221-aa PRP.
Identity of QnrC with QnrA1, QnrB1, QnrS1, and other PRPs.
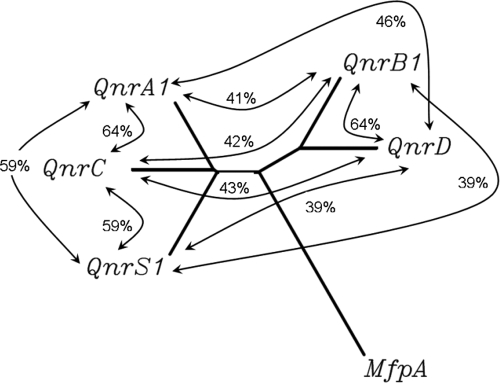
qnrC had 60%, 45%, 59%, and 32% nucleotide identity with qnrA1, qnrB1, qnrS1, and qnrD, respectively, while QnrC shared 64%, 42%, 59%, and 43% amino acid identity with QnrA1, QnrB1, QnrS1, and QnrD, respectively, by Clustal W alignment (http://align.genome.jp/) (Fig. 3).
FIG. 3.
Amino acid relationships of QnrC with QnrA1, QnrB1, QnrS1, and QnrD. Sequence identities are shown in the figure. MfpA is found in Mycobacterium tuberculosis. The unrooted dendrogram was generated using Clustal W (http://align.genome.jp/).
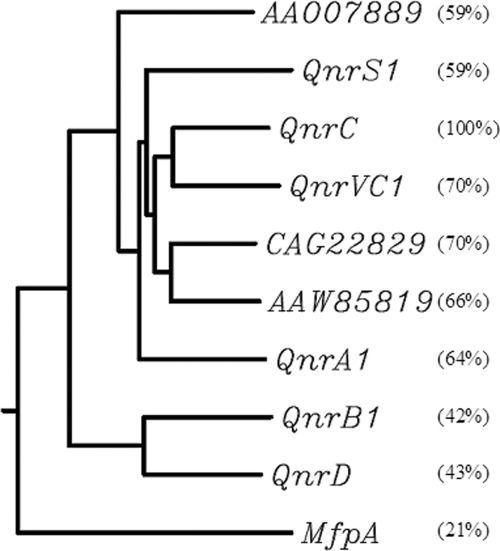
qnrC had 58 to 68% identities with DNA sequences found in the chromosome or plasmid in Vibrio cholerae, P. profundum, Vibrio fischeri, and Vibrio vulnificus (GenBank accession no. EU436855, CR378678, CP000020, and AE016796, respectively), while QnrC shared 59 to 70% amino acid identities with the PRPs found in the above strains (ACC54440, CAG22829, AAW85819, and AAO07889, respectively) (Fig. 4).
FIG. 4.
Clustal W analysis of QnrC and other PRPs known to affect DNA gyrase. Amino acid sequence identities are indicated in parentheses. QnrD (ACG70184), QnrVC1 (ACC54440), CAG22829, AAW85819, AAO07889, and MfpA are found in Salmonella enterica, V. cholerae strain VC627, P. profundum SS9, V. fischeri ES114, V. vulnificus CMCP6, and M. tuberculosis, respectively. The dendrogram was generated using Clustal W (http://align.genome.jp/).
Genetic environment of qnrC.
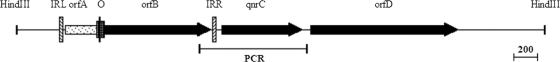
There were four ORFs found in the 4.4-kb HindIII DNA fragment containing qnrC (Fig. 5). qnrC was found downstream from orfA and orfB, which contained a new insertion sequence belonging to the IS51 group of the IS3 family. orfA and orfB were partially overlapping and arranged in reading phases 0 and −1, respectively. The coding potential of orfA was 107 aa and that of orfB was 318 aa, and the putative OrfAB protein was 405 aa long. OrfAB was a fusion protein with an identity of 58% to IS51 (M14365), which has transposase activity produced by the −1 programmed ribosomal frameshift (PRF-1) (4). A predicted −1 translational frameshift signal (TTTTG) associated with an apical loop-internal loop pseudoknot was deduced near the 3′ end of orfA and the 5′ end of orfB, suggesting the existence of frameshifted products that could be responsible for transposition of this new IS element (5, 16).
FIG. 5.
Genetic environment of the qnrC gene in the 4.4-kb HindIII fragment of plasmid pHS11. E. coli TOP10 cells containing pHS11 had a ciprofloxacin MIC of 0.125 μg/ml and a levofloxacin MIC of 0.19 μg/ml. ISPmi1 is enclosed by two inverted repeats (hatched boxes), labeled IRL (inverted repeat left) and IRR (inverted repeat right). The grid box labeled “O” indicates the partially overlapping ORFs orfA and orfB, and the black stick within the grid box is the location of the putative −1 translational frameshift signal, TTTTG. orfD encodes an amidase-like protein. The marked PCR region is the PCR amplification region containing qnrC.
The new insertion sequence has been designated ISPmi1 according to guidelines for IS nomenclature for different bacterial species (http://www-is.biotoul.fr/). ISPmi1 is 1,306 bp in length and has a pair of 26-bp imperfect inverted repeats at its termini, with a conserved 5′-TG—-CA-3′ sequence. A 3-bp direct repeat (ATA) flanks the element, possibly as the result of a transposition event. In ISPmi1, a putative Shine-Dalgarno sequence was found upstream from the initiation codon of orfA, but no such sequence was present in the upstream region of orfB.
Downstream from qnrC was orfD, which encoded a protein with highest identity (42%) to an amidase family protein from Methylococcus capsulatus (YP113430) (Fig. 5).
Effect of QnrC on quinolone susceptibility.
Like QnrA, QnrB, and QnrS, QnrC provided low-level resistance to quinolones, with a ciprofloxacin MIC of 0.25 μg/ml, in E. coli J53 (Table 2).
Prevalence of qnrC.
Unexpectedly, no qnrC PCR products were detected by PCR amplification of DNAs from 2,020 clinical strains of Enterobacteriaceae.
DISCUSSION
In this study, qnrC was discovered from a clinical strain of P. mirabilis. qnrC is a new plasmid-borne qnr gene, in addition to three existing families, qnrA, qnrB, and qnrS. The name qnrC was designated according to the recently published qnr numbering proposal by Jacoby et al. (12). qnrC differs substantially from existing families, with >30% (40 to 68%) differences in comparison to qnrA, qnrB, qnrS, and qnrD and also >30% (36 to 58%) differences in derived amino acid sequences. Like other Qnr determinants, QnrC provides low-level quinolone resistance, with a ciprofloxacin MIC of 0.25 μg/ml for E. coli J53.
Like other Qnr proteins found in gram-negative species, QnrC has a consensus sequence of (A,C)(D,N)(L,F)XX and contains two domains, of 11 and 32 units each, connected by a single glycine (G56), which is not conserved in PRPs from the gram-positive species studied (Fig. 3) (22, 26). AUG is the initiation codon used most frequently in prokaryotic genes. However, there are other non-AUG initiation codons, such as GUG and UUG, with frequencies of 8% and 1% in E. coli, respectively. Protein synthesis is still thought to be initiated with methionine because these codons are all decoded by the initiator fMet-tRNAfMet and translated as formylmethionine (14). Previous studies showed that initiation at AUG in E. coli is more efficient than that at the non-AUG codons (25). QnrC was deduced to initiate at UUG, an uncommon start codon, but it conferred low-level ciprofloxacin resistance (MIC, 0.25 μg/ml) similar to that of QnrA1, QnrB1, and QnrS1 (9, 11, 15).
qnrA and sometimes qnrB are associated with ISCR1 (IS common region 1; previously also called orf513), which is embedded in class 1 integrons (10, 13, 29). Some qnrB alleles are linked to orf1005, which encodes another putative integrase (11). A novel qnrB19 allele is associated with an ISEcp1-like insertion element that is able to mobilize the qnrB19 gene (2). Although qnrS has not been found on an integron thus far, it has been linked to insertion sequences such as IS26, an IS2 homolog, or ISEcl2, a novel insertion element belonging to the IS3 family (6, 10, 19, 20). qnrC is associated with ISPmi1, a member of the IS3 family, as well.
Members of the IS3 family are spread widely in more than 40 bacterial species and are characterized by lengths of between 1,200 and 1,550 bp. This family is divided into five subgroups, IS3, IS51, IS150, IS2, and IS407, based on alignment of the various OrfB sequences (4). OrfB and OrfAB of ISPmi1 contain a highly conserved DDE motif, a catalytic site of IS3 transposase. The spacing between the second catalytic aspartic acid (D) and glutamic acid (E) is conserved (35 aa), and a lysine (K) is present 7 aa downstream of the glutamic acid.
As a characteristic of the IS3 family, PRF-1 is a nonconventional translation phenomenon induced by a stimulatory signal. It involves the backward slippage of the ribosome by one nucleotide at a given point on the message. ISPmi1 shares a common genetic organization with members of the IS51 group, in which a potential tetrameric frameshift motif is accompanied by an elaborate frameshift stimulator, the apical loop-internal loop pseudoknot. Conserved structural elements similar to those in IS3411 and IS629 have been demonstrated experimentally to be involved in the control of gene expression by translational frameshifting, and the OrfAB transposase is indeed synthesized via PRF-1 on the predicted motif (5, 16).
Although no other qnrC gene was detected among 2,020 clinical strains of Enterobacteriaceae, two homologs, named qnrVC1 and qnrVC2 (8), are found in GenBank from two strains of V. cholerae O1, isolated from Brazil and Vietnam, both with 67% nucleotide identity to qnrC. qnrVC1 was located in gene cassettes of a class 1 integron and downstream of aadA2 in the Brazil strain (GenBank accession no. EU436855). qnrVC2 (GenBank accession no. AB200915) was also located on an integron, in plasmid pVN84, in the Vietnam strain, upstream from repA, orf1, and intI and downstream from dhfr6. QnrVC1 (GenBank accession no. ACC54440) has a high amino acid identity of 70% with QnrC. qnrVC2 could not be translated into a Qnr protein, since it has three nucleotide insertions and one nucleotide deletion compared to functional qnr genes.
It has been shown that Shewanella algae and Vibrio splendidus are probably the progenitors of qnrA- and qnrS-like genes (1, 18). QnrB-like proteins were recently found in Stenotrophomonas maltophilia (23) and members of the Vibrionaceae family (17). The 4.4-kb HindIII DNA fragment in this study showed strong nucleotide homology with chromosomal or plasmid sequences in the Vibrionaceae family, indicating that waterborne Vibrionaceae organisms might be the source of QnrC. Notably, the −1 to 206 nucleotide sequence of ISPmi1 showed 69% identity to the Shewanella sp. genome (GenBank accession no. CP000469), qnrC showed high identities with genomic sequences found in waterborne Vibrionaceae, such as P. profundum, V. cholerae, V. fischeri, and V. vulnificus, and the 3′ end of the fragment from nucleotides 3723 to 4409 showed over 80% identity with genomes of Vibrio harveyi (CP000790), several Shewanella spp., and P. profundum SS9. These findings suggest that qnrC might be acquired from a chromosomal source in the Vibrionaceae family by the ISPmi1 insertion sequence. Gene movement and exchange, augmented by transduction, transformation, and conjugation in aquatic environments, might have occurred in association with the increased use of antimicrobials in fish farming. Aquatic environments carrying resistant bacteria are not only reservoirs of clinical resistance genes but also media for the spread and evolution of resistance genes (30).
Acknowledgments
This work was supported by grant 2005CB0523101 (to M.W.) from the National Basic Research Program of China from the Ministry of Science and Technology, China, by grant 30572229 (to M.W.) from the National Natural Science Foundation of China, by grant LJ06052 (to M.W.) from the Shanghai Municipal Health Bureau, and by grant AI57576 (to D.C.H.) from the National Institutes of Health, U.S. Public Health Service.
Footnotes
Published ahead of print on 2 March 2009.
REFERENCES
- 1.Cattoir, V., L. Poirel, D. Mazel, C. J. Soussy, and P. Nordmann. 2007. Vibrio splendidus as the source of plasmid-mediated QnrS-like quinolone resistance determinants. Antimicrob. Agents Chemother. 51:2650-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cattoir, V., P. Nordmann, J. Silva-Sanchez, P. Espinal, and L. Poirel. 2008. ISEcp1-mediated transposition of qnrB-like gene in Escherichia coli. Antimicrob. Agents Chemother. 52:2929-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cavaco, L. M., H. Hasman, S. Xia, and F. M. Aarestrup. 2009. qnrD, a novel gene conferring transferable quinolone resistance in Salmonella enterica serovars Kentucky and Bovismorbificans of human origin. Antimicrob. Agents Chemother. 53:603-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chandler, M., and J. Mahillon. 2002. Insertion sequences revisited, p. 305-366. In N. L. Craig, R. Craigie, M. Gellert, and A. Lambowitz (ed.), Mobile DNA, vol. 2. ASM Press, Washington, DC. [Google Scholar]
- 5.Chen, C. C., and S. T. Hu. 2006. Two frameshift products involved in the transposition of bacterial insertion sequence IS629. J. Biol. Chem. 281:21617-21628. [DOI] [PubMed] [Google Scholar]
- 6.Chen, Y.-T., H.-Y. Shu, L.-H. Li, T.-L. Liao, K.-M. Wu, Y.-R. Shiau, J.-J. Yan, I.-J. Su, S.-F. Tsai, and T.-L. Lauderdale. 2006. Complete nucleotide sequence of pK245, a 98-kilobase plasmid conferring quinolone resistance and extended-spectrum-β-lactamase activity in a clinical Klebsiella pneumoniae isolate. Antimicrob. Agents Chemother. 50:3861-3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute. 2007. Performance standards for antimicrobial susceptibility testing; seventeenth informational supplement M100-S17, vol. 27, no. 1. Clinical and Laboratory Standards Institute, Wayne, PA.
- 8.Fonseca, E. L., F. S. Freitas, V. V. Vieira, and C. P. Vicente. 2008. New qnr gene cassettes associated with superintegron repeats in Vibrio cholerae O1. Emerg. Infect. Dis. 14:1129-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hata, M., M. Suzuki, M. Matsumoto, M. Takahashi, K. Sato, S. Ibe, and K. Sakae. 2005. Cloning of a novel gene for quinolone resistance from a transferable plasmid in Shigella flexneri 2b. Antimicrob. Agents Chemother. 49:801-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu, F., X. Xu, D. Zhu, and M. Wang. 2008. Coexistence of qnrB4 and qnrS1 in a clinical strain of Klebsiella pneumoniae. Acta Pharmacol. Sin. 29:320-324. [DOI] [PubMed] [Google Scholar]
- 11.Jacoby, G. A., K. E. Walsh, D. M. Mills, V. J. Walker, H. Oh, A. Robicsek, and D. C. Hooper. 2006. qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob. Agents Chemother. 50:1178-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacoby, G., V. Cattoir, D. Hooper, L. Martínez-Martínez, P. Nordmann, A. Pascual, L. Poirel, and M. Wang. 2008. qnr gene nomenclature. Antimicrob. Agents Chemother. 52:2297-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lascols, C., I. Podglajen, C. Verdet, V. Gautier, L. Gutmann, C. J. Soussy, E. Collatz, and E. Cambau. 2008. A plasmid-borne Shewanella algae gene, qnrA3, and its possible transfer in vivo between Kluyvera ascorbata and Klebsiella pneumoniae. J. Bacteriol. 190:5217-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laursen, B. S., H. P. Sorensen, K. K. Mortensen, and H. U. Sperling-Petersen. 2005. Initiation of protein synthesis in bacteria. Microbiol. Mol. Biol. Rev. 69:101-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez-Martinez, L., A. Pascual, and G. A. Jacoby. 1998. Quinolone resistance from a transferable plasmid. Lancet 351:797-799. [DOI] [PubMed] [Google Scholar]
- 16.Mazauric, M. H., P. Licznar, M. F. Prére, I. Canal, and O. Fayet. 2008. Apical loop-internal loop RNA pseudoknots: a new type of stimulator of −1 translational frameshifting in bacteria. J. Biol. Chem. 283:20421-20432. [DOI] [PubMed] [Google Scholar]
- 17.Poirel, L., A. Liard, J. M. Rodriguez-Martinez, and P. Nordmann. 2005. Vibrionaceae as a possible source of Qnr-like quinolone resistance determinants. Antimicrob. Agents Chemother. 56:1118-1121. [DOI] [PubMed] [Google Scholar]
- 18.Poirel, L., J. M. Rodriguez-Martinez, H. Mammeri, A. Liard, and P. Nordmann. 2005. Origin of plasmid-mediated quinolone resistance determinant QnrA. Antimicrob. Agents Chemother. 49:3523-3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poirel, L., T. V. Nguyen, A. Weintraub, C. Leviandier, and P. Nordmann. 2006. Plasmid-mediated quinolone resistance determinant qnrS in Enterobacter cloacae. Clin. Microbiol. Infect. 12:1021-1023. [DOI] [PubMed] [Google Scholar]
- 20.Poirel, L., V. Cattoir, A. Soares, C.-J. Soussy, and P. Nordmann. 2007. Novel Ambler class A β-lactamase LAP-1 and its association with the plasmid-mediated quinolone resistance determinant QnrS1. Antimicrob. Agents Chemother. 51:631-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robicsek, A., G. A. Jacoby, and D. C. Hooper. 2006. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect. Dis. 6:629-640. [DOI] [PubMed] [Google Scholar]
- 22.Rodríguez-Martínez, J. M., C. Velasco, A. Briales, I. García, M. C. Conejo, and A. Pascual. 2008. Qnr-like pentapeptide repeat proteins in gram-positive bacteria. J. Antimicrob. Chemother. 61:1240-1243. [DOI] [PubMed] [Google Scholar]
- 23.Sánchez, M. B., A. Hernández, J. M. Rodríguez-Martínez, L. Martínez-Martínez, and J. L. Martínez. 2008. Predictive analysis of transmissible quinolone resistance indicates Stenotrophomonas maltophilia as a potential source of a novel family of Qnr determinants. BMC Microbiol. 8:148-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimizu, K., K. Kikuchi, T. Sasaki, N. Takahashi, M. Ohtsuka, Y. Ono, and K. Hiramatsu. 2008. Smqnr, a new chromosome-carried quinolone resistance gene in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 52:3823-3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sussman, J. K., E. L. Simons, and R. W. Simons. 1996. Escherichia coli translation initiation factor 3 discriminates the initiation codon in vivo. Mol. Microbiol. 21:347-360. [DOI] [PubMed] [Google Scholar]
- 26.Tran, J. H., and G. A. Jacoby. 2002. Mechanism of plasmid-mediated quinolone resistance. Proc. Natl. Acad. Sci. USA 99:5638-5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tran, J. H., G. A. Jacoby, and D. C. Hooper. 2005. Interaction of the plasmid-encoded quinolone resistance protein Qnr with Escherichia coli DNA gyrase. Antimicrob. Agents Chemother. 49:118-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tran, J. H., G. A. Jacoby, and D. C. Hooper. 2005. Interaction of the plasmid-encoded quinolone resistance protein QnrA with Escherichia coli topoisomerase IV. Antimicrob. Agents Chemother. 49:3050-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang, M., J. H. Tran, G. A. Jacoby, Y. Zhang, F. Wang, and D. C. Hooper. 2003. Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob. Agents Chemother. 47:2242-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Young, H. K. 1993. Antimicrobial resistance spread in aquatic environments. J. Antimicrob. Chemother. 31:627-635. [DOI] [PubMed] [Google Scholar]