Abstract
In vitro, telaprevir selects subtype-specific resistance pathways for hepatitis C virus GT-1a and GT-1b, as described to have occurred in patients. In GT-1a, the HCV-796 resistance mutation C316Y has low replication capacity (7%) that can be compensated for by the emergence of the mutation L392F or M414T, resulting in an increase in replication levels of ≥10-fold.
The current standard of care for hepatitis C virus (HCV)-infected patients involves a treatment regimen of pegylated alpha interferon in combination with ribavirin, which results in a sustained viral response of approximately 50% for genotype 1 (GT-1)-infected patients (1, 11). There is a clear medical need for more efficacious therapies, and to this effect, a number of novel specific antiviral compounds are currently in preclinical and clinical development. A majority of these compounds inhibit the enzymatic activity of either the NS3/4A serine protease or the NS5B RNA-dependent RNA polymerase.
One factor that may limit the clinical efficacy of specific HCV antiviral drugs is the development of resistance. HCV presents a number of features that make drug resistance likely to occur upon treatment, such as the following: (i) the NS5B polymerase lacks proofreading activity, which results in the introduction of random mutations during the replication of the genomic RNA; (ii) HCV replicates as a genetic population known as a quasispecies that allows quick adaptation of the viral population upon changes in the environment (12); (iii) HCV produces a large number of infectious particles (up to 1012) per day, which means that each genetic variant made during RNA replication may be packaged into an infectious viral particle and can quickly spread (15); and (iv) the short half-life of the HCV genome, as estimated for the circulating virus (14) and calculated for the HCV replicon (3), is such that a variant present at low prevalence within the quasispecies can quickly become the dominant sequence if it offers a selective advantage. Resistance to specific HCV inhibitors in vitro has been well characterized through the use of the HCV GT-1b replicon system, and these studies have been predictive of the amino acid substitution(s) selected in HCV-infected patients upon drug treatment (4, 7-10, 13). For example, for the NS3/4A protease inhibitor telaprevir and the nonnucleoside polymerase inhibitor HCV-796, the resistance mutations identified in vitro (NS3 substitutions at residues T54 and A156 for telaprevir and an NS5B substitution at residue C316 for HCV-796) were also identified in GT-1b-treated patients (4, 5, 16).
One limitation of the majority of the replicon resistance studies reported to date is that only a single HCV subtype, GT-1b, has been used. HCV subtypes can vary by up to 25% at the nucleotide level, and this variability may lead to subtype-specific differences in the resistance profiles. In fact, subtype-specific resistance profiles for HCV-infected patients treated with telaprevir have been described previously. Substitutions at NS3 residues V36 and R155 were identified only in GT-1a-infected patients treated with telaprevir and not in GT-1b-infected patients (5, 16). As a result, the findings of the in vitro replicon resistance studies of telaprevir, which used a GT-1b replicon, were predictive for the GT-1b-infected patients but did not identify the emergence of substitutions at V36 or R155. Therefore, in this study, we determined if the HCV replicon system could be used to identify subtype-specific resistance mutations. For these experiments, we treated both a GT-1b replicon and a GT-1a replicon with either the protease inhibitor telaprevir (synthesized at Acme Bioscience, Inc.) or the nonnucleoside polymerase inhibitor HCV-796 (synthesized at Roche Palo Alto) at 15 times the 50% effective concentration (EC50), which for both compounds approximates the EC99, and monitored the emergence of resistance mutations in the NS3 protease or NS5B polymerase gene, respectively. Four independent selection experiments were performed for GT-1b, and two were performed for GT-1a. The GT-1b and GT-1a replicons are both bicistronic replicons in which the first open reading frame (driven by the HCV internal ribosome entry site) contains the Renilla luciferase gene fused with the neomycin phosphotransferase II gene and the second open reading frame (driven by the encephalomyocarditis virus internal ribosome entry site) contains the HCV nonstructural genes with engineered cell culture-adaptive mutations (2, 6). We monitored the kinetics of the development of telaprevir resistance in both GT-1b (n = 4) and GT-1a (n = 2) replicon-bearing cells and characterized the telaprevir resistance profiles to determine if the differences in resistance profiles identified in patients infected with either GT-1a or GT-1b HCV would also be observed in the replicon system. The stably transfected replicon cells bearing either a GT-1b or GT-1a replicon (GT-1a replicon encodes 75 amino acid residues of NS3 protease from the GT-1b Con 1 strain, as described by Gu et al. [2]) were incubated for a maximum of 21 days with 15 times the EC50 of telaprevir (Table 1) as described previously (13), with the exception that sampling was performed on days 3, 6, 9, 13, 16, and 21. Consistent with the data in previous reports (9, 10, 13), the incubation of GT-1b replicon cells with telaprevir resulted in the emergence of an amino acid substitution at NS3 position 156, and this substitution was identified as early as 3 days after the beginning of treatment (Table 2) by comparing the NS3 protease sequence from the untreated replicon cells with that from replicon cells taken at the specified treatment time points. The incubation of GT-1a replicon cells with telaprevir resulted in the selection of a mixture of sequences with wild-type (WT) and mutant R155R/K residues on day 3, with additional substitutions present at positions 156 (day 6) and 54 (day 9) (Table 2). The selection of R155K in GT-1a replicon cells, but not in GT-1b replicon cells, is consistent with the resistance profile described for treated patients. We were unable to detect the emergence of a resistance amino acid substitution at NS3 position 36 given the GT-1a/1b chimeric nature of the NS3 protease region used in this study, reinforcing the observation that V36M occurs only in GT-1a HCV.
TABLE 1.
Inhibitory activity and cytotoxicity against stable HCV GT-1b and GT-1a replicon cells
| Compound | EC50 (μM)a for cells bearing:
|
CC50 (μM)b for cells bearing:
|
||
|---|---|---|---|---|
| GT-1b | GT-1a | GT-1b | GT-1a | |
| Telaprevir | 0.56 ± 0.11 | 0.50 ± 0.18 | 26.7 ± 3.1 | 31.6 ± 2.9 |
| HCV-796 | 0.017 ± 0.005 | 0.033 ± 0.007 | >100 | >100 |
The inhibition of HCV replicon-encoded Renilla luciferase reporter activity after 3 days of incubation is presented as the mean ± the standard deviation of results from at least four independent experiments.
CC50, 50% cytotoxicity concentration. Cell viability was determined using a water-soluble tetrazolium salt assay (WST-1; Roche Applied Science, Indianapolis, IN) and is presented as the mean ± the standard deviation of results from at least four independent experiments.
TABLE 2.
Genotypic characterization of the NS3 protease from GT-1b and GT-1a replicons after telaprevir treatment
| Replicon (no. of expts)a | Residue(s) at position(s) with substitution(s) in NS3 proteaseb on telaprevir treatment day:
|
|||||
|---|---|---|---|---|---|---|
| 3 | 6 | 9 | 13 | 16 | 21 | |
| GT-1b (4) | A156A/S | A156A/S/T | A156A/S/T | A156A/S/T/V | A156A/S/T/V | A156A/S/T/V |
| GT-1a (2) | R155R/K | R155R/K, A156A/T | T54T/A, R155R/K, | T54T/A, R155R/K, A156A/T | R155R/K, A156A/T | R155R/K, A156A/T |
Stable GT-1b and GT-1a replicon cell lines were treated with 15 times the EC50 of telaprevir.
Results were obtained from direct sequencing of PCR products representing the major population. Only amino acid substitutions known to confer resistance and/or substitutions identified consistently in the independent experiments and not observed in untreated controls are reported. Data were derived from at least two independent cell culture selection experiments.
To monitor the kinetics of resistance development for the nonnucleoside polymerase inhibitor HCV-796 and to investigate whether this compound also demonstrates a subtype-specific resistance profile, GT-1b (n = 4) and GT-1a (n = 2) replicon cells were incubated for up to 21 days with 15 times the EC50 of HCV-796 (Table 1). The incubation of GT-1b and GT-1a replicon cells with HCV-796 resulted in the selection of a substitution in NS5B (C316Y) after 3 days (Table 3), as determined by comparing the NS5B sequence from the untreated replicon cells with that from replicon cells taken at the specified treatment time points. In the GT-1b replicon cells selected with HCV-796, the tyrosine substitution at NS5B position 316 was the only change observed and tyrosine completely replaced the WT cysteine by day 21 (Fig. 1). However, selective pressure from HCV-796 on a GT-1a replicon rendered a mixture of sequences with the WT cysteine and the mutant tyrosine at all time points, with the WT population being predominant over the 316Y mutant population. This mixture of sequences with different residues at position 316 was always present alongside mixtures of sequences with amino acid substitutions at L392 and M414 (Table 3). This finding suggests that the C316Y substitution may have a lower selective advantage in the genetic context of the GT-1a replicon than in that of the GT-1b replicon and may require compensatory mutations. To investigate whether this is the case, transient GT-1b and GT-1a replicons encoding the C316Y substitution were generated. The C316Y substitution in both the GT-1a and GT-1b replicons conferred high-level resistance, approximately 400- and 100-fold reductions in sensitivity, respectively; however, the replication capacity for the C316Y mutant in the GT-1a genetic context was 7% compared to that for the WT, approximately 10-fold lower than the 60% observed for the C316Y mutant in the GT-1b genetic context (Table 4) (13). To assess whether double mutants involving C316Y, L392F, or M414T existed in the population, clonal analyses of amino acid positions 220 to 430 of the NS5B region in the population at day 21 were performed and confirmed the population sequence findings, with substitutions C316Y, L392F, or M414T observed either alone or in combination. Given that the double mutations C316Y/L392F, C316Y/M414T, and L392F/M414T were identified, these amino acid substitutions were introduced into the transient GT-1a replicon and their effects on the sensitivity to HCV-796 and the replication capacities of the mutants were determined. As described above, the C316Y substitution resulted in an approximately 400-fold reduction in the sensitivity to HCV-796, while the substitutions L392F and M414T individually had no effect on the sensitivity to the compound (Table 4). The sensitivity to NNI-1, a thiophene-2-carboxylic acid (8) which binds to the thumb II site, was assessed as a control and found to be unaffected by these NS5B substitutions (Table 4). Double mutation L392F/M414T conferred a low level of resistance (5.5-fold reduction in sensitivity to HCV-796 compared to that of the WT). Interestingly, the double mutation C316Y/L392F or C316Y/M414T did not have an effect on the sensitivity to HCV-796 compared to that seen with the C316Y single substitution, but the double mutants showed significantly increased replication capacities compared to that of the C316Y single mutant, suggesting that the changes at positions 392 and 414 have a compensatory effect in the GT-1a genetic context (Table 4).
TABLE 3.
Sequence analysis of NS5B from GT-1b and GT-1a replicons after HCV-796 treatment
| Replicona | Residue(s) at position(s) with substitution(s) in NS5Bb on HCV-796 treatment day:
|
|||||
|---|---|---|---|---|---|---|
| 3 | 6 | 9 | 13 | 16 | 21 | |
| GT-1b | C316C/Y | C316C/Y | C316C/Y | C316C/Y | C316C/Y | C316Y |
| GT-1a | C316C/Y | C316C/Y | C316C/Y | C316C/Y | C316C/Y | C316C/Y |
| L392L/F | L392L/F | L392L/F | L392L/F | L392L/F | L392L/F | |
| M414M/V/T | M414M/V/T | M414M/V/T | M414M/T | M414M/V/T | M414M/T | |
Stable GT-1b (n = 4) and GT-1a (n = 2) replicon cell lines were treated with 15 times the EC50 of HCV-796.
Results were obtained from direct sequencing of PCR products representing the major population. Only amino acid substitutions known to confer resistance and/or substitutions identified consistently in the independent experiments and not observed in untreated controls are reported. Data were derived from at least two independent cell culture selection experiments.
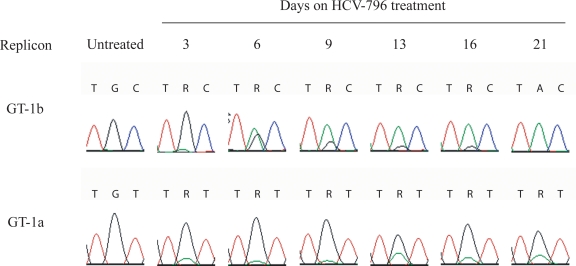
FIG. 1.
GT-1b and GT-1a replicon cells treated with HCV-796 select for the C316Y substitution in NS5B. Huh7 cells that stably maintain either a GT-1b or GT-1a subgenomic replicon carrying the neomycin phosphotransferase II gene were incubated with HCV-796 at 15 times the EC50 in the presence of G418. Results for both selections derive from four independent experiments for GT-1b and from two independent experiments for GT-1a. At the indicated times, the cellular RNA was extracted and the NS5B gene was amplified and then sequenced. The sequencing chromatograms obtained from each time point are shown for NS5B position 316. The codon utilized for cysteine 316 is UGC (i.e., TGC in the sequencing chromatogram) in GT-1b and UGU in GT-1a. After incubation with HCV-796, tyrosine emerges and the codon is UAC in GT-1b and UAU in GT-1a.
TABLE 4.
Phenotypic characterization of HCV-796-resistant variants of GT-1b or GT-1a
| Replicon background and NS5B phenotype or mutation(s) | Replication capacitya | EC50 (μM ± SD)b of:
|
|
|---|---|---|---|
| HCV-796 | NNI-1 (thumb II inhibitor) | ||
| GT-1b background | |||
| WT | 1 | 0.006 ± 0.002 | 0.16 ± 0.03 |
| C316Y | 0.6 ± 0.2 | 0.56 ± 0.1 | 0.20 ± 0.04 |
| GT-1a background | |||
| WT | 1 | 0.004 ± 0.001 | 0.63 ± 0.12 |
| L392F | 2.1 ± 0.5 | 0.008 ± 0.001 | 0.58 ± 0.11 |
| M414T | 2.2 ± 1.1 | 0.007 ± 0.0004 | 0.53 ± 0.17 |
| C316Y | 0.07 ± 0.03 | 1.7 ± 0.5 | 0.39 ± 0.10 |
| C316Y/L392F | 0.71 ± 0.12 | 3.2 ± 0.8 | 0.34 ± 0.13 |
| C316Y/M414T | 1.2 ± 0.4 | 2.8 ± 1.1 | 0.30 ± 0.07 |
| L392F/M414T | 0.9 ± 0.5 | 0.022 ± 0.009 | 0.39 ± 0.1 |
The replication capacities were determined as the ratio of the firefly luciferase signal at 4 days postelectroporation to the luciferase signal at 4 h postelectroporation. The replication capacities of the mutants were expressed as their normalized replication efficiencies compared to that of the WT, set at a value of 1. The values are presented as means ± standard deviations of results from at least three independent experiments.
EC50 values are presented as means ± standard deviations of results from at least four independent experiments.
This study provides evidence that in vitro HCV replicon resistance experiments performed with HCV GT-1a and GT-1b subtype replicons can provide important data to predict compounds' resistance profiles. For telaprevir, the previously reported replicon resistance studies were performed using GT-1b replicon cells and did not identify substitutions that emerged in GT-1a-treated patients. By performing selection of GT-1a replicon cells with telaprevir, we were able to confirm the different resistance pathway in GT-1a HCV, through the R155K substitution in NS3, as observed in HCV-infected patients. The R155K substitution does not emerge in GT-1b strains likely because of a codon usage bias (16). In GT-1b strains, the arginine codon requires two nucleotide changes in order for lysine to be encoded, while a different arginine codon that requires only a single nucleotide change to encode lysine is utilized in GT-1a strains.
Importantly, similarly to that from telaprevir, selective pressure from HCV-796 on GT-1b and GT-1a replicon cells identified subtype-specific resistance profiles for this nonnucleoside polymerase inhibitor. In the GT-1b replicon system, the C316Y NS5B substitution was identified as the primary HCV-796 resistance substitution, which confers a high level of resistance and produces a mutant with a good replication capacity of 60% that of the WT (4, 13). However, in the GT-1a replicon system, the C316Y substitution, which also confers a high level of resistance, results in a >10-fold reduction in the replication capacity (to 7% that of the WT). This reduction in replication capacity most likely limits the emergence of the C316Y substitution in the GT-1a genetic backbone compared to the complete emergence of C316Y in the GT-1b replicon system. C316Y is the main HCV-796 resistance mutation (4), and therefore, the selection of compensatory substitutions L392F and/or M414T that restore the replication capacity seems to be required in GT-1a. This finding would suggest a higher genetic barrier to the development of resistance to HCV-796 in HCV GT-1a-infected patients than in GT-1b-infected patients. It would be interesting to know if compensatory mutations were also identified in patients infected with a GT-1a subtype that were treated with HCV-796. In conclusion, these results provide evidence that in vitro replicon selection experiments can identify subtype-specific resistance profiles in vitro that can be predictive of the amino acid substitutions identified in treated patients.
Footnotes
Published ahead of print on 9 March 2009.
REFERENCES
- 1.Fried, M. W., M. L. Shiffman, K. R. Reddy, C. Smith, G. Marinos, F. L. Goncales, Jr., D. Haussinger, M. Diago, G. Carosi, D. Dhumeaux, A. Craxi, A. Lin, J. Hoffman, and J. Yu. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975-982. [DOI] [PubMed] [Google Scholar]
- 2.Gu, B., A. T. Gates, O. Isken, S. E. Behrens, and R. T. Sarisky. 2003. Replication studies using genotype 1a subgenomic hepatitis C virus replicons. J. Virol. 77:5352-5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo, J. T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howe, A. Y., H. Cheng, S. Johann, S. Mullen, S. K. Chunduru, D. C. Young, J. Bard, R. Chopra, G. Krishnamurthy, T. Mansour, and J. O'Connell. 2008. Molecular mechanism of hepatitis C virus replicon variants with reduced susceptibility to a benzofuran inhibitor, HCV-796. Antimicrob. Agents Chemother. 52:3327-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kieffer, T. L., C. Sarrazin, J. S. Miller, M. W. Welker, N. Forestier, H. W. Reesink, A. D. Kwong, and S. Zeuzem. 2007. Telaprevir and pegylated interferon-alpha-2a inhibit wild-type and resistant genotype 1 hepatitis C virus replication in patients. Hepatology 46:631-639. [DOI] [PubMed] [Google Scholar]
- 6.Klumpp, K., V. Leveque, S. Le Pogam, H. Ma, W. Jiang, H. Kang, C. Granycome, M. Singer, C. Laxton, J. Q. Hang, K. Sarma, D. B. Smith, D. Heindl, C. J. Hobbs, J. H. Merrett, J. Symons, N. Cammack, J. A. Martin, R. Devos, and I. Najera. 2006. The novel nucleoside analog R1479 (4′-azidocytidine) is a potent inhibitor of NS5B-dependent RNA synthesis and hepatitis C virus replication in cell culture. J. Biol. Chem. 281:3793-3799. [DOI] [PubMed] [Google Scholar]
- 7.Le Pogam, S., W. Jiang, V. Leveque, S. Rajyaguru, H. Ma, H. Kang, S. Jiang, M. Singer, S. Ali, K. Klumpp, D. B. Smith, J. Symons, N. Cammack, and I. Najera. 2006. In vitro selected Con1 subgenomic replicons resistant to 2′-C-methyl-cytidine or to R1479 show lack of cross resistance. Virology 351:349-359. [DOI] [PubMed] [Google Scholar]
- 8.Le Pogam, S., H. Kang, S. F. Harris, V. Leveque, A. M. Giannetti, S. Ali, W. R. Jiang, S. Rajyaguru, G. Tavares, C. Oshiro, T. Hendricks, K. Klumpp, J. Symons, M. F. Browner, N. Cammack, and I. Najera. 2006. Selection and characterization of replicon variants dually resistant to thumb- and palm-binding nonnucleoside polymerase inhibitors of the hepatitis C virus. J. Virol. 80:6146-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin, C., C. A. Gates, B. G. Rao, D. L. Brennan, J. R. Fulghum, Y. P. Luong, J. D. Frantz, K. Lin, S. Ma, Y. Y. Wei, R. B. Perni, and A. D. Kwong. 2005. In vitro studies of cross-resistance mutations against two hepatitis C virus serine protease inhibitors, VX-950 and BILN 2061. J. Biol. Chem. 280:36784-36791. [DOI] [PubMed] [Google Scholar]
- 10.Lin, C., K. Lin, Y. P. Luong, B. G. Rao, Y. Y. Wei, D. L. Brennan, J. R. Fulghum, H. M. Hsiao, S. Ma, J. P. Maxwell, K. M. Cottrell, R. B. Perni, C. A. Gates, and A. D. Kwong. 2004. In vitro resistance studies of hepatitis C virus serine protease inhibitors, VX-950 and BILN 2061: structural analysis indicates different resistance mechanisms. J. Biol. Chem. 279:17508-17514. [DOI] [PubMed] [Google Scholar]
- 11.Manns, M. P., J. G. McHutchison, S. C. Gordon, V. K. Rustgi, M. Shiffman, R. Reindollar, Z. D. Goodman, K. Koury, M. Ling, and J. K. Albrecht. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958-965. [DOI] [PubMed] [Google Scholar]
- 12.Martell, M., J. I. Esteban, J. Quer, J. Genesca, A. Weiner, R. Esteban, J. Guardia, and J. Gomez. 1992. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J. Virol. 66:3225-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCown, M. F., S. Rajyaguru, S. Le Pogam, S. Ali, W. R. Jiang, H. Kang, J. Symons, N. Cammack, and I. Najera. 2008. The hepatitis C virus replicon presents a higher barrier to resistance to nucleoside analogs than to nonnucleoside polymerase or protease inhibitors. Antimicrob. Agents Chemother. 52:1604-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neumann, A. U., N. P. Lam, H. Dahari, M. Davidian, T. E. Wiley, B. P. Mika, A. S. Perelson, and T. J. Layden. 2000. Differences in viral dynamics between genotypes 1 and 2 of hepatitis C virus. J. Infect. Dis. 182:28-35. [DOI] [PubMed] [Google Scholar]
- 15.Neumann, A. U., N. P. Lam, H. Dahari, D. R. Gretch, T. E. Wiley, T. J. Layden, and A. S. Perelson. 1998. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science 282:103-107. [DOI] [PubMed] [Google Scholar]
- 16.Sarrazin, C., T. L. Kieffer, D. Bartels, B. Hanzelhalka, U. Muh, M. Welker, D. Wincheringer, Y. Zhou, H.-M. Chu, C. Lin, C. Weegink, H. Reesink, S. Zeuzem, and A. D. Kwong. 2007. Dynamic hepatitis C virus genotypic and phenotypic changes in patients treated with the protease inhibitor telaprevir. Gastroenterology 132:1767-1777. [DOI] [PubMed] [Google Scholar]